Abstract
Hematopoiesis is the hierarchical process in which all lineages of blood cells are produced by self-renewing hematopoietic stem cells (HSCs) in the bone marrow (BM). While the regulatory factors that maintain proper HSC function and lineage output under normal conditions are well understood, significantly less is known about how HSC fate is regulated in response to inflammation or disease. As many blood disorders are associated with overproduction of pro-inflammatory cytokines, significant interest has emerged in understanding the impact of these factors on HSC function. In this review we highlight key advances demonstrating the impact of pro-inflammatory cytokines on the biology of HSCs and the BM niche, and address ongoing questions regarding their role in normal and pathogenic hematopoiesis.
Keywords: hematopoietic stem cell, cytokine, inflammation, interferon, interleukin, myeloproliferative neoplasm
Introduction
Hematopoiesis is a highly organized process in which all lineages of blood are produced by a population of rare hematopoietic stem cells (HSCs) residing in the bone marrow (BM) (Orkin and Zon, 2008). In adult organisms, this process is tightly regulated to maintain homeostatic blood production while ensuring lifelong maintenance of the HSC pool. Thus, HSCs are kept in a quiescent, or dormant state, occasionally activating and entering the cell cycle to replenish mature blood cells as they turn over (Pietras et al., 2011). Such HSCs may also undergo alternative fates including egress from the BM, induction of apoptosis, or self-renewal to maintain proper blood production and the overall functional integrity of the HSC pool.
HSC fate choices can result from the interplay of several cell-intrinsic regulatory networks (Orkin and Zon, 2008). Thus, HSC survival is dictated in part by the balance of pro- and anti-apoptotic Bcl2 family proteins (Mohrin et al., 2010), while differentiation is regulated by the stochastic activation of lineage-specific transcription factor networks such as the antagonistic GATA-1/PU.1 axis driving erythroid and myeloid differentiation (Laslo et al., 2008). Other transcriptional and epigenetic factors, including Bmi-1, p53, Ikaros and C/EBPα also play key roles in directing HSC fate decisions (He et al., 2009, Laslo et al., 2008). Moreover, the specialized niches in which HSCs reside produce cell-extrinsic factors including Notch ligands, TGF-β, SCF, and CXCL12 that further regulate these networks (Pietras et al., 2011, Warr et al., 2011). Thus, HSC fate is tightly regulated by a complex interplay between cell-intrinsic and cell-extrinsic factors.
Although HSCs are maintained in a predominantly quiescent state, they can rapidly enter the cell cycle and differentiate, often preferentially along the myeloid lineage, in response to infection or injury (Manz and Boettcher, 2014). Under such dynamic conditions, additional signals related to the insult may intercede to regulate HSC fate. It is well known that mature immune cells and HSCs alike can activated either by direct activation of pathogen recognition receptors (PRRs) such as the Toll-like receptors (TLRs), or via pro-inflammatory cytokine signals (Kawai et al., 2011, Zhao et al., 2014). Indeed, a range of pro-inflammatory cytokines produced during infection and injury impact the size and shape of the hematopoietic system (Manz and Boettcher, 2014). These cytokines likely represent a critical line of communication by which the presence of danger is relayed to HSCs and progenitors, along with instructions on what cell types to produce in response to the threat. While the integration of such signals with hematopoietic output may promote effective host defense in acute conditions, prolonged exposure to them can be detrimental to HSC function. Indeed, hematopoietic failure associated with overproduction of pro-inflammatory cytokines is often a feature of chronic inflammatory diseases, hematological malignancies and bone marrow failure syndromes (Anand et al., 1998, Ishihara and Hirano, 2002, Braun et al., 2013). Thus, identifying how inflammation regulates HSC fate and function in normal and disease conditions represents a crucial frontier for our understanding and treatment of chronic inflammation and blood disorders.
In this review we will illustrate how pro-inflammatory cytokines regulate normal and diseased hematopoiesis, highlighting recent advances that have provided new insights into outstanding questions and controversies in the field. We will provide a critical examination of the role of interferons (IFNs) in regulating HSC fate, and discuss the evolving role of pro-inflammatory cytokines, particularly interleukin-6 (IL-6), as key regulators of myeloid lineage output in normal and disease conditions. Moreover, we will address emerging findings demonstrating how crosstalk between pro-inflammatory cytokines and the BM niche impacts the health and function of the HSC pool. Lastly, we will identify important new questions raised by these studies and their implications in understanding the interaction between HSCs and inflammation.
Interferons: paradoxical regulators of HSC function
The role of IFNs in HSC biology appears both complex and contradictory. IFNs are a family of over 13 cytokines produced in response to intracellular pathogens, categorized based on biochemical characteristics, surface receptor affinity and biological activity (Platanias, 2005). Type I IFNs (IFN-Is) include multiple IFNα species and a single IFNβ, and are broadly expressed by many cell types. Conversely, expression of the single type II IFN (IFN-II), IFNγ, is limited primarily to NK and T cells. Both classes of IFNs induce an anti-proliferative, pro-apoptotic ‘antiviral state’ that prevents cellular hijacking by intracellular pathogens (Platanias, 2005).
