Abstract
Nitric oxide (NO) synthase 2 (NOS2), a major inflammatory protein, modulates disease progression via NO in a number of pathologies, including cancer. The role of NOS2-derived NO is not only flux-dependent, which is higher in mouse vs. human cells, but also varies based on spatial and temporal distribution both within tumor cells and in the tumor microenvironment. NO donors have been utilized to mimic NO flux conditions and to investigate the effects of varied NO concentrations. As a wide range of effects mediated by NO and other nitrogen oxides such as nitroxyl (HNO) have been elucidated, multiple NO- and HNO-releasing compounds have been developed as potential therapeutics, including as tumor modulators. One of the challenges is to determine differences in biomarker expression from extracellular vs. intracellular generation of NO or HNO. Taking advantage of new NO and HNO releasing agents, we have characterized the gene expression profile of estrogen receptor-negative human breast cancer (MDA-MB-231) cells following exposure to aspirin, the NO donor DEA/NO, the HNO donor IPA/NO and their intracellularly-activated prodrug conjugates DEA/NO-aspirin and IPA/NO-aspirin. Comparison of the gene expression profiles demonstrated that several genes were uniquely expressed with respect to NO or HNO, such as miR-21, HSP70, cystathionine γ-lyase and IL24. These findings provide insight into targets and pathways that could be therapeutically exploited by the redox related species NO and HNO.
Keywords: nitric oxide, nitroxyl, nitric oxide synthase, breast cancer, cell signaling, cancer biology
NOS2 in Disease
Nitric oxide (NO) synthase 2 (NOS2; aka inducible NOS, iNOS) is an inflammatory protein in humans and mice. NOS2 modulates disease initiation and progression in pathologies including asthma, neurological disorders and cancer [1–6]. While the specific activities of human and mouse NOS2 are nearly identical, the promoter regions differ dramatically, leading to significantly higher levels of NO generated by mouse vs. human cells [7–10]. These observations implicate differences in the physiological roles of NOS2-derived NO in the two species. For example, elevated NOS2-derived NO generated by murine immune cells acts as an immunotoxin [10,11]. In contrast, human NOS2-derived NO is generated at a much lower flux and mediates pro-survival signaling and cancer progression [7,12]. The flux-dependent role of NO suggests that care must be taken when extrapolating data from murine models to humans. Nonetheless, understanding of the chemical biology of NO has relied heavily on analysis in both humans and rodents.
In the 1980s, Hibbs and coworkers implicated a role for NO in the tumoricidal effects of activated murine macrophages [13,14]. Mechanistically, Stuehr and Nathan later reported the induction of apoptosis in leukemic cells by NOS2-derived NO [15]. Generation of NOS2-derived NO by cytokine stimulated rodent vascular cells also imparted cytotoxic effects in a human erythroleukemia cell co-culture model, through the involvement of nonheme iron-nitrosyl complex formation and inhibition of mitochondrial complexes I and II [16]. Similarly, Xie and coworkers demonstrated anti-tumor activity of transfected murine NOS2 in a variety of murine cancer cell lines [17]. This tumoricidal activity involved high NO fluxes, leading to nitrosation of amines to form carcinogenic nitrosamines in murine macrophages [18].
Tannenbaum and coworkers first reported elevated nitrite/nitrate levels in the urine of a subset of healthy volunteers who were immune stimulated due to a temporary infection, which suggested the involvement of NO during human response to disease and inflammation [19]. Subsequent studies demonstrated increased nitrite and nitrate in human hepatocytes, which lead to the cloning of human NOS2 [20–22]. Moncada and coworkers went on to assess NOS2 activity in human tumors and cancer cells [23–25]. They observed that while human tumor cells with high NOS2 expression grew slowly in culture, tumor xenografts of these cells exhibited more aggressive tumor characteristics, suggesting an oncogenic role of human NOS2 in disease progression. Ambs and Harris also showed that elevated NOS2 expression in human colon adenomas correlated with elevated tumor progression and angiogenesis [26–28]. They went on to show NOS2 regulation by p53 [29]; while high NO fluxes induced p53 stabilization, p53 inhibited NOS2 expression, indicating p53 regulation of NOS2 via negative feedback [30,31]. Thus, mutations in p53 can lead to chronic NOS2 expression in the tumor epithelium.
Jeannin and coworkers explored the role of NOS2 expression in tumor progression using two murine syngeneic models [32]. Two clonal populations were isolated from EMT-6 breast cancer cells; one clone (EMT-6J) expressed constitutive NOS2 and secreted high levels of NO while the other clone (EMT-6H) did not. Cytokine stimulation (IL-1β + TNF-α) induced NOS2 expression and nitrite/nitrate production in both clones with EMT-6J cells producing 15-fold higher NO than EMT-6H cells. In vivo studies demonstrated increased survival of EMT-6J tumor-bearing BALB/c mice when compared to the EMT-6H model, suggesting that high fluxes of NO abate tumor progression. Interestingly, when the same study was repeated in wild type and NOS2 knockout animals, tumor-bearing NOS2 knockout mice survived longer, independent of the cell clone xenograft. While elevated tumor NOS2 (15-fold greater) mediated cytotoxic and cytostatic effects on tumor growth, the authors also suggested that FAS-mediated cell death, and the involvement of CTL and NK cells may be important in the anti-tumor effects of high-flux NO. On a similar note, other reports have shown that NO donors can promote FAS and TRAIL apoptotic pathways through inhibition of nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) [33–37]. Thus, the impact of NO on cellular and disease processes is temporally-, spatially-, and concentration-dependent.
In contrast, our in vitro model indicated that NOS2 induction in human breast cancer cells produced an NO flux of <300 nM [7]. This level of NO promoted cell migration and drug resistance, which was abated by the NOS2 inhibitor aminoguanidine [7]. Importantly, aminoguanidine also dramatically decreased growth rates of MB-231 tumor xenografts and brain metastases [7]. Moreover, while the same level of NO activated NFκB, NO fluxes exceeding this level has inhibitory effects, as demonstrated by a bell shaped NO dose response. These results suggest that NO can inhibit pro-survival signaling through nitrosative mechanisms [38]. Similarly, reactive nitrogen species (RNS) associated with high NO flux mediated anti-tumor activity by inhibiting both EGFR and NFκB signaling [39]. Importantly, examination of the gene expression profile of high NOS2 expressing breast tumors suggests that this level of NO induces tumor biomarker expression that leads to increased metastasis and poor outcome [7,12]. Thus, when compared to human NOS2, fully activated murine NOS2 produces a higher localized flux of NO with vastly different effects.
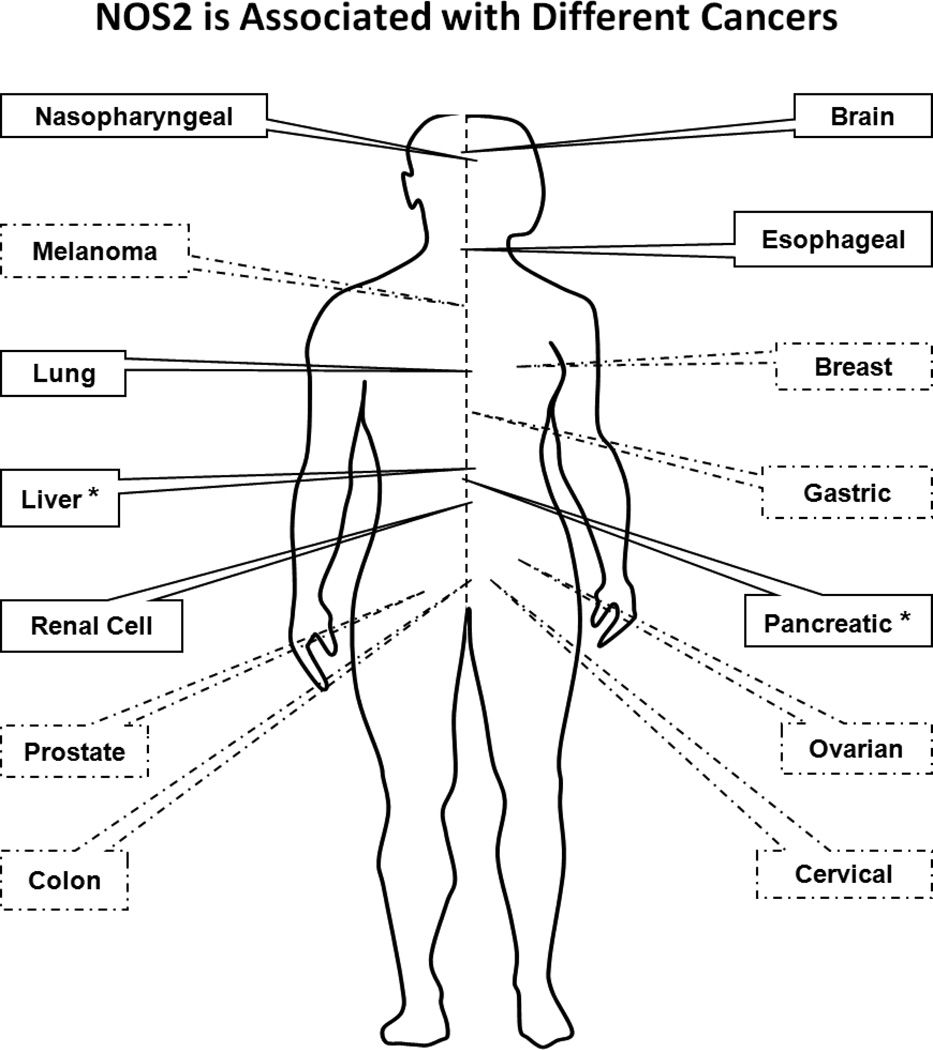
Elevated tumor NOS2 expression in human tumors now correlates with increased aggressiveness in brain, lung, pancreas, breast and colon cancers [5,12,40–42] (Figure 1). Recent reports have shown that NOS2 expression in tumor versus leukocytes is a key determining factor that dictates tumor progression and immunosuppression. Elevated levels of NO in leukocytes increase cellular toxicity, which either directly or indirectly orchestrates the tumor-host immune response [43–45]. While there are contradicting reports with respect to the role of NOS2 in tumor progression, for a diagnostic point of view, NOS2 is not a single prognostic marker but rather should be employed in conjunction with other tumor biomarkers to yield detailed information about cancer phenotype. Such information may lead to improved treatment regimens and disease outcomes. Clinical studies demonstrating localization of NOS2 in the tumor cell vs. stroma or leukocyte have shown that elevated NOS2 expression in the tumor epithelium correlates with reduced disease-specific survival. Thus, clinical studies implicate a key role of the spatial dependence of NO in modulating tumor behavior and tumor-host immune response.