Consistent with their antiviral effects, IFNs exert anti-proliferative effects on immature hematopoietic cells in vitro (Verma et al., 2002). Indeed, IFN-Is were thus widely used as a therapy to suppress aberrant hematopoiesis in patients with myeloproliferative neoplasms (MPNs) including chronic myelogenous leukemia (CML), a blood malignancy in which granulocytes are overproduced by transformed leukemic HSCs (LSCs) carrying the BCR/ABL fusion gene (Kiladjian et al., 2011). Although IFN-Is reduced disease burden, they were not effective in eliminating LSCs and were supplanted by targeted therapies such as the BCR/ABL kinase inhibitor Imatinib, which yielded better results with significantly less toxicity (Kiladjian et al., 2011).
In vivo studies have demonstrated the paradoxical ability of IFNs to directly induce HSC proliferation in response to acute stimulation in mice (Essers et al., 2009, Baldridge et al., 2010). Nevertheless, in vitro culture of purified HSCs yields the expected anti-proliferative effects (de Bruin et al., 2013, Prendergast and Essers, 2014), suggesting that IFNs activate HSC proliferation via means other than direct engagement of the cell cycle machinery. Indeed, analyses of quiescent versus proliferating HSCs harvested from IFN-I-exposed mice do not show a strong induction of pro-proliferative cell cycle genes (Pietras et al., 2014). Rather, IFN-Is suppress transcription of key HSC quiescence-enforcing mechanisms including the cyclin kinase inhibitor (CKI) Cdkn1c (p57) and the transcription factor Foxo3a, as well as components of the Notch and TGFβ pathways (Pietras et al., 2014). Moreover, acute exposure to IFN-Is in vivo promotes increased Myc protein levels in HSCs, which may also contribute to their proliferative phenotype (Ehninger et al., 2014). Furthermore, recent work has shown that proliferating IFN-I-exposed HSCs move away from quiescence-associated periarteriolar niches (Kunisaki et al, 2013), suggesting the regulatory links between the BM niche and HSCs are severed during IFN-I-induced proliferation. Collectively, these findings describe a new model in which HSCs are ‘licensed’ by IFNs to proliferate by suppression of quiescence-enforcing mechanisms (Fig. 1). On the other hand, HSCs removed from the BM niche and cultured in vitro deterministically enter the cell cycle, likely masking the IFN-induced ‘licensing’ effect (de Bruin et al., 2013, Pietras et al., 2014, Pendergast and Essers, 2014), thus highlighting the shortcomings of in vitro culture assays in replicating the contribution of the BM niche to HSC biology.
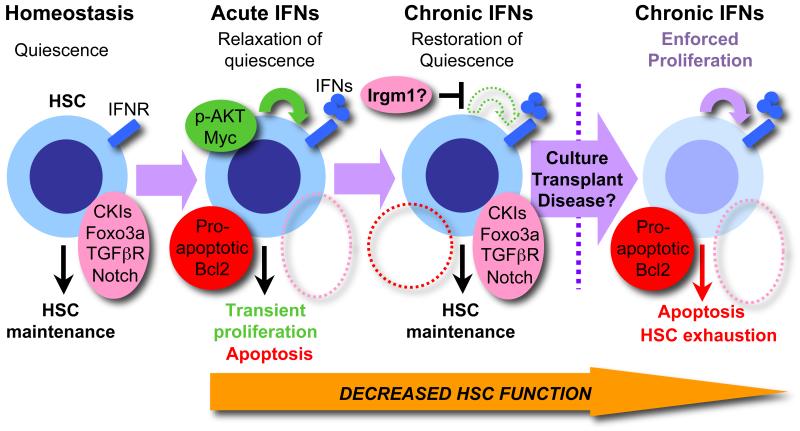
Figure 1. Complex effects of IFNs on HSCs.
Under homeostatic conditions HSCs reside in a quiescent state regulated by the concerted activity of cell-intrinsic and cell-extrinsic mechanisms including high expression of cyclin kinase inhibitors (CKIs), the transcription factor Foxo3a, and signaling via the TGFβR and Notch pathways. In response to IFN activation via interferon receptors (IFNR), these mechanisms are transiently repressed, hence licensing HSCs to proliferate in the context of elevated AKT activity, but also rendering them vulnerable to apoptosis due to activation of a Bcl2 pro-apoptotic gene program. During chronic IFN exposure, HSC quiescence-enforcing mechanisms are re-established, possibly through suppression of AKT signaling by Irgm1, and the HSC pool returns to a quiescent state, thus protecting HSCs from IFN-induced apoptosis. However, if IFN-exposed HSCs are forced back into the cell cycle due to transplantation, ex vivo culture, or cell cycle activation following a myeloablative treatment, they rapidly re-activate the Bcl2 pro-apoptotic gene program and undergo apoptosis, hence impairing HSC function and rendering the HSC pool vulnerable to exhaustion. Similar effects may underlie the effects of IFNs on diseased HSCs with deregulated cell cycle activity.
Interestingly, IFN-driven proliferation is an acute and brief event. Indeed, HSCs rapidly re-enter a quiescent state even during chronic exposure to IFN-Is (Pietras et al., 2014) that is associated with restoration of quiescence-enforcing genes, implying the action of a cellular ‘braking’ mechanism. While this mechanism has not been clearly identified, HSCs from mice deficient in the GTPase Irgm1 exhibit chronic loss of quiescence that is rescued by blockade of IFNγR (King et al., 2011), implicating Irgm1 as a candidate brake factor in response to IFNs. Notably, returning to quiescence spares HSCs from the killing effects of IFNs. In particular, IFN-I-exposed HSCs forced back into the cell cycle by in vitro culture, transplantation, or 5-FU-mediated myeloablation rapidly upregulate pro-apoptotic Bcl2 genes and undergo apoptosis (Pietras et al., 2014). Thus, apoptosis of IFN-exposed HSCs forced back into cycle likely accounts for the decreased engraftment observed upon transplantation of IFN-exposed HSCs (Baldridge et al., 2010, de Bruin et al., 2013, Pietras et al., 2014). Importantly, the protection quiescence provides against IFN-mediated killing likely explains the limited effectiveness of IFN-Is in eliminating quiescent CML LSCs, as well as the relative absence of bone marrow failure in patients chronically exposed to IFNs absent other complications (Ionnou et al., 2010). Altogether these findings indicate that HSC quiescence acts as a protective mechanism that limits IFN-induced apoptosis during chronic exposure (Fig. 1). Interestingly, HSCs display a similarly rapid but transient cell cycle activation during chronic in vivo G-CSF exposure, suggesting that similar ‘licensing’ and ‘braking’ mechanisms may be engaged in response to many inflammatory stimuli (Schuttpueltz et al., 2014). Such a mechanism could ensure response to acute need, while preserving the HSC pool from exhaustion during chronic insults. Further work is needed to identify the relative contribution of individual quiescence-enforcing mechanisms in regulating HSC cell cycle activity in response to IFNs and other cytokines, and to identify how quiescence is re-established during chronic exposure to these factors.
A number of studies have demonstrated that IFNs induce expansion of the LSK compartment (Pietras et al., 2014), which has been interpreted as expansion of multipotent progenitors (MPPs) downstream of HSCs (Zhang et al., 2013). However, IFNs strongly induce Sca-1 expression in myeloid progenitors, hence contaminating the phenotypic LSK compartment and confounding analysis of these populations (Pietras et al., 2014). Furthermore, only the most stringent phenotypic definitions, such as the SLAM markers CD48 and CD150, can properly distinguish HSCs from contaminating myeloid progenitors, making interpretation of data using less stringent HSC definitions problematic (Pietras et al., 2014) and highlighting the challenges inherent to studying hematopoietic populations under activating conditions, when their biological and phenotypic properties may be significantly altered.
Due to their proliferative effects on HSCs, IFN-Is have re-emerged as a therapeutic option for CML and other MPNs, based on the rationale that their ability to drive quiescent LSCs into cycle will render these cells vulnerable to killing when combined with Imatinib or other drugs (Kiladjian, et al, 2011). However, the transient nature of this effect suggests the therapeutic window would be brief (Essers et al., 2009, Pietras et al., 2014). On the other hand, JAK2V617F LSCs are selectively depleted by IFN-Is alone in a mouse model of human polycythemia vera (PV), presumably due to elevated steady-state cell cycle activity relative to normal HSCs (Mullally et al., 2013). Thus, LSCs with increased basal cell cycle activity may be uniquely sensitive to the killing effect of IFNs, providing further rationale for their use alongside targeted therapies in MPN. In addition, IFN-mediated HSC killing may underlie the pathogenesis of bone marrow failure syndromes. This is particularly true for conditions like Fanconi anemia (FA) where HSC cell cycle control is disrupted and high IFN levels are present (Li, et al., 2003, de Bruin et al., 2013). Collectively, these data demonstrate that IFNs play a critical, if complex, role in regulating HSC fate that underlies both their therapeutic and pathogenic effects.
Importantly, while the effects of both IFN-Is and IFN-II on HSCs appear broadly similar, most studies to date have focused exclusively on one IFN type or the other, resulting in methodologically heterogeneous analyses with limited overlap. As IFN-Is and IFN-II have some distinct transcriptional activities (Platanais, 2005), it is possible that they also have some differential effects on HSCs. Interestingly, recent evidence indicates IFN-II exposure impairs HSC self-renewal by selectively inducing differentiation of myeloid-biased HSC subsets, which express elevated levels of IFNγR (Matatall et al., 2014). These results suggest that IFN-II may regulate HSC fate distinctly from IFN-Is, though parallel analyses on IFN-I-exposed HSCs have not been published. Thus, a comprehensive side-by-side analysis of the effects and underlying mechanisms of both IFN types on HSCs using a uniform set of assays will be required to confirm potential similarities and differences in the effect of both IFN types on the biology of HSCs.