Figure 1.
NOS2 is associated with different cancers. Dashed lines: predict poor prognosis. Solid lines: weak to not predictive or not studied. Asterisk: some studies indicate that NOS2 is associated with poor prognosis.
Mimics of NO-flux Profile In Vivo
To understand the mechanisms of NO in cellular function and the dependence on NO concentration, various donors have been used to mimic NO flux conditions at the cellular level [46]. Toward this end, NONOates, due to their controlled release profiles with little influence on extraneous conditions, have emerged as indispensable tools to assess or mimic the effects of intracellular NOS-derived NO [47]. Using different NONOates at varying concentrations, our laboratory and others have identified NO flux-dependent signaling profiles that mediate different biological mechanisms. Brune and coworkers identified that HIF-1α and p53 levels are increased by different NO concentrations, thus demonstrating the NO flux-dependence of cell signaling [48–50]. Thomas et al. showed that low NO levels (< 50 nM steady state NO) increased cGMP-dependent mechanisms, while higher levels (500–800 nM steady state NO) led to increased HIF-1α and p53 [51,52]. Importantly, this work demonstrated a pro-tumorigenic level of NO at lower concentrations, while higher NO fluxes induced stress responses involving p53. Similar responses have been observed in a variety of cells including endothelial cells [53]. Taken together, NO-induced signaling occurs at concentrations ranging from high picomolar to low micromolar. This five-order of magnitude concentration range provides insight into the versatility of NO signaling while emphasizing the importance of concentration, spatial and temporal constraints, In particular, chromic exposure to elevated NO fluxes profoundly affects cancer phenotype [54,55].
Nitric Oxide as a Cancer Therapeutic
A wide range of NO-releasing compounds have been developed as potential therapeutic agents to exploit the diverse biological roles of NO. These donors range from organic nitrates such as glyceryl trinitrate or derivatized NSAIDs to S-nitrosothiols and NONOates [47,56,57]. Macromolecular NO-releasing scaffolds are also promising due to their ability to store and deliver larger NO payloads in a more controlled and effective manner compared to low molecular weight donors [58,59]. In addition to NO donors, there are numerous NOS inhibitors and scavengers that can be used for modulation of NO flux [60,61]. Strategically combining these donors and inhibitors may provide unique tools for exploring potential therapeutic applications of NO [62]. Also, drug resistance and the toxicity of therapeutics are both clinical hurdles. In the face of these obstacles, NO modulation has emerged as a powerful adjuvant for the hypersensitization of tumors to more traditional chemo- and radiation therapies. Furthermore, emerging evidence indicates that NO donors have the potential to function independently in the clinical management of cancer [63].
Nitric oxide donors of the NONOate class are routinely used as reliable sources of NO in laboratory settings [64,65]. Such donors have been instrumental in exploration of the concentration and temporal profiles of NO with respect to tumor response. However, using these compounds in vivo to treat solid tumors is problematic due to systemic hypotension, which has limited the amount of NO that can be delivered to the tumor. However, clinical application of sub-vasoactive levels of glyceryl trinitrate has shown some success in limiting recurrence of prostate and lung cancers [66–68]. In addition, a new generation of prodrugs have emerged based on the strategy of activation at the lesion site to abate the adverse cardiovascular effects of systemically administered drugs that modulate NO flux [69].
Another area of research involves development of hybrid compounds of functional groups with potential therapeutic value conjugated with NO donors (e.g., ester nitrates, furoxans, benzofuroxans, NONOates, S-nitrosothiols, metal nitrosyl complexes) designed to release NO while maintaining the native drug activity. This approach has proved useful in targeting cardiovascular, inflammatory, bacterial, fungal, viral, parasitic, and ocular diseases as well as cancer [70]. Potent and selective NOS inhibitors are also being designed, often through an enzyme structure based process. However, the high homology shared by the NOS isoforms, is a challenge in this pursuit.
Several reports have suggested that NOS may generate nitroxyl (HNO) [71–74] in addition to NO, in a condition-dependent manner. In cardiovascular models, HNO donors were shown to elicit unique effects when compared to NO [75–77], thus suggesting related but distinct targeting mechanisms [78,79]. Donors of HNO has also been shown to have anticancer activity [80], in part due to modification of a critical thiol of the glycolytic enzyme GAPDH [80]. As with NO, several classes of HNO donor have been developed, including NONOates [81,82].
We recently demonstrated that the NONOate-based prodrugs DEA/NO-aspirin and IPA/NO-aspirin, which donate NO and HNO, respectively, were activated by esterase cleavage within the cell [83]. These prodrugs thus facilitate intracellular delivery of these redox modulators compared to the parent NONOates. Interestingly, the intracellular release of HNO by IPA/NO-aspirin and, to a lesser extent, NO by DEA/NO-aspirin inhibited proliferation of A549 non-small cell lung cancer cells. In contrast, the same prodrugs were not appreciable toxic toward primary human endothelial cells (HUVECs). This observation suggests that IPA/NO-aspirin and DEA/NO-aspirin induce cancer-specific anti-proliferative pathways. The prodrugs also were protective against the gastrointestinal toxicity associated with aspirin in rats.
Chemical Induction of NO and HNO Inside and Outside of the Cell
Our recent work has demonstrated a key role of NOS2 in estrogen receptor negative (ER(-)) breast cancer disease progression [7]. Here, we employed MB-231 ER(-) breast cancer cells to examine the presence of NOS2 responsive gene set. We also were sought to explore the effects of intracellular and extracellular delivery of HNO and NO. MB-231 cells were exposed to 50 µM aspirin, DEA/NO, IPA/NO, DEA/NO-aspirin, or IPA/NO-aspirin or to 10 mM NaOH (vehicle control) for 24 h. DEA/NO and IPA/NO are short-lived donors (half-lives of ∼5 min under physiological conditions [84,85]). Thus, NO/HNO levels will rise quickly to a maximum and then decay [86]. The aspirin derivatives are much more stable to spontaneous hydrolysis, with half-lives of 7.5 and ∼36 h [87]. In the presence of cellular esterases, hydrolysis is accelerated, such that complete dissociation of IPA/NO aspirin occurs in ∼150 min. DEA/NO-aspirin is somewhat longer-lived in cells. Slower donation will lead to lower, but more steady fluxes of HNO or NO.
Following treatment, RNAs were harvested with TRIzol, followed by reverse transcription and in vitro transcription to generate sufficient aRNA fragments, and then hybridized to an Affymetrix GeneChip Human Gene ST Array for transcriptome analysis. High throughput gene expression data were exported to BRB ArrayTools, Partek Genomic Analysis Suite for data mining and Ingenuity Pathway Analysis for pathway analysis.
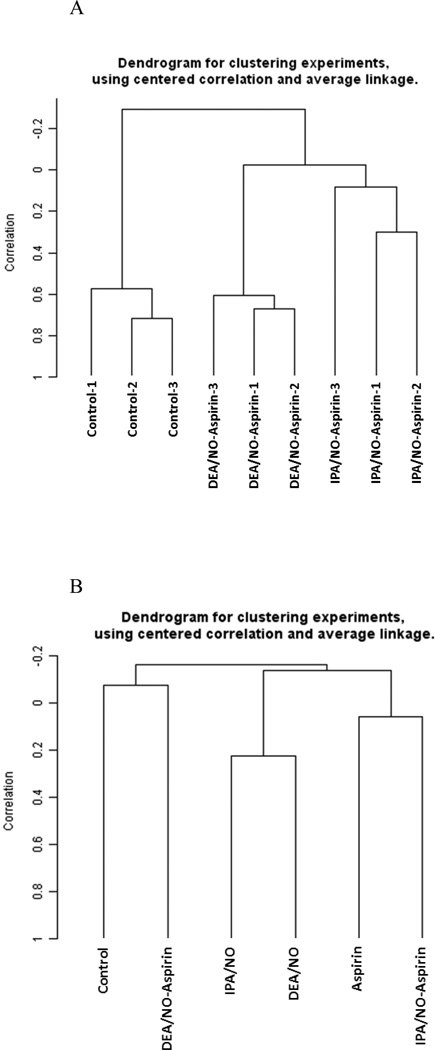
For a global view of the RNA fingerprinting data, multivariate analysis was adopted. Non-supervised hierarchical clustering analysis using different combinations of linkage methods and distance measuring algorithms confirmed that the gene expression fingerprinting for different treatment groups is not only consistent but also unique (Figure 2). In order to identify unique gene subsets that can differentiate each treatment group, class comparison analysis was adopted to identify subsets of genes that are statistically significant and uniquely expressed among the treatment groups (Tables 1–6). Genes identified in the comparison analyses are components of various pathways, which as described below.
Figure 2.
Hierarchical Clustering Analysis. Non-supervised learning hierarchical clustering analysis computed with different combinations of single, complete and average linkage methods and Euclidean, centered correlation and uncentered correlation distance measurement algorithms. Figure 2A showed the clear separation of control, IPA/NO-aspirin and DEA/NO-aspirin groups. Figure 2B showed the clustering patterns when all six treated groups were analyzed together.
Table 1.
Class Comparison Analysis: Differentially expressed genes in control vs IPA/NO vs DEA/NO
| Parametric p-value |
FDR | Control | IPA/NO aspirin |
DEA/NO aspirin |
Symbol | Name |
|---|---|---|---|---|---|---|
| 5.00E-07 | 0.00044 | 158.29 | 182.26 | 357.41 | MIR21 | microRNA 21 |
| 1.20E-06 | 0.00044 | 134.74 | 374.85 | 168.1 | HSPA1A | heat shock 70kDa protein 1A |
| 1.60E-06 | 0.00044 | 178.03 | 440.57 | 229.01 | HSPA1B | heat shock 70kDa protein 1B |
| 2.60E-06 | 0.000557 | 691.18 | 307.05 | 532.37 | CTH | cystathionase (cystathionine gamma-lyase) |
| 5.40E-06 | 0.000946 | 550.46 | 259.01 | 346.26 | GTPBP2 | GTP binding protein 2 |
| 6.70E-06 | 0.00108 | 69.32 | 37.01 | 52.14 | SYT1 | synaptotagmin I |
| 7.70E-06 | 0.00114 | 276.85 | 470.1 | 427.43 | MASTL | microtubule associated serine/threonine kinase- like |
| 9.00E-06 | 0.00122 | 129.41 | 71.47 | 103.75 | ZNF643 | zinc finger protein 643 |
| 1.31E-05 | 0.00146 | 47.71 | 80.12 | 76.08 | KATNAL1 | katanin p60 subunit A-like 1 |
| 1.85E-05 | 0.00178 | 113.37 | 183.65 | 130.16 | ARRB1 | arrestin, beta 1 |
| 2.53E-05 | 0.00212 | 60.59 | 125.97 | 70.11 | MIR27B | microRNA 27b |
| 3.31E-05 | 0.00251 | 27.11 | 48.35 | 26.25 | ID2 | inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| 3.39E-05 | 0.00251 | 43 | 81.96 | 62.54 | CSNK1G1 | casein kinase 1, gamma 1 |
| 3.73E-05 | 0.00255 | 141.68 | 255.44 | 216.93 | CDCA3 | cell division cycle associated 3 |
| 3.84E-05 | 0.00255 | 310.44 | 136.8 | 270.54 | DDIT3 | DNA-damage-inducible transcript 3 |
| 4.50E-05 | 0.00289 | 1031.9 | 1113.08 | 1691.28 | MMP1 | matrix metallopeptidase 1 (interstitial collagenase) |
| 4.79E-05 | 0.00298 | 26.13 | 37.57 | 49.8 | C5orf54 | chromosome 5 open reading frame 54 |
| 5.81E-05 | 0.00334 | 79.86 | 77.24 | 132.14 | MIR100 | microRNA 100 |
| 5.89E-05 | 0.00334 | 488.23 | 274.58 | 356.01 | UFM1 | ubiquitin-fold modifier 1 |
| 6.35E-05 | 0.0034 | 1770.85 | 826.61 | 894.51 | TXNIP | thioredoxin interacting protein |
| 6.84E-05 | 0.0034 | 289.23 | 156.84 | 189.52 | MXD1 | MAX dimerization protein 1 |
| 7.23E-05 | 0.0034 | 54 | 111.12 | 75.49 | TGFBR3 | transforming growth factor, beta receptor III |
| 8.39E-05 | 0.00385 | 145.67 | 216.93 | 105.31 | SNORD14E | small nucleolar RNA, C/D box 14E |
| 8.80E-05 | 0.00394 | 97.67 | 135.92 | 148.06 | FAM36A | family with sequence similarity 36, member A |
| 0.0001013 | 0.00438 | 16.26 | 41.79 | 25.53 | KLRC4-KLRK1 | KLRC4-KLRK1 readthrough |
| 0.0001033 | 0.00438 | 186.29 | 219.91 | 320.13 | CPA4 | carboxypeptidase A4 |
| 0.0001045 | 0.00438 | 819.14 | 396.85 | 530.51 | SLFN5 | schlafen family member 5 |
| 0.0001127 | 0.00456 | 44.67 | 70.34 | 49.72 | CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 |
| 0.0001186 | 0.00466 | 151.53 | 241.16 | 184.47 | SOS1 | son of sevenless homolog 1 (Drosophila) |
| 0.0001275 | 0.00491 | 125.65 | 56.4 | 94.63 | SLC16A1 | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) |
| 0.0001711 | 0.00611 | 37.19 | 56.42 | 33.29 | KLRC4 | killer cell lectin-like receptor subfamily C, member 4 |
| 0.0001782 | 0.00624 | 32.9 | 55.11 | 35.37 | CYP1A1 | cytochrome P450, family 1, subfamily A, |
| polypeptide 1 | ||||||
| 0.0002038 | 0.00687 | 95.02 | 163.46 | 105.71 | TFAP2C | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| 0.0002069 | 0.00687 | 145.58 | 217.73 | 265.69 | HIST1H2AK | histone cluster 1, H2ak |
| 0.0002325 | 0.00747 | 30.48 | 42 | 53.64 | MIR15A | microRNA 15a |
| 0.0002495 | 0.00788 | 1318.22 | 731.84 | 1096.74 | ASNS | asparagine synthetase (glutamine-hydrolyzing) |
| 0.0002608 | 0.00811 | 55.23 | 70.89 | 63.14 | TAS2R31 | taste receptor, type 2, member 31 |
| 0.0002742 | 0.00826 | 321.72 | 184.71 | 240.24 | TRIML2 | tripartite motif family-like 2 |
| 0.0003194 | 0.00947 | 40.67 | 19.71 | 27.67 | OLAH | oleoyl-ACP hydrolase |
| 0.0003389 | 0.0096 | 143.33 | 262.36 | 199.65 | FAM102B | family with sequence similarity 102, member B |
| 0.0003919 | 0.0103 | 110.28 | 73.16 | 72.47 | ANKRD11 | ankyrin repeat domain 11 |
| 0.0004377 | 0.011 | 191.39 | 349.72 | 290.22 | GMNN | geminin, DNA replication inhibitor |
| 0.0004424 | 0.011 | 722.45 | 438.41 | 647.09 | CASP4 | caspase 4, apoptosis-related cysteine peptidase |
| 0.0004676 | 0.011 | 470.21 | 848.73 | 689.63 | HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| 0.0004948 | 0.0111 | 123.33 | 272.9 | 202.96 | FAM111B | family with sequence similarity 111, member B |
| 0.0005848 | 0.0122 | 80.38 | 103.67 | 96.64 | ZNF699 | zinc finger protein 699 |
| 0.0007473 | 0.0147 | 83.91 | 164 | 155.71 | CENPI | centromere protein I |
| 0.0007983 | 0.0151 | 442.05 | 561.76 | 586.57 | CDK1 | cyclin-dependent kinase 1 |
| 0.0008052 | 0.0151 | 41.62 | 17.11 | 27.66 | C12orf39 | chromosome 12 open reading frame 39 |
| 0.0008272 | 0.0152 | 276.03 | 131.2 | 174.89 | GDF15 | growth differentiation factor 15 |
| 0.0009894 | 0.0177 | 129.32 | 183.21 | 159.07 | PLK4 | polo-like kinase 4 |
| 0.0010473 | 0.0182 | 190.18 | 247.79 | 233.37 | GPR125 | G protein-coupled receptor 125 |
| 0.0010698 | 0.0184 | 101.54 | 51.49 | 55.33 | UFM1 | ubiquitin-fold modifier 1 |
| 0.0011657 | 0.0197 | 36.87 | 36.84 | 26.34 | SNORA2B | small nucleolar RNA, H/ACA box 2B |
| 0.0011932 | 0.02 | 25.76 | 18.03 | 16.58 | TNFSF18 | tumor necrosis factor (ligand) superfamily, member 18 |
| 0.0013126 | 0.0213 | 78.71 | 152.05 | 121.04 | ORC1 | origin recognition complex, subunit 1 |
| 0.0013441 | 0.0216 | 32.29 | 50.95 | 43.56 | DSEL | dermatan sulfate epimerase-like |
| 0.0013774 | 0.0219 | 81.09 | 110.77 | 97.89 | SERTAD4 | SERTA domain containing 4 |
| 0.0014926 | 0.0233 | 161.4 | 206.74 | 119.33 | SNORD12C | small nucleolar RNA, C/D box 12C |
| 0.0015019 | 0.0233 | 184.74 | 180.78 | 124.2 | EIF4A2 | eukaryotic translation initiation factor 4A2 |
| 0.001705 | 0.0246 | 35.9 | 54.09 | 43.2 | CDRT1 | CMT1A duplicated region transcript 1 |
| 0.0018118 | 0.0255 | 140.04 | 220.86 | 208.61 | MMS22L | MMS22-like, DNA repair protein |
| 0.0018598 | 0.026 | 362.63 | 592.15 | 516.16 | SKP2 | S-phase kinase-associated protein 2 (p45) |
| 0.0019382 | 0.0267 | 33.46 | 57.16 | 41.69 | PIK3R3 | phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| 0.0022295 | 0.0296 | 148.85 | 208.38 | 179.76 | TANC2 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| 0.002407 | 0.0309 | 122.27 | 209.4 | 183.63 | ESCO2 | establishment of cohesion 1 homolog 2 (S. cerevisiae) |
| 0.0024782 | 0.0316 | 299.78 | 478.11 | 464.06 | WDHD1 | WD repeat and HMG-box DNA binding protein 1 |
| 0.0026491 | 0.0333 | 81.77 | 114.13 | 75.33 | S100A4 | S100 calcium binding protein A4 |
| 0.0027266 | 0.0338 | 53.48 | 91.61 | 72.04 | SLC38A4 | solute carrier family 38, member 4 |
| 0.002739 | 0.0338 | 62.92 | 40.49 | 47.37 | SLC5A4 | solute carrier family 5 (low affinity glucose cotransporter), member 4 |
| 0.0028887 | 0.0349 | 834.01 | 538.55 | 516.07 | SNORD15B | small nucleolar RNA, C/D box 15B |
| 0.0030393 | 0.0353 | 66.68 | 88.7 | 91.01 | LRRCC1 | leucine rich repeat and coiled-coil domain containing 1 |
| 0.0034645 | 0.0384 | 69.46 | 107.45 | 91.53 | BMP4 | bone morphogenetic protein 4 |
| 0.0034884 | 0.0384 | 919.74 | 1180.48 | 863.52 | SNORD30 | small nucleolar RNA, C/D box 30 |
| 0.0039179 | 0.0412 | 241.05 | 341.9 | 337.09 | HSPB11 | heat shock protein family B (small), member 11 |
| 0.0039544 | 0.0412 | 121.73 | 189.61 | 186.52 | CCNE2 | cyclin E2 |
| 0.003966 | 0.0412 | 100.5 | 161.62 | 144.65 | HIST1H3A | histone cluster 1, H3a |
| 0.0045144 | 0.044 | 127.21 | 82.32 | 100.69 | U2AF1L4 | U2 small nuclear RNA auxiliary factor 1-like 4 |
| 0.0045306 | 0.044 | 89.42 | 133.28 | 131.31 | NMU | neuromedin U |
| 0.0047269 | 0.0451 | 79.93 | 37.91 | 57.23 | IL24 | interleukin 24 |
| 0.00486 | 0.0456 | 76.16 | 97.2 | 108.87 | HSD17B7P2 | hydroxysteroid (17-beta) dehydrogenase 7 pseudogene 2 |
| 0.0053066 | 0.0478 | 42.77 | 27.46 | 30.84 | RAB7A | RAB7A, member RAS oncogene family |
| 0.0055138 | 0.0488 | 34.57 | 49.41 | 39.87 | OVOS | ovostatin |
| 0.0059611 | 0.052 | 83.56 | 108.25 | 85.13 | SNORD4B | small nucleolar RNA, C/D box 4B |
| 0.0061285 | 0.0527 | 52.16 | 32.28 | 28.13 | NOL7 | nucleolar protein 7, 27kDa |
| 0.0068846 | 0.0562 | 69.47 | 97.5 | 98.3 | FANCB | Fanconi anemia, complementation group B |
| 0.0070186 | 0.0568 | 23.73 | 22.28 | 26.93 | MC3R | melanocortin 3 receptor |
| 0.0071233 | 0.0574 | 301.44 | 496.32 | 409.56 | CLSPN | claspin |
| 0.0081236 | 0.0616 | 195.84 | 213.56 | 150.45 | SNORA62 | small nucleolar RNA, H/ACA box 62 |
| 0.0088207 | 0.0646 | 73.42 | 109.89 | 61.49 | FABP4 | fatty acid binding protein 4, adipocyte |
| 0.0092606 | 0.0661 | 49.63 | 34.77 | 31.84 | LLPH | LLP homolog, long-term synaptic facilitation (Aplysia) |
| 0.0097317 | 0.0679 | 118.67 | 147.13 | 149.61 | SCAI | suppressor of cancer cell invasion |
| 0.0098834 | 0.0685 | 32.32 | 50.91 | 45.37 | GPR137C | G protein-coupled receptor 137C |
Table 6.