IL-6: a critical mediator of hematopoietic lineage choice
While some cytokines, particularly M-CSF, GM-CSF, and G-CSF are hematopoietic-specific growth factors that play critical roles in myeloid lineage specification (Manz and Boettcher, 2014), pleiotropic pro-inflammatory cytokines such as IL-6 act on many cell types, including hematopoietic cells (Ishihara and Hirano, 2002). IL-6 has been implicated as a critical activator of myelopoiesis in response to pathogen infection and chronic inflammatory disorders (Ishihara and Hirano, 2002) and inhibits lymphopoiesis as well, suggesting it regulates lineage choice in uncommitted progenitors (Maeda et al., 2005). Although IL-6 is dispensable in establishing steady state levels of myeloid and lymphoid cells (Maeda et al., 2005), it is a key regulator of hematopoietic lineage output in the context of inflammation. Consistently, IL-6 receptor (IL-6R) is robustly expressed by MPPs, though not by HSCs (Reynaud et al., 2011), and in vitro culture of purified MPPs with IL-6 promotes myeloid cell production while abrogating lymphoid lineage output (Reynaud et al., 2011, Schurch et al., 2014). On the other hand, lineage-committed myeloid progenitors do not have increased output when cultured with IL-6 (Schurch et al., 2014). These data suggest that MPPs represent a highly ‘instructable’ progenitor population whose lineage output can be reprogrammed in response to inflammatory cues such as IL-6.
Interestingly, recent work has demonstrated that IFNγ induces IL-6 production by mesenchymal stem cells (MSCs) in the BM niche, hence promoting myelopoiesis during viral infection (Schurch et al., 2014). These findings also may suggest a mechanism by which IFNs could activate myeloid cell production (Schurch et al., 2014), favoring a model in which IFNs directly suppress HSC function, while secondary factors such as IL-6 activate myeloid cell production by downstream progenitors. Such population-specific responses could facilitate rapid myeloid cell production for host defense, while limiting the potential for the HSC pool to become infected. Intriguingly, IL-6 is also produced by a subpopulation of MPPs in response to direct TLR stimulation (Zhao et al., 2014). While the in vivo relevance of this finding remains to be fully assessed, it suggests that immature hematopoietic progenitors might produce IL-6 to trigger myeloid differentiation in response to pathogens or injury, perhaps in locations outside the BM. Such data highlight the inherent challenges of studying the roles of individual cytokines on hematopoiesis in vivo, when multiple cytokines and PRR pathways may be concurrently activated in different cell populations.
Notably, elevated IL-6 levels are found in CML patients (Anand et al., 1998). Similar IL-6 overproduction was traced to myeloid cells using a tetracycline-inducible transgenic BCR/ABL (BAScl-tTA) mouse model that reproduces the hematological features of human CML (Reynaud et al., 2011). Strikingly, loss of IL-6 signaling in BAScl-tTA:Il-6−/− mice substantially extends survival of diseased mice and restores lymphoid cell production (Reynaud et al., 2011). Thus, IL-6-producing MPN myeloid cells create a pathogenic feed-forward loop in which IL-6 reprograms normal and transformed MPPs to produce more myeloid cells that secrete additional IL-6 (Reynaud et al., 2011). These findings demonstrate the importance of pro-inflammatory cytokines in driving diseased hematopoiesis and implicate blockade of IL-6 as a therapeutic approach for CML and other MPNs.
Importantly, other pro-inflammatory cytokines, including IL-1 and IL-17, also activate myelopoiesis in response to infection and inflammatory disease (Nagareddy et al., 2014, Tan et al., 2008). On the other hand, tumor necrosis factor (TNF) signaling is largely thought to restrict HSC potential (Pronk et al., 2011). Moreover, TNF may play a role, alongside IFN-Is, in the HSC dysfunction underlying FA and myelodysplastic syndrome (MDS) (Geiselhart et al., 2012, Braun et al., 2013). However, other evidence suggests TNF signaling may enhance HSC function (Rebel et al., 1999, Rezzoug et al., 2008), again highlighting the potentially complex effects of pro-inflammatory cytokines on blood production in different contexts. Further investigations are needed to identify the direct cellular targets and regulatory roles of pro-inflammatory factors in normal and pathogenic hematopoiesis.
Inflammatory cytokines as critical regulators of the BM niche
The effect of pro-inflammatory cytokines on the biology of the BM niche is an emergent area of study with significant implications for our understanding of normal and diseased hematopoiesis. Many cellular components of the BM niche have now been identified, including endothelial cells (EC), perivascular MSCs, and osteolineage cells (OBCs) associated with endosteal bone (Morrison and Scadden, 2014). In addition, mature hematopoietic cells, including CD169+ bone marrow macrophages and T cells, also regulate HSC activity (Winkler et al., 2012, Schurch et al., 2014) (Fig. 2A). The BM niche promotes HSC maintenance by producing SCF and CXCL12, as well as quiescence and fate regulators including Notch ligands and TGF-β (Pietras et al., 2011). Perturbation of these signals or the cells generating them by pro-inflammatory cytokines can directly impact HSCs. Notably, G-CSF depletes CD169+ BM macrophages and osteoblasts, inhibiting production of SCF and CXCL12 and impairing HSC function (Winkler et al., 2012). Crosstalk between pro-inflammatory cytokines and the BM niche may also play a critical role in the pathogenesis and/or progression of hematological diseases. MPNs such as CML and primary myelofibrosis (PMF) originate from transformed LSCs, but also lead to significant changes in the bone marrow environment, characterized by excessive deposition of collagen by fibroblasts in the bone marrow, resulting in fibrosis (Abdel-Wahab and Levine, 2009). These diseases, as well as BM fibrosis arising from autoimmune disorders, are associated with overproduction of inflammatory cytokines, and recent work using the BAScl-tTA CML model demonstrates that these factors can directly reprogram MSC fate toward overproduction of inflammatory OBCs, resulting in BM fibrosis (Schepers et al., 2013). Indeed, elevated production of TPO, the chemokine CCL3 (MIP-1α) and contact-dependent signals by MPN myeloid cells directly induce aberrant differentiation and expansion of OBCs from MSCs (Fig. 2B). On the other hand, while IL-6 reprograms MPPs (Reynaud et al., 2011), it does not affect MSC differentiation. Thus, distinct subsets of cytokines overproduced by MPN myeloid cells can simultaneously activate aberrant differentiation programs in hematopoietic and stromal populations (Schepers et al., 2013).
Figure 2. Disease-mediated remodeling of the BM niche impairs HSC maintenance.
(A) HSCs reside in specialized niches in the BM comprised of many specialized cell types, including endothelial cells, perivascular multipotent stromal cells (MSCs) which differentiate into osteoblast-lineage cells (OBC), and hematopoietic populations including CD169+ macrophages and T-lymphocytes. Collectively, these cell populations produce a variety of soluble and membrane-bound factors, which contribute to HSC maintenance and regulate their activity and output. (B) During the development of blood disease such as MPNs, LSCs aberrantly overproduce granulocytes. These MPN myeloid cells in turn produce high levels of IL-6, which reprogram MPPs to overproduce myeloid cells at the expense of the lymphoid lineage, hence promoting disease progression. In addition, MPN myeloid cells drive aberrant MSC differentiation into OBCs via elevated production of CCL3, TPO, and additional contact-dependent signals. These overproduced OBCs display features of remodeled fibrotic cells, and produce elevated levels of inflammatory factors at the expense of HSC maintenance factors such as SCF and CXCL12. Collectively, this leads to impaired maintenance of normal HSCs, whereas LSCs are insensitive to these alterations, leading to the establishment of a self-reinforcing leukemic niche that favors the expansion of LSCs and hence disease progression.
Strikingly, the OBCs overproduced in BAScl-tTAmice are themselves reprogrammed by this environment into fibrotic inflammatory cells, expressing high levels of extracellular matrix proteins, metalloproteases, and inflammatory cytokines at the expense of HSC maintenance factors including SCF and CXCL12 (Schepers et al, 2013), hence compromising HSC maintenance. Strikingly, however, LSC maintenance is unaffected, demonstrating that inflammatory feedback from MPN cells alters the BM niche into a self-reinforcing leukemic niche favoring the maintenance of transformed LSCs over normal HSCs and facilitating disease progression (Fig. 2B). Studies using models of acute myelogenous leukemia (AML) and multiple myeloma (MM) have also demonstrated that disease-mediated BM remodeling inhibits normal hematopoiesis, in these cases through CCL3-dependent osteoblast depletion (Vallet et al., 2011, Frisch et al., 2012). Together these findings are a critical illustration of how pro-inflammatory cytokines can significantly impact the composition of the BM niche and its ability to maintain the HSC pool. Moreover they suggest that targeting signals driving such remodeling may rebalance the BM niche to favor normal hematopoiesis, thereby slowing or halting disease progression.
Conclusions: an expanding cast of players in HSC regulation
Although long considered primarily as activators of immune cell function, a compelling body of evidence now demonstrates that pro-inflammatory cytokines strongly impact the size and lineage distribution of the blood system via direct reprogramming of HSCs, immature hematopoietic progenitors and the BM niche that supports it. While our understanding of the function of inflammatory cytokines on blood development has advanced, a number of outstanding questions remain to be addressed. In particular, understanding the direct cellular target(s) of individual pro-inflammatory cytokines is a critical next step in uncovering the anatomy of inflammatory responses and their implications for the functionality of the hematopoietic system. Indeed, while IFNs act on HSCs and IL-6 acts on MPPs, other cytokines such as IL-1 and IL-17, which have similar myelopoietic effects, may have different target cells. Hence, recent work has suggested that IL-1 may target CMPs, whereas IL-17 may act indirectly through induction of G-CSF (Nagareddy et al., 2014, Forlow et al., 2001). Collectively this demonstrates that inflammatory responses can produce cytokines that target the hematopoietic hierarchy at multiple levels, with potential consequences for blood regeneration. Moreover, the potential impact of secondary factors produced in response to these cytokines cannot be overlooked (Schurch et al., 2014).