Class Comparison Analysis: Differentially expressed genes in aspirin vs DEA/NO-aspirin
| Parametric p-value |
FDR | Aspirin | DEA/NO aspirin |
Symbol | Name |
|---|---|---|---|---|---|
| 0.0003854 | 0.195 | 129.96 | 357.21 | MIR21 | microRNA 21 |
| 0.0039924 | 0.403 | 2063.12 | 894 | TXNIP | thioredoxin interacting protein |
Nitrogen Oxide-Modulated Genes
microRNA 21 (miRNA21)
Micro RNAs (miRNA) are short non-coding RNAs that are involved in post-translational regulation of gene expression in multicellular organisms by affecting both the stability and translation of mRNA. miR21 (aka oncomiR21) expression has been reported in various types of cancers including breast, colorectal and brain and is believed to be oncogenic [88–91]. In this regard, miR21 negatively regulates the tumor suppressor PTEN [92]. Moreover, Harris and coworkers identified enhanced miR21 and PI3K signaling in association with elevated NOS2 expression and KRAS activation in lung cancer cells [93]. miR21 can be detected in human serum and has predicted poor therapeutic outcome in cancer patients [94–99].
When compared to control cells, IPA/NO caused roughly a three-fold increase in miR21, while IPA/NO-aspirin only modestly increased miR21. This may indicate a localization effect of HNO delivery. Interestingly, while miR21 was induced by both DEA/NO-aspirin and IPA/NO-aspirin, our earlier studies have demonstrated cytotoxic activity of these prodrugs [100]. These contrasting observations may implicate miR21 as a potential candidate for modulating drug resistance by these prodrugs.
Cystathionase (cystathionine γ-lyase; CTH)
Cystathionase catalyzes the last step in the trans-sulfuration pathway from methionine to cysteine and also can generate the endogenous signaling molecule hydrogen sulfide (H2S). Pupo et al. reported that H2S is pro-angiogenic and promotes migration in a tumor environment [101,102]. CTH also acts as a cysteine-protein sulfhydrase by mediating sulfhydration of target proteins. The sulfhydration reaction regulates target proteins including the NFκB subunit RelA, PTPN1 and GAPDH by converting -SH groups on specific cysteine residues to -SSH.
Our study demonstrated significant decreased CTH expression in the IPA/NO-aspirin treatment group when compared to control, which may in part explain the anti-angiogenic and anti-metastatic properties of HNO-releasing drugs. Fukuto and colleagues have demonstrated HNO-mediated inhibition of GAPDH activity in human breast cancer cells and tumor xenografts, as well as reduced blood vessel density and growth of tumor xenografts [80]. The authors suggested that the anti-tumor mechanisms of HNO may stem from the inhibition of GAPDH to impede glycolysis and reduce HIF-1α levels and tumor angiogenesis. Importantly, this work demonstrated that the anti-cancer mechanisms of HNO are different from those of current clinically available anti-cancer therapeutics. Collectively, these observations suggest that HNO drugs, including IPA/NO-aspirin, may improve therapeutic tumor response.
Heat Shock 70 kDa Protein 1A/1B (HSPA1A/1B)
Heat shock 70 kDa proteins 1A and 1B are members of the heat shock protein 70 family. In conjunction with other heat shock proteins, these proteins stabilize existing protein aggregates and mediate the folding of newly translated proteins in the cytosol and in organelles. Both HSPA1A and HSPA1B are expressed in a cell-type-specific manner, and certain human tissues constitutively express varying levels of HSPA1 and HSPA2 proteins in a highly differentiated way [103]. Various diseases demonstrate elevation of HSP70 family proteins, such as human breast cancer, murine triple negative mammary cancer, metastatic breast cancer stem cells, hyperthermia, and adult T-cell leukemia [104–108]. Since silencing the HSP1A1 is cytotoxic to transformed but not normal cells, various HSP70 inhibitors are in development in the hope of triggering cell cycle arrest in cancer cells [109]. Unexpectedly, elevated HSP70 was found to be beneficial to therapy as reported in Nakatsu’s breast cancer study, as increased HSPA1A enhanced sensitivity to anti-cancer drugs [110].
We found that HSP70 family proteins are significantly up-regulated in the IPA/NO-aspirin group and slightly up-regulated in the DEA/NO-aspirin and DEA/NO groups. To our surprise, IPA/NO itself down-regulates both HSPA1A and HSPA1B expression. This finding is in accordance with Nakatsu’s work and strongly suggests that overexpressed HSP70 proteins triggers different downstream pathways than the commonly reported anti-apoptotic pathway.
Thioredoxin Interacting Protein (TXNIP)
Thioredoxin interacting protein mediates oxidative stress by inhibiting thioredoxin activity or by limiting its bioavailability [111]. Increased reactive oxygen species (ROS) production increases cancer progression in some tumors, including pancreatic cancer, while ROS inhibits disease progression in others such as hepatocellular carcinoma [112]. In general, TXNIP is thought to be a tumor suppressor. TXNIP functions as a transcriptional repressor, possibly by acting as a bridge molecule between transcription factors and corepressor complexes, and its over-expression induces G0/G1 cell cycle arrest [113]. In addition, TXNIP is required for the maturation of natural killer cells. It inhibits the proteasomal degradation of DDIT4 and thereby contributes to the inhibition of the mammalian target of rapamycin complex 1 (mTORC1). Elevated TXNIP has been associated with poor histological grade in phyllodes tumors [114].
Our analysis demonstrated significantly reduced TXNIP expression levels in only the IPA/NO-aspirin and DEA/NO-aspirin treatment groups, suggesting that IPA/NO-aspirin or DEA/NO-aspirin combined with mTOR inhibitors may further improve the cytotoxic responses of these prodrugs.
DNA-Damage-Inducible Transcript 3 (DDIT3)
DNA-damage-inducible-transcript 3 is a multifunctional transcription factor in endoplasmic reticulum (ER) stress response. It plays an essential role in the response to a wide variety of cellular stress and induces cell cycle arrest and apoptosis in response to ER stress [115]. DDIT3 positively regulates transcription of TRIB3, IL6, IL8, IL23, TNFRSF10B/DR5, PPP1R15A/GADD34, BBC3/PUMA, BCL2L11/BIM and ERO1L and negatively regulates expression of BCL2 and MYOD1, ATF4-dependent transcriptional activation of asparagine synthetase (ASNS), CEBPA-dependent transcriptional activation of hepcidin (HAMP) and CEBPB-mediated expression of peroxisome proliferator-activated receptor gamma (PPARG) [115]. DDIT3 plays a regulatory role in the inflammatory response through the induction of caspase-11 (CASP4/CASP11), which induces the activation of caspase-1 (CASP1) [115]. Both caspases increase activation of pro-IL1β to mature IL-1β, which mediates inflammatory response. NO-induced apoptosis has been reported in pancreatic β cells, p53-deficient microglial cells and RAW 264.7 cells [116–118], which are triggered by DDIT3 through ER stress. Also, DDIT3 inhibits the canonical WNT signaling pathway by binding to TCF7L2/TCF4, impairing its DNA-binding properties and repressing its transcriptional activity.
Our study demonstrated decreased DDIT3 by IPA/NO-aspirin. This may indicate that IPA/NO-aspirin and WNT inhibitors may be attractive drug combinations.
Nitric Oxide Driven Pathways
Using the Canonical Pathway Analysis module in the Ingenuity Pathway Analysis portal reveals the most significantly affected cellular pathways to be: (1) endothelin-1 (ET1) signaling, (2) NO signaling in the cardiovascular system, (3) relaxin signaling, (4) G-protein coupled receptor signaling and (5) HER2 signaling in breast cancer. Given that ET1 and NO signaling are known to play key roles in the cardiovascular system, it is not surprising to see that these two pathways are among the highest-scoring in our analysis.