Recent advances in single-cell tracking technology have opened important avenues in understanding how pro-inflammatory cytokines regulate the fate of their target cells. M-CSF and G-CSF were first demonstrated to have true ‘instructive’ (inducing a molecular program driving lineage choice in an uncommitted cell) effects on GMP differentiation using continuous single-cell tracking (Reiger et al., 2009). A similar approach was then employed to demonstrate an instructive effect of M-CSF on HSC differentiation, allowing the identification of a discrete Pu.1-driven molecular program (Mossadegh-Keller et al., 2013). While such in vitro studies must be validated using in vivo assays, they provide critical insights on how inflammatory cytokines reprogram HSCs under normal and pathogenic conditions.
As we begin mapping the interaction between the inflammatory cytokine milieu and the hematopoietic hierarchy, it also becomes essential to identify how chronic exposure to these factors alters the functionality of the HSC compartment in vivo. Indeed, chronic exposure of mice to pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) severely impacts HSC repopulating potential (Esplin et al., 2011, Zhao et al., 2013). LPS itself induces a wide array of pro-inflammatory cytokines including IL-6, TNF and IFN-Is in addition to direct signaling to HSCs themselves, hence reiterating the complexity of physiological inflammatory responses (Zhao et al., 2013, Zhao et al, 2014). While stimuli such as LPS or pathogen infection are useful immunological study tools, they may not recapitulate the unique pro-inflammatory cytokine milieus associated with different BM failure syndromes, chronic infections, obesity, and other diseases, and in turn may not address the diverse hematological manifestations associated with these conditions. Thus, to more clearly understand how different types chronic inflammation impairs HSC function, the distinct contribution(s) of individual cytokines and/or direct signals in different inflammatory conditions must be clearly identified, ideally through combinatorial study of how individual cytokines affect HSC function in vivo and in vitro, and through the use of models that faithfully recapitulate the inflammatory and hematological manifestations of human disease conditions. Moreover, such work must discern whether the effects of chronic inflammation on HSCs are reversible, or result in heritable (and cumulative) epigenetic or genomic changes that could promote leukemic transformation or disease evolution. This type of studies will significantly enhance our understanding of how inflammation and HSC biology intersect, and may serve as powerful pre-clinical testbeds for therapies that limit or reverse the detrimental effects of chronic inflammation on HSC function and blood production.
While our understanding of the cellular components of the BM niche has improved substantially in recent years, relatively little is known about their participation in inflammatory responses and the effects of such responses on the integrity and ability of the BM niche to support HSC maintenance. However, a growing body of evidence suggests that stromal cells in the BM niche play a fundamental role in regulating inflammatory responses (Bernardo and Fibbe, 2013, Schurch et al., 2014). As approaches for the isolation and analysis of BM stromal populations have improved, comprehensive analyses of cytokine production by BM niche cells in the context of inflammation or hematopoietic disease is increasingly feasible. Moreover, the widening range of lineage-specific conditional knockout mouse models (Joseph et al., 2013) should provide significant flexibility in using genetic approaches to demonstrate the relative importance of individual pro-inflammatory cytokines and the cells producing them. Thus, the impact of pro-inflammatory cytokines on the cellular makeup and function of the BM niche must be considered, particularly in understanding the pathogenesis of blood dysfunction in the context of diseases with chronic inflammatory components.
Taken together, pro-inflammatory cytokines are emerging as novel and fundamental regulators of hematopoiesis, hence expanding the cast of players regulating the function of HSCs, progenitors and the BM niche. Moreover, they highlight the important point that blood malignancies are often more complex than the cell-intrinsic genetic lesions that initiate cellular transformation, with critical and potentially targetable roles for the cell-extrinsic pro-inflammatory environment in disease development. Future studies should therefore continue to uncover the roles these cytokines play in regulating acute HSC responses to infection and injury, and in the pantheon of debilitating hematological, autoimmune, and chronic inflammatory diseases affecting human patients.
Highlights.
Emerging work identifies how pro-inflammatory cytokines impact HSC biology.
Significant interest lies in how inflammation remodels the BM niche in disease.