(1&2) Endothelin-1
ET1 is a 21-amino acid vasoconstrictor peptide, which is able to induce cardiac hypertrophy. ET1 synthesis includes NO and its intracellular effectors cGMP, prostacyclin, atrial natriuretic peptides, and steroid hormones [119]. ET1 is considered a stress-responsive regulator working in a paracrine and autocrine fashion in a variety of organs, with both beneficial and detrimental roles in mammals [120]. In addition to its potent cardiovascular actions, ET1 causes contraction of nonvascular smooth muscle; stimulation of the release of neuropeptides, pituitary hormones, and atrial natriuretic peptide; biosynthesis of aldosterone; modulation of neurotransmitter release; and increase of bone resorption [121,122]. Furthermore, ET1 has mitogenic properties, causing proliferation and hypertrophy of vascular smooth muscle, cardiac myocytes, mesangium, bronchial smooth muscle, and fibroblasts. ET1 also induces expression of several proto-oncogenes, including c-Fos, c-Jun, and c-Myc [123–125]. These actions are of potential significance in chronic congestive heart failure, renal disease, hypertension, cerebral vasospasm, and pulmonary hypertension, all of which are conditions commonly associated with increased expression of ET1 [126].
(3) Relaxin
Relaxin is a polypeptide hormone that is secreted by the corpus luteum into the circulation during the menstrual cycle and throughout pregnancy [127]. Binding of relaxin to its receptor also activates a tyrosine kinase pathway that inhibits the activity of a phosphodiesterase (PDE) that degrades cAMP. The consequent rise in cAMP levels activates protein kinase A (PKA), which leads to activation of transcription factors like the cAMP response-element binding protein and NFκB. PKA phosphorylates and inactivates I-kappa-B-α, the inhibitor subunit of the transcription factor NFκB, thus allowing NFκB to translocate into the nucleus and bind the NOS promoter, resulting in expression of NOS2. Relaxin can act on several of its targets by increasing the expression and/or activity of NOS isoenzymes, thereby promoting generation of NO. NOS3 can be activated by direct stimulation of the β and γ subunits of the g-proteins. This is accomplished by means of stimulation of PI3-kinase, followed by Akt/PKB, which in turn activates NOS3 by phosphorylation at Ser-1179 [128]. By stimulation of distinct signal transduction pathways, relaxin can have pleiotropic downstream effects, many of which may have useful clinical applications [129]. Relaxin elicits a vasodilatory response in several target organs. This response is mediated by the stimulation of intrinsic NO generation. In the uterus NO production via NOS is up-regulated during pregnancy by relaxin. NO induces uterine quiescence, which is deemed necessary for the maintenance of pregnancy. Relaxin increases intracellular cGMP levels in a concentration-related fashion. This effect of relaxin is likely a consequence of stimulation of NO production by this hormone. In fact, NO binds to the heme iron of soluble guanylate cyclase and thereby activates the synthesis of cGMP [130,131]. In turn, increased production of cGMP plays an important role in vasorelaxation. Moreover, relaxin induces changes in cell shape and the actin cytoskeleton that are consistent with cell relaxation [128].
(4) G-protein Coupled Receptors (GPCRs)
G-protein-coupled receptors are activated by a wide variety of external stimuli. Upon receptor activation, the G-protein exchanges GDP for GTP, causing dissociation of the GTP-bound α and β/γ subunits and triggering diverse signaling cascades. Receptors coupled to different heterotrimeric G-protein subtypes can utilize different scaffolds to activate the small G-protein/MAPK cascade, employing at least three different classes of tyrosine kinases. Src family kinases are recruited following activation of PI3Kγ by the β/γ subunits. They are also recruited by receptor internalization, cross-activation of receptor Tyr kinases, or by signaling through an integrin scaffold involving Pyk2 and/or FAK. GPCRs can also employ PLCβ to mediate activation of PKC and CaMKII, which can have either stimulatory or inhibitory consequences for the downstream MAPK pathway [132–134].
(5) HER2 signaling in breast cancer
Dysregulation of HER-mediated signaling pathways results in the growth and spread of cancer cells [135]. Inappropriate signaling may occur as a result of receptor overexpression or dysregulation of receptor activation, which may lead to increased or uncontrolled cell proliferation, decreased apoptosis, enhanced cancer cell motility, and angiogenesis [136–138].
Future for Diagnosis
Although the molecular and genetic determinants of most sporadic breast cancers remain unclear, significant advances in the understanding of events that contribute to breast cancer formation have been made. Deactivation mutations in tumor suppressor genes, such as p53, BRCA1, BRCA2, PTEN, or ATM, epigenetic functional inactivation of other tumor suppressor genes such as SYK and NES1, or activation of proto-oncogenes, such as HER2/Neu, can all play important roles in breast carcinogenesis [139]. Breast cancer is a clinically heterogeneous disease, and there is evidence that the varied clinical courses of patients with histologically similar tumors are due to molecular differences among cancers. Therefore, detailed molecular analysis of the cancer could yield important diagnostic and prognostic information as well as aid in the design of treatment regimens. Recent advances in molecular analytical techniques have led to a rapid expansion of new diagnostics designed to personalize breast cancer care [140]. However, approximately one-quarter of patients with lymph node-negative disease and one half of patients with lymph node-positive tumors will ultimately develop distant recurrent breast cancer. Standard treatment of metastatic breast cancer generally includes systemic treatment and surgery or radiation as needed and when indicated for palliation of localized symptomatic metastases [141]. The immediate challenge is to learn how to utilize the molecular characteristics of an individual and their tumor to improve tumor detection rate, enhance treatment effectiveness, reduce metastasis, and ultimately to prevent the development of breast cancer recurrence.
To better predict the clinical outcome of malignant disease, researchers have tried to use NOS2 to predict prognosis in various diseases. Toward this end, Huang has shown that NOS2 expression was higher in lymph node metastasis and recurrent groups in supraglottic squamous cell carcinoma [142]. Later, Hara and Qiu reported that elevated NOS2 expression predicted poor survival in human astrocytic glioma and immunoglobulin A nephropathy (IgAN) [143,144]. In addition, Li reported that increased NOS2 and nitrotyrosine (NT) predicted poor survival of gastric adenocarcinoma patients [145]. Also Pinlaor reported that the co-activation and co-localization of HIF1A and NOS2 was associated with poor survival of intrahepatic cholangiocarcinoma [146]. As mentioned earlier, Glynn has shown that NOS2 expression predicted poor survival of ER- breast cancer patients [12,147]. Zhang and Wang added that NOS2 expression also correlated with poor prognosis in gastric cancer patients and salivary gland adenoid cystic carcinoma [95,148]. Evidence supports the strategic combination of NOS2 with other relevant biomarkers, which could greatly improve the prognostic accuracy of various malignancies.
Besides genetically engineered mice, xenograft transplantation is another common tool adopted for cancer research. Xenograft transplantation via intravenous, intraperitoneal, subcutaneous or orthotopic injection is a well-defined approach for breast cancer metastasis research and has been utilized for years. Since metastasis is a multi-step process, and cancer interact differently with its surrounding microenvironment is a crucial step, spontaneous metastasis from primary tumor provides an improved model to mimic human breast cancer [149]. Interestingly, tumor cells targeting different target organs have distinct gene expression profiles, and this in vivo selection experiment allows efficient identification of target genes and specific drugs to treat metastatic breast cancer patients. In our murine model, xenograft transplantation (231-GFP) under the mammary fat pad leads to spontaneous brain metastasis and some lung metastasis, thus providing an excellent orthotopic model for breast cancer metastasis research. Moreover, our NO/HNO prodrugs provided a class of compound that may be used together with other therapeutic agents (chemotherapy, radiation, etc.) to enhance tumor killing and reduce metastasis.
Conclusion
In this study, we identified distinct molecular signatures from NO- and HNO-donor treated breast cancer cells. Given that other cancer cells may be activated/modulated by these molecular changes, we suggest that other cell types may show similar cellular responses in the presence of NO- and HNO-donors. We are in the process of testing this hypothesis in lung and pancreatic cancer cells. However, due to the heterogeneity and sporadic nature of cancer, we may not expect that other cell types will show the exact molecular signatures as breast cancer cells. Nonetheless, such comparisons across cell types would assist in discovery and identification of key gene sets that governing the molecular response to NO and HNO.
In order to accurately assess the effects of NO production, it is important to have a basic understanding of the spatial and temporal distributions of NO at the cellular level. The latest developments in NONOate derivatives and animal models are expected to facilitate deciphering of the mechanisms by which NO modulates cellular responses under different stress conditions. Our group identified uniquely expressed genes such as mir21, CTH, HSP70, TXNIP and DDIT3, and key pathways such as ET-1 and relaxin signaling, in response to NO. These findings enable the fine mapping of the cellular responses modulated by NO. Strikingly, we demonstrated that the HNO releasing donor, IPA/NO, is able to suppress HSP70 gene expression which functions like a HSP70 inhibitor while the IPA/NO derivative IPA/NO-aspirin can elevate HSP70 gene expression to a much higher level. This could trigger an alternate cellular pathway that also leads to cancer cell death. Taken together, our findings provide insight in evaluating and improving the cytotoxicity of the NONOate derivatives especially IPA/NO-aspirin in cancer therapy.
Table 2.
Class Comparison Analysis: Differentially expressed genes in aspirin vs Control vs DEA/NO vs DEA/NO- aspirin vs IPA/NO vs IPA/NO-aspirin
| Parametric p-value |
FDR | Aspirin | Control | DEA/NO | DEA/NO aspirin |
IPA/NO | IPA/NO aspirin |
Symbol | Name |
|---|---|---|---|---|---|---|---|---|---|
| < 1e-07 | < 1e-07 | 157.71 | 134.74 | 180.66 | 168.1 | 138.36 | 374.85 | HSPA1A | heat shock 70kDa protein 1A |
| 1.00E-07 | 3.85E-05 | 217.58 | 178.76 | 253.16 | 228.54 | 196.13 | 441.1 | HSPA1B | heat shock 70kDa protein 1B |
| 2.00E-07 | 6.42E-05 | 129.96 | 158.29 | 212.72 | 357.41 | 336.45 | 182.26 | MIR21 | microRNA 21 |
| 3.70E-06 | 0.000732 | 2063.12 | 1770.85 | 1458.8 | 894.51 | 1869.4 | 826.61 | TXNIP | thioredoxin interacting protein |
| 3.80E-06 | 0.000732 | 520.53 | 550.46 | 407.47 | 346.26 | 479.93 | 259.01 | GTPBP2 | GTP binding protein 2 |
| 4.50E-06 | 0.000788 | 648.79 | 691.18 | 598.54 | 532.37 | 628.32 | 307.05 | CTH | cystathionase (cystathionine gamma- lyase) |
| 6.18E-05 | 0.00791 | 817.15 | 819.14 | 695 | 530.51 | 772.75 | 396.85 | SLFN5 | schlafen family member 5 |
| 7.29E-05 | 0.00791 | 34.78 | 28.38 | 58.9 | 22.48 | 27.46 | 32.25 | FLJ16171 | FLJ16171 protein |
| 7.36E-05 | 0.00791 | 1230.98 | 1318.22 | 1113.71 | 1096.74 | 1284.57 | 731.84 | ASNS | asparagine synthetase (glutamine- hydrolyzing) |
| 7.59E-05 | 0.00791 | 123.22 | 113.37 | 113.62 | 130.16 | 132.96 | 183.65 | ARRB1 | arrestin, beta 1 |
| 9.90E-05 | 0.00954 | 55.15 | 54 | 71.05 | 75.49 | 69.29 | 111.12 | TGFBR3 | transforming growth factor, beta receptor III |
| 0.0001065 | 0.00977 | 344.03 | 310.44 | 239.07 | 270.54 | 401.34 | 136.8 | DDIT3 | DNA-damage- inducible transcript 3 |
| 0.000115 | 0.0101 | 60.95 | 69.32 | 64.71 | 52.14 | 63.82 | 37.01 | SYT1 | synaptotagmin I |
| 0.0001325 | 0.0109 | 109.96 | 125.65 | 116.2 | 94.63 | 103.04 | 56.4 | SLC16A1 | solute carrier family 16, member 1 (monocarboxylic acid transporter 1) |
| 0.0001818 | 0.014 | 275.67 | 289.23 | 173.28 | 189.52 | 241.75 | 156.84 | MXD1 | MAX dimerization protein 1 |
| 0.000231 | 0.0167 | 174.29 | 186.29 | 176.02 | 320.13 | 172.25 | 219.91 | CPA4 | carboxypeptidase A4 |
| 0.0002426 | 0.0167 | 30.09 | 27.11 | 32.82 | 26.25 | 25.56 | 48.35 | ID2 | inhibitor of DNA binding 2, dominant negative helix-loop- helix protein |
| 0.0003971 | 0.0225 | 116.22 | 145.67 | 199.52 | 105.31 | 103.5 | 216.93 | SNORD14E | small nucleolar RNA, C/D box 14E |
| 0.0004856 | 0.026 | 113.64 | 97.67 | 192.34 | 148.06 | 165.15 | 135.92 | FAM36A | family with sequence similarity 36, member A |
| 0.0005102 | 0.0262 | 46.31 | 44.67 | 50.76 | 49.72 | 42.76 | 70.34 | CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 |
| 0.0006235 | 0.0308 | 28.1 | 37.96 | 22.55 | 30.14 | 28.77 | 28.42 | CXCL2 | chemokine (C-X-C motif) ligand 2 |
| 0.0007044 | 0.0335 | 343.39 | 321.72 | 293.69 | 240.24 | 288.81 | 184.71 | TRIML2 | tripartite motif family- like 2 |
| 0.0007155 | 0.0335 | 163.87 | 184.74 | 198.14 | 124.2 | 103.51 | 180.78 | EIF4A2 | eukaryotic translation initiation factor 4A2 |
| 0.0007603 | 0.0335 | 242.43 | 276.03 | 160.58 | 174.89 | 269.25 | 131.2 | GDF15 | growth differentiation factor 15 |
| 0.0007652 | 0.0335 | 46.17 | 43 | 67.92 | 62.54 | 49.22 | 81.96 | CSNK1G1 | casein kinase 1, gamma 1 |
| 0.0009788 | 0.0355 | 471.51 | 488.23 | 502.42 | 356.01 | 449.83 | 274.58 | UFM1 | ubiquitin-fold modifier 1 |
| 0.0009939 | 0.0355 | 79.45 | 89.42 | 95.62 | 131.31 | 83.43 | 133.28 | NMU | neuromedin U |
| 0.001165 | 0.0401 | 101.31 | 95.02 | 114.88 | 105.71 | 110.36 | 163.46 | TFAP2C | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| 0.0016524 | 0.0506 | 662.91 | 722.45 | 664.69 | 647.09 | 696.38 | 438.41 | CASP4 | caspase 4, apoptosis- related cysteine peptidase |
| 0.0018161 | 0.0538 | 149.69 | 141.68 | 216.36 | 216.93 | 156.4 | 255.44 | CDCA3 | cell division cycle associated 3 |
| 0.0019214 | 0.0553 | 137.44 | 123.33 | 219.24 | 202.96 | 130.18 | 272.9 | FAM111B | family with sequence similarity 111, member B |
| 0.0019549 | 0.0554 | 98.22 | 83.91 | 121.46 | 155.71 | 95.21 | 164 | CENPI | centromere protein I |
| 0.0021586 | 0.0562 | 29.63 | 26.13 | 32.67 | 49.8 | 39.36 | 37.57 | C5orf54 | chromosome 5 open reading frame 54 |
| 0.0022755 | 0.0585 | 53.28 | 47.71 | 62.76 | 76.08 | 49.37 | 80.12 | KATNAL1 | katanin p60 subunit A- like 1 |
| 0.0026234 | 0.0647 | 32.01 | 30.48 | 56.94 | 53.64 | 50.44 | 42 | MIR15A | microRNA 15a |
| 0.0026532 | 0.0647 | 37.97 | 41.62 | 37.8 | 27.66 | 38.07 | 17.11 | C12orf39 | chromosome 12 open reading frame 39 |
| 0.002932 | 0.0689 | 87.1 | 79.86 | 121.93 | 132.14 | 150.85 | 77.24 | MIR100 | microRNA 100 |
| 0.0033175 | 0.0726 | 211.91 | 191.39 | 272.01 | 290.22 | 212.27 | 349.72 | GMNN | geminin, DNA replication inhibitor |
| 0.0033797 | 0.0732 | 151.81 | 151.53 | 145.21 | 184.47 | 157.86 | 241.16 | SOS1 | son of sevenless homolog 1 (Drosophila) |
| 0.0034924 | 0.0748 | 98.82 | 78.71 | 148.21 | 121.04 | 86.69 | 152.05 | ORC1 | origin recognition complex, subunit 1 |
| 0.0035543 | 0.0753 | 87.19 | 79.93 | 88.71 | 57.23 | 78.89 | 37.91 | IL24 | interleukin 24 |
| 0.0036557 | 0.0757 | 133.48 | 129.41 | 135.11 | 103.75 | 114.46 | 71.47 | ZNF643 | zinc finger protein 643 |
| 0.0040681 | 0.08 | 19.66 | 28.53 | 13.67 | 19.35 | 23.01 | 19.85 | RNF185 | ring finger protein 185 |
| 0.0042597 | 0.0822 | 34.63 | 37.19 | 35.04 | 33.29 | 48.72 | 56.42 | KLRC4 | killer cell lectin-like receptor subfamily C, member 4 |
| 0.0046958 | 0.0892 | 190.64 | 145.58 | 233.96 | 265.69 | 261.36 | 217.73 | HIST1H2AK | histone cluster 1, H2ak |
| 0.0050098 | 0.0919 | 145.49 | 143.33 | 192.08 | 199.65 | 143.97 | 262.36 | FAM102B | family with sequence similarity 102, member B |
| 0.0055921 | 0.0971 | 33.08 | 32.32 | 39.31 | 45.37 | 32.02 | 50.91 | GPR137C | G protein-coupled receptor 137C |
| 0.0063822 | 0.109 | 87.31 | 60.59 | 112.04 | 70.11 | 85.08 | 125.97 | MIR27B | microRNA 27b |
| 0.0068906 | 0.115 | 320.62 | 276.85 | 414.25 | 427.43 | 314.83 | 470.1 | MASTL | microtubule associated serine/threonine kinase-like |
| 0.0069605 | 0.115 | 26.56 | 16.26 | 25.57 | 25.53 | 38.79 | 41.79 | KLRC4- KLRK1 |
KLRC4-KLRK1 readthrough |
| 0.0070361 | 0.115 | 1166.34 | 1031.9 | 1010.08 | 1691.28 | 1032.54 | 1113.08 | MMP1 | matrix metallopeptidase 1 (interstitial collagenase) |
| 0.0073541 | 0.116 | 120.93 | 127.21 | 85.92 | 100.69 | 121.26 | 82.32 | U2AF1L4 | U2 small nuclear RNA auxiliary factor 1-like 4 |
| 0.0091362 | 0.131 | 416.41 | 362.63 | 588.42 | 516.16 | 370.17 | 592.15 | SKP2 | S-phase kinase- associated protein 2 (p45) |
| 0.0093547 | 0.133 | 71.86 | 76.16 | 123.41 | 108.87 | 101.19 | 97.2 | HSD17B7P2 | hydroxysteroid (17- beta) dehydrogenase 7 pseudogene 2 |
| 0.0098301 | 0.134 | 38.32 | 32.29 | 38.33 | 43.56 | 31.37 | 50.95 | DSEL | dermatan sulfate epimerase-like |
| 0.009931 | 0.134 | 67.62 | 62.92 | 55.15 | 47.37 | 60.1 | 40.49 | SLC5A4 | solute carrier family 5 (low affinity glucose cotransporter), member 4 |
Table 3.
Class Comparison Analysis: Differentially expressed genes in IPA/NO vs IPA/NO-aspirin
| Parametric p-value |
FDR | IPA/NO | IPA/NO aspirin |
Symbol | Name |
|---|---|---|---|---|---|
| 0.0004702 | 0.0745 | 401.34 | 136.8 | DDIT3 | DNA-damage-inducible transcript 3 |
| 0.0006614 | 0.0745 | 336.45 | 182.26 | MIR21 | microRNA 21 |
| 0.0028398 | 0.145 | 103.5 | 216.93 | SNORD14E | small nucleolar RNA, C/D box 14E |
| 0.0034745 | 0.145 | 38.07 | 17.11 | C12orf39 | chromosome 12 open reading frame 39 |
| 0.0036564 | 0.145 | 1869.4 | 826.61 | TXNIP | thioredoxin interacting protein |
| 0.0098684 | 0.262 | 269.25 | 131.2 | GDF15 | growth differentiation factor 15 |
Table 4.
Class Comparison Analysis: Differentially expressed genes in DEA/NO vs DEA/NO-aspirin
| Parametric p-value |
FDR | DEA/NO | DEA/NO aspirin |
Symbol | Name |
|---|---|---|---|---|---|
| 6.19E-05 | 0.0253 | 192.41 | 357.21 | MIR21 | microRNA 21 |
| 0.0023481 | 0.118 | 1319.53 | 894 | TXNIP | thioredoxin interacting protein |
| 0.0028045 | 0.118 | 52.17 | 55.3 | UFM1 | ubiquitin-fold modifier 1 |
| 0.0083349 | 0.234 | 53.27 | 22.46 | FLJ16171 | FLJ16171 protein |
| 0.0099746 | 0.265 | 22.21 | 28.11 | NOL7 | nucleolar protein 7, 27kDa |
Table 5.
Class Comparison Analysis: Differentially expressed genes in aspirin vs IPA/NO-aspirin
| Parametric p-value |
FDR | Aspirin | IPA/NO- aspirin |
Symbol | Name |
|---|---|---|---|---|---|
| 1.73E-05 | 0.00392 | 344.03 | 136.8 | DDIT3 | DNA-damage-inducible transcript 3 |
| 0.0001249 | 0.0158 | 37.97 | 17.11 | C12orf39 | chromosome 12 open reading frame 39 |
| 0.0002115 | 0.0178 | 2063.12 | 826.61 | TXNIP | thioredoxin interacting protein |
| 0.0003061 | 0.0221 | 87.19 | 37.91 | IL24 | interleukin 24 |
| 0.0072281 | 0.121 | 52.92 | 109.89 | FABP4 | fatty acid binding protein 4, adipocyte |
Highlights.
-
-
The biological effects of NO and HNO are governed by their cellular concentrations, plus spatial and temporal distribution both within tumor cells and in the tumor microenvironment.
-
-
Various NO- and HNO-releasing compounds have been developed as potential therapeutics, including as tumor modulators.
-
-
Whole genome gene expression profiling analysis indicates NO and HNO are driven different molecular pathways.
Acknowledgments
This research has been funded with federal funds from the National Cancer Institute, National Institutes of Health as well as by a National Institutes of Health grant (R01-GM076247 to KMM).
Abbreviations
- 231-GFP
MDA-MB-231 cells stably transfected with green fluorescent protein
- AKT
V-Akt murine thymoma viral oncogene homolog 1
- aRNA
amplified RNA
- ASNA
asparagine synthetase
- ATM
ataxia telangiectasia mutated
- BBC3
BCL2 binding component 3
- BCL2
B-cell CLL/lymphoma 2
- BCL2L11
BCL2-like 11 (apoptosis facilitator)
- BIM
BCL2-interacting mediator
- BRCA1
breast cancer 1, early onset
- BRCA2
breast cancer 2, early onset
- CaMKII
calcium/calmodulin-dependent protein kinase II inhibitor 1
- cAMP
cyclic adenosine monophosphate
- CASP1
caspase 1, apoptosis-related cysteine peptidase
- CASP11
SR-related CTD-associated factor 1 (SCAF1)
- CASP4
caspase 4, apoptosis-related cysteine peptidase
- CEBPA
CCAAT/enhancer binding protein (C/EBP), alpha
- CEBPB
CCAAT/enhancer binding protein (C/EBP), beta
- c-Fos
FBJ murine osteosarcoma viral oncogene homolog
- c-Jun
jun proto-oncogene
- c-Myc
v-myc avian myelocytomatosis viral oncogene homolog
- cGMP
cyclic guanosine monophosphate
- CTC
circulating tumor cells
- CTH
cystathionase (cystathionine-gamma-lyase)
- CTL
cytotoxic T cell
- DDIT3/4
DNA-damage-inducible transcript ¾
- DEA/NO
diethylaminamine NONOate
- DEA/NO-aspirin
DEA/NO-aspirin conjugate
- DR5
death receptor 5
- EGFR
pidermal growth factor receptor
- ER
estrogen receptor
- ERO1L
endoplasmic oxidoreductin-1-like protein \ (S. cerevisiae); ET1,endothelin-1
- FAK
protein tyrosine kinase 2
- FAS
Fas cell surface death receptor
- GADD34
growth arrest and DNA damage-inducible protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GDP
cyclic guanosine diphosphate
- GPCR
G-protein-coupled receptor
- GTP
cyclic guanosine triphosphate
- HAMP
hepcidin antimicrobial peptide
- HER2
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2
- HIF1 α
hypoxia inducible factor 1 α
- HNO
nitroxyl
- H2S
hydrogen sulfide
- HSPA1A/1B
heat shock 70 kDa protein 1A/1B
- HSP70
heat shock protein 70
- IgAN
immunoglobulin A nephropathy
- IL1β
interleukin 1 beta
- IL23
interleukin 23
- IL24
interleukin 24
- IL6
interleukin 6
- IL8
interleukin 8
- IPA/NO
isopropylamine NONOate
- IPA/NO-aspirin
IPA/NO-aspirin conjugate
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- MAPK
mitogen-activated protein kinase
- MB-231
MDA-MB-231 cells
- mir21
microRNA 21
- MMTV
mouse mammary tumor virus
- mTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MYOD1
myogenic differentiation 1
- NES1
kallikrein-related peptidase 10 (KLK10)
- Neu
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2
- NSAID
on-steroidal anti-inflammatory drug
- NFκB
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
- NK
natural killer
- NO
nitric oxide
- NOS
nitric oxide synthase
- NOS2
nitric oxide synthase 2 (inducible NOS)
- NOS3
nitric oxide synthase 3 (endothelial NOS)
- NT
nitrotyrosine
- PDE
phosphodiesterase
- PI3K
phosphoinositide-3-kinase
- PLCβ
phospholipase C, beta
- PKA
protein kinase A
- PKB
protein kinase B
- PKC
proetin kinase C
- PPARG
peroxisome proliferator-activated receptor gamma
- PPP1R15 A
protein phosphatase 1, regulatory subunit 15A
- PTEN
phosphatase and tensin homolog
- PTPN1
protein tyrosine phosphatase, non-receptor type 1
- PUMA
P53 upregulated modulator of apoptosis
- Pyk2
PITPNM family member 3 (PITPNM3)
- RelA
v-rel avian reticuloendotheliosis viral oncogene homolog A
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- Src
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog
- SYK
spleen tyrosine kinase
- TCF4
transcription factor 4
- TCF7L2
transcription factor 7-like 2 (T-cell specific, HMG-box)
- TNFa
tumor necrosis factor alpha
- TNFRSF10B
tumor necrosis factor receptor superfamily, member 10b
- TRAIL
TNF-related apoptosis-inducing ligand
- TRIB3
tribbles pseudokinase 3
- TXNIP
thioredoxin interacting protein
- WAP
whey acidic protein
- WNT
wingless-type MMTV integration site family
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Obermajer N, Wong JL, Edwards RP, Odunsi K, Moysich K, et al. PGE(2)-driven induction and maintenance of cancer-associated myeloid-derived suppressor cells. Immunol Invest. 2012;41:635–657. doi: 10.3109/08820139.2012.695417. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez PM, Ardavin C. Differentiation and function of mouse monocyte-derived dendritic cells in steady state and inflammation. Immunol Rev. 2010;234:90–104. doi: 10.1111/j.0105-2896.2009.00876.x. [DOI] [PubMed] [Google Scholar]
- 3.Ikonomidis I, Michalakeas CA, Parissis J, Paraskevaidis I, Ntai K, et al. Inflammatory markers in coronary artery disease. Biofactors. 2012;38:320–328. doi: 10.1002/biof.1024. [DOI] [PubMed] [Google Scholar]
- 4.Ridnour LA, Cheng RY, Switzer CH, Heinecke JL, Ambs S, et al. Molecular pathways: toll-like receptors in the tumor microenvironment--poor prognosis or new therapeutic opportunity. Clin Cancer Res. 2013;19:1340–1346. doi: 10.1158/1078-0432.CCR-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi M, Mutoh M, Ishigamori R, Fujii G, Imai T. Involvement of inflammatory factors in pancreatic carcinogenesis and preventive effects of anti-inflammatory agents. Semin Immunopathol. 2013;35:203–227. doi: 10.1007/s00281-012-0340-x. [DOI] [PubMed] [Google Scholar]
- 6.Sugiura H, Ichinose M. Nitrative stress in inflammatory lung diseases. Nitric Oxide. 2011;25:138–144. doi: 10.1016/j.niox.2011.03.079. [DOI] [PubMed] [Google Scholar]
- 7.Heinecke JL, Ridnour LA, Cheng RY, Switzer CH, Lizardo MM, et al. Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression; Proceedings of the National Academy of Sciences of the United States of America; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geller DA, Billiar TR. Molecular biology of nitric oxide synthases. Cancer Metastasis Rev. 1998;17:7–23. doi: 10.1023/a:1005940202801. [DOI] [PubMed] [Google Scholar]
- 9.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, et al. The chemical biology of nitric oxide: implications in cellular signaling. Free radical biology & medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, et al. Nitric oxide and redox mechanisms in the immune response. Journal of leukocyte biology. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bermudez LE. Differential mechanisms of intracellular killing of Mycobacterium avium and Listeria monocytogenes by activated human and murine macrophages. The role of nitric oxide. Clin Exp Immunol. 1993;91:277–281. doi: 10.1111/j.1365-2249.1993.tb05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. The Journal of clinical investigation. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hibbs JB, Jr, Taintor RR, Vavrin Z. Macrophage cytotoxicity: role for L-arginine deiminase and imino nitrogen oxidation to nitrite. Science. 1987;235:473–476. doi: 10.1126/science.2432665. [DOI] [PubMed] [Google Scholar]
- 14.Hibbs JB, Jr, Taintor RR, Vavrin Z, Rachlin EM. Nitric oxide: a cytotoxic activated macrophage effector molecule. Biochem Biophys Res Commun. 1988;157:87–94. doi: 10.1016/s0006-291x(88)80015-9. [DOI] [PubMed] [Google Scholar]