Identifying how chronic inflammation alters HSC function is a critical new frontier.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Wahab OI, Levine RL. Primary myelofibrosis: update on definition, pathogenesis, and treatment. Annu Rev Med. 2009;60:233–245. doi: 10.1146/annurev.med.60.041707.160528. [DOI] [PubMed] [Google Scholar]
- Anand M, Chodda SK, Parikh PM, Nadkarni JS. Abnormal levels of proinflammatory cytokines in serum and monocyte cultures from patients with chronic myeloid leukemia in different stages, and their role in prognosis. Hematol Oncol. 1998;16:143–154. doi: 10.1002/(sici)1099-1069(199812)16:4<143::aid-hon628>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo ME, Fibbe WE. Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell. 2013;13:392–402. doi: 10.1016/j.stem.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Bryder D, et al. Self-renewal of multipotent long-term repopulating hematopoietic stem cells is negatively regulated by Fas and tumor necrosis factor receptor activation. J. Exp. Med. 2001;194:941–952. doi: 10.1084/jem.194.7.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin AM, Demirel O, Hooibrink B, Brandts CH, Nolte MA. Interferon-gamma impairs proliferation of hematopoietic stem cells in mice. Blood. 2013;121:3578–3585. doi: 10.1182/blood-2012-05-432906. [DOI] [PubMed] [Google Scholar]
- Dybedal I, Bryder D, Fossum A, Rusten LS, Jacobsen SE. Tumor necrosis factor (TNF)-mediated activation of the p55 TNF receptor negatively regulates maintenance of cycling reconstituting human hematopoietic stem cells. Blood. 2001;98:1782–1791. doi: 10.1182/blood.v98.6.1782. [DOI] [PubMed] [Google Scholar]
- Ehninger A, Boch T, Uckelmann H, Essers MA, Mudder K, Sleckman BP, Trumpp A. Post-transcriptional regulation of c-Myc expression in adult murine HSCs during homeostasis and interferon-α induced stress response. Blood. 2014 doi: 10.1182/blood-2013-10-531038. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Esplin BJ, Shimazu T, Welner RS, Garrett KP, Nie L, Zhang Q, Humphrey MB, Yang Q, Borghesi LA, Kincade PW. Chronic Exposure to a TLR LIgand Injures Hematopoietic Stem Cells. J Immunol. 2011;186:5367–5375. doi: 10.4049/jimmunol.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M, Offner S, Blanco-Bose W, Waibler Z, Kalinke U, Duchosal M, Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Forlow SB, Schurr JR, Kolls JK, Bagby GJ, Schwarzenberger PO, Ley K. Increased granulopoiesis through interleukin-17 and granulocyte colony-stimulating factor in leukocyte adhesion molecule-deficient mice. Blood. 2001;98:3309–3314. doi: 10.1182/blood.v98.12.3309. [DOI] [PubMed] [Google Scholar]
- Frisch BJ, Ashton JM, Xing L, Becker MW, Jordan CT, Calvi LM. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119:540–550. doi: 10.1182/blood-2011-04-348151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiselhart A, Lier A, Walter D, Milsom MD. Disrupted Signaling through the Fanconi Anemia Pathway Leads to Dysfunctional Hematopoietic Stem Cell Biology: Underlying mechanisms and Potential Therapeutic Strategies. Anemia. 2012;2012:265790. doi: 10.1155/2012/265790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Ioannou S, Hatzis G, Vlahadami I, Voulgarelis M. Aplastic anemia associated with interferon alpha 2a in a patient with chronic hepatitis C virus infection: a case report. J Med Case Reports. 2010;4:268. doi: 10.1186/1752-1947-4-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Hirano T. IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev. 2002;13:357–368. doi: 10.1016/s1359-6101(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Joseph C, Quach JM, Walkley CR, Lane SW, Lo Celso C, Purton LE. Deciphering hematopoietic stem cells and their niches: a critical appraisal of genetic models, lineage tracing, and imaging strategies. Cell Stem Cell. 2013;13:520–533. doi: 10.1016/j.stem.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kiladjian JJ, Mesa RA, Hoffman R. The renaissance of interferon therapy for the treatment of myeloid malignancies. Blood. 2011;18:4706–4715. doi: 10.1182/blood-2010-08-258772. [DOI] [PubMed] [Google Scholar]
- King KY, Baldridge MT, Weksberg DC, Chambers SM, Lukov GL, Wu S, Boles NC, Jung SY, Qin J, Liu D, Songyang Z, Eissa NT, Taylor GA, Goodell MA. Irgm1 protects hematopoietic stem cells by negative regulation of IFN signaling. Blood. 2011;118:525–1533. doi: 10.1182/blood-2011-01-328682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y, Bruns I, Scheiermann C, Amhed J, Pinho S, Zhang D, Mizogunchi T, Wei Q, Lucas D, Ito K, Mar JC, Bergman A, Frenette PS. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Pongubala JMR, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin Immunology. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Li X, Plett PA, Yang Y, Hong P, Freie B, Srour EF, Orschell CM, Clapp DW, Haneline LS. Fanconi anemia type C-deficient hematopoietic stem/progenitor cells exhibit aberrant cell cycle control. Blood. 2003;102:2081–2084. doi: 10.1182/blood-2003-02-0536. [DOI] [PubMed] [Google Scholar]