- 15.Stuehr DJ, Gross SS, Sakuma I, Levi R, Nathan CF. Activated murine macrophages secrete a metabolite of arginine with the bioactivity of endothelium-derived relaxing factor and the chemical reactivity of nitric oxide. The Journal of experimental medicine. 1989;169:1011–1020. doi: 10.1084/jem.169.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geng YJ, Hellstrand K, Wennmalm A, Hansson GK. Apoptotic death of human leukemic cells induced by vascular cells expressing nitric oxide synthase in response to gamma-interferon and tumor necrosis factor-alpha. Cancer research. 1996;56:866–874. [PubMed] [Google Scholar]
- 17.Xie K, Huang S, Dong Z, Juang SH, Gutman M, et al. Transfection with the inducible nitric oxide synthase gene suppresses tumorigenicity and abrogates metastasis by K-1735 murine melanoma cells. The Journal of experimental medicine. 1995;181:1333–1343. doi: 10.1084/jem.181.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyengar R, Stuehr DJ, Marletta MA. Macrophage synthesis of nitrite, nitrate, and N-nitrosamines: precursors and role of the respiratory burst. Proc Natl Acad Sci U S A. 1987;84:6369–6373. doi: 10.1073/pnas.84.18.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, et al. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 20.Geller DA, Di Silvio M, Nussler AK, Wang SC, Shapiro RA, et al. Nitric oxide synthase expression is induced in hepatocytes in vivo during hepatic inflammation. J Surg Res. 1993;55:427–432. doi: 10.1006/jsre.1993.1164. [DOI] [PubMed] [Google Scholar]
- 21.Geller DA, Lowenstein CJ, Shapiro RA, Nussler AK, Di Silvio M, et al. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc Natl Acad Sci U S A. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nussler AK, Geller DA, Sweetland MA, Di Silvio M, Billiar TR, et al. Induction of nitric oxide synthesis and its reactions in cultured human and rat hepatocytes stimulated with cytokines plus LPS. Biochem Biophys Res Commun. 1993;194:826–835. doi: 10.1006/bbrc.1993.1896. [DOI] [PubMed] [Google Scholar]
- 23.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, et al. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomsen LL, Lawton FG, Knowles RG, Beesley JE, Riveros-Moreno V, et al. Nitric oxide synthase activity in human gynecological cancer. Cancer research. 1994;54:1352–1354. [PubMed] [Google Scholar]
- 25.Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, et al. Roles of nitric oxide in tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4392–4396. doi: 10.1073/pnas.92.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambs S, Merriam WG, Bennett WP, Felley-Bosco E, Ogunfusika MO, et al. Frequent nitric oxide synthase-2 expression in human colon adenomas: implication for tumor angiogenesis and colon cancer progression. Cancer Res. 1998;58:334–341. [PubMed] [Google Scholar]
- 27.Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, et al. Vascular endothelial growth factor and nitric oxide synthase expression in human lung cancer and the relation to p53. Br J Cancer. 1998;78:233–239. doi: 10.1038/bjc.1998.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, et al. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J Natl Cancer Inst. 1999;91:86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 29.Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TR, et al. Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice. Proc Natl Acad Sci U S A. 1998;95:8823–8828. doi: 10.1073/pnas.95.15.8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrester K, Ambs S, Lupold SE, Kapust RB, Spillare EA, et al. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofseth LJ, Saito S, Hussain SP, Espey MG, Miranda KM, et al. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:143–148. doi: 10.1073/pnas.0237083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauthier N, Lohm S, Touzery C, Chantome A, Perette B, et al. Tumour-derived and host-derived nitric oxide differentially regulate breast carcinoma metastasis to the lungs. Carcinogenesis. 2004;25:1559–1565. doi: 10.1093/carcin/bgh158. [DOI] [PubMed] [Google Scholar]
- 33.Garban HJ, Bonavida B. Nitric oxide sensitizes ovarian tumor cells to Fas-induced apoptosis. Gynecol Oncol. 1999;73:257–264. doi: 10.1006/gyno.1999.5374. [DOI] [PubMed] [Google Scholar]
- 34.Huerta S, Chilka S, Bonavida B. Nitric oxide donors: novel cancer therapeutics (review) Int J Oncol. 2008;33:909–927. [PubMed] [Google Scholar]
- 35.Huerta-Yepez S, Vega M, Escoto-Chavez SE, Murdock B, Sakai T, et al. Nitric oxide sensitizes tumor cells to TRAIL-induced apoptosis via inhibition of the DR5 transcription repressor Yin Yang 1. Nitric Oxide. 2009;20:39–52. doi: 10.1016/j.niox.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Huerta-Yepez S, Vega M, Jazirehi A, Garban H, Hongo F, et al. Nitric oxide sensitizes prostate carcinoma cell lines to TRAIL-mediated apoptosis via inactivation of NF-kappa B and inhibition of Bcl-xl expression. Oncogene. 2004;23:4993–5003. doi: 10.1038/sj.onc.1207655. [DOI] [PubMed] [Google Scholar]
- 37.Rapozzi V, Della Pietra E, Zorzet S, Zacchigna M, Bonavida B, et al. Nitric oxide-mediated activity in anti-cancer photodynamic therapy. Nitric Oxide. 2013;30:26–35. doi: 10.1016/j.niox.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 38.Switzer CH, Cheng RY, Ridnour LA, Murray MC, Tazzari V, et al. Dithiolethiones inhibit NF-kappaB activity via covalent modification in human estrogen receptor-negative breast cancer. Cancer Res. 2012;72:2394–2404. doi: 10.1158/0008-5472.CAN-11-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonavida B, Baritaki S. Dual role of NO donors in the reversal of tumor cell resistance and EMT: Downregulation of the NF-kappaB/Snail/YY1/RKIP circuitry. Nitric Oxide. 2011;24:1–7. doi: 10.1016/j.niox.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Rao CV. Nitric oxide signaling in colon cancer chemoprevention. Mutat Res. 2004;555:107–119. doi: 10.1016/j.mrfmmm.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Lala PK, Chakraborty C. Role of nitric oxide in carcinogenesis and tumour progression. Lancet Oncol. 2001;2:149–156. doi: 10.1016/S1470-2045(00)00256-4. [DOI] [PubMed] [Google Scholar]
- 42.Jahani-Asl A, Bonni A. iNOS: a potential therapeutic target for malignant glioma. Curr Mol Med. 2013;13:1241–1249. doi: 10.2174/1566524011313080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gannot G, Buchner A, Keisari Y. Interaction between the immune system and tongue squamous cell carcinoma induced by 4-nitroquinoline N-oxide in mice. Oral Oncol. 2004;40:287–297. doi: 10.1016/j.oraloncology.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 44.Chu M, Su YX, Wang L, Zhang TH, Liang YJ, et al. Myeloid-derived suppressor cells contribute to oral cancer progression in 4NQO-treated mice. Oral Dis. 2012;18:67–73. doi: 10.1111/j.1601-0825.2011.01846.x. [DOI] [PubMed] [Google Scholar]
- 45.Mardente S, Zicari A, Consorti F, Mari E, Di Vito M, et al. Cross-talk between NO and HMGB1 in lymphocytic thyroiditis and papillary thyroid cancer. Oncol Rep. 2010;24:1455–1461. doi: 10.3892/or_00001005. [DOI] [PubMed] [Google Scholar]
- 46.Thomas DD, Miranda KM, Espey MG, Citrin D, Jourd'heuil D, et al. Guide for the use of nitric oxide (NO) donors as probes of the chemistry of NO and related redox species in biological systems. Methods in enzymology. 2002;359:84–105. doi: 10.1016/s0076-6879(02)59174-6. [DOI] [PubMed] [Google Scholar]
- 47.Keefer LK. Fifty years of diazeniumdiolate research. From laboratory curiosity to broad-spectrum biomedical advances. ACS Chem Biol. 2011;6:1147–1155. doi: 10.1021/cb200274r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Schmid T, Brune B. HIF-1alpha and p53 as targets of NO in affecting cell proliferation, death and adaptation. Current molecular medicine. 2004;4:741–751. doi: 10.2174/1566524043359926. [DOI] [PubMed] [Google Scholar]
- 49.Brune B, von Knethen A, Sandau KB. Nitric oxide (NO): an effector of apoptosis. Cell Death Differ. 1999;6:969–975. doi: 10.1038/sj.cdd.4400582. [DOI] [PubMed] [Google Scholar]
- 50.Sarti P, Avigliano L, Gorlach A, Brune B. Superoxide and nitric oxide--participation in cell communication. Cell Death Differ. 2002;9:1160–1162. doi: 10.1038/sj.cdd.4401099. [DOI] [PubMed] [Google Scholar]
- 51.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, et al. Hypoxic inducible factor 1alpha, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas DD, Ridnour LA, Espey MG, Donzelli S, Ambs S, et al. Superoxide fluxes limit nitric oxide-induced signaling. The Journal of biological chemistry. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 53.Ridnour LA, Isenberg JS, Espey MG, Thomas DD, Roberts DD, et al. Nitric oxide regulates angiogenesis through a functional switch involving thrombospondin-1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13147–13152. doi: 10.1073/pnas.0502979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garthwaite J. New insight into the functioning of nitric oxide-receptive guanylyl cyclase: physiological and pharmacological implications. Mol Cell Biochem. 2010;334:221–232. doi: 10.1007/s11010-009-0318-8. [DOI] [PubMed] [Google Scholar]
- 55.Ridnour LA, Thomas DD, Donzelli S, Espey MG, Roberts DD, et al. The biphasic nature of nitric oxide responses in tumor biology. Antioxidants & redox signaling. 2006;8:1