- Maeda K, Baba Y, Nagai Y, Miyazaki K, Malykhin A, Nakamura K, Kincade PW, Sakaguchi N, Coggeshall KM. IL-6 blocks a discrete early step in lymphopoiesis. Blood. 2005;106:879–885. doi: 10.1182/blood-2005-02-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz MG, Boettcher S. Emergency granulopoiesis. Nature Rev Immunol. 2014;14:302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- Matatall KA, Shen CC, Challen GA, King KY. Type II Interferon Promotes Differentiation of Myeloid-Biased Hematopoietic Stem Cells. Stem Cells. 2014 doi: 10.1002/stem.1799. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, Morrison CG, Passegué E. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:74–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–34. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, Moore J, Sieweke MH. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497:239–243. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally A, Bruedigam C, Poveromo L, Heidel FH, Purdon A, Vu T, Austin R, Heckl D, Breyfogle LJ, Kuhn CP, Kalaitzidis D, Armstrong SA, Williams DA, Hill GR, Ebert BL, Lane SW. Depletion of Jak2V617F myeloproliferative neoplasm-propagating stem cells by interferon-α in a murine model of polycythemia vera. Blood. 2013;121:3692–3702. doi: 10.1182/blood-2012-05-432989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagareddy PR, Kraakman M, Masters SL, Stirzaker RA, Gorman DJ, Grant RW, Dragoljevic D, Hong ES, Abdel-Latif A, Smyth SS, Choi SH, Korner J, Bornfeldt KE, Fisher EA, Dixit VD, Tall AR, Goldberg IJ, Murphy AJ. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metabolism. 2014;19:821–835. doi: 10.1016/j.cmet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Lakshminarasimhan R, Techner JM, Fong S, Flach J, Binnewies M, Passegué E. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 2014;211:245–262. doi: 10.1084/jem.20131043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Prendergast AM, Essers MAG. Hematopoietic stem cells, infection, and the niche. Ann N.Y. Acad Sci. 2014;1310:51–57. doi: 10.1111/nyas.12400. [DOI] [PubMed] [Google Scholar]
- Pronk CJH, Veiby OP, Bryder D, Jacobsen SEW. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J. Exp. Med. 2011;208:1563–1570. doi: 10.1084/jem.20110752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel VI, Hartnett S, Hill GR, Lazo-Kallanian SB, Ferrara JL, Sieff CA. Essential role for the p55 tumor necrosis factor receptor in regulating hematopoiesis at a stem cell level. J Exp Med. 1999;190:1493–1504. doi: 10.1084/jem.190.10.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP, Passegué E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzoug F, Huang Y, Tanner MK, Wysoczynski M, Schanie CL, Chilton PM, Ratajczak MZ, Fugier Vivier, I.J., Ilstad ST. TNF-alpha is critical to facilitate hemopoietic stem cell engraftment and function. J. Immunol. 2008;180:49–57. doi: 10.4049/jimmunol.180.1.49. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T, Wagers AJ, Hsiao EC, Passegué E. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettpelz LG, Borgerding JN, Christopher MJ, Gopalan PK, Romine MP, Herman AC, Woloszynek JR, Greenbaum AM, Link DC. G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia. 2014 doi: 10.1038/leu.2014.68. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch CM, Riether C, Ochsenbein AF. Cytotoxic CD8(+) T cells stimulate hematopoietic progenitors by promoting cytokine release from bone marrow mesenchymal stromal cells. Cell Stem Cell. 2014;14:460–472. doi: 10.1016/j.stem.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Vallet S, Pozzi S, Patel K, Vaghela N, Fulciniti MT, Veiby P, Hideshima T, Santo L, Cirstea D, Scadden DT, et al. A novel role for CCL3 (MIP-1α) in myeloma-induced bone disease via osteocalcin downregulation and inhibition of osteoblast function. Leukemia. 2011;25:1174–81. doi: 10.1038/leu.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Deb DK, Sassano A, Uddin S, Varga J, Wickrema A, Platanias LC. Activation of the p38 Mitogen-activated Protein Kinase Mediates the Suppressive Effects of Type I Interferons and Transforming Growth Factor-β on Normal Hematopoiesis. J Biol Chem. 2002;277:7726–7735. doi: 10.1074/jbc.M106640200. [DOI] [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med. 2011 doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- Winkler IG, Pettit AR, Raggatt LJ, Jacobsen RN, Forristal CE, Barbier V, Nowlan B, Cisterne A, Bendall LJ, Sims NA, Levesque J-P. Hematopoietic stem cell mobilizing agents G-CSF, cyclophosphamide or AMD3100 have distinct mechanisms of action on bone marrow HSC niches and bone formation. Leukemia. 2012;26:1594–1601. doi: 10.1038/leu.2012.17. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jones M, McCabe A, Winslow GM, Avram D, MacNamara KC. MyD88 signaling in CD4 T cells promotes IFN-γ production and hematopoietic progenitor cell expansion in response to intracellular bacterial infection. J Immunol. 2013:4725–4735. doi: 10.4049/jimmunol.1203024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JL, Ma C, O’Connell RM, Mehta A, DiLoreto R, Heath JR, Baltimore D. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell. 2014;14:445–459. doi: 10.1016/j.stem.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ling F, Wang HC, Sun XH. Chronic TLR signaling impairs the long-term repopulating potential of hematopoietic stem cells of wild type but not Id1 deficient mice. PLoS One. 2013;8:e55552. doi: 10.1371/journal.pone.0055552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Fenaux P. Myelodysplastic syndromes (MDS) and autoimmune disorders (AD): cause or consequence? Best Prac Res Clin Haematol. 2013;4:327–336. doi: 10.1016/j.beha.2013.09.003. [DOI] [PubMed] [Google Scholar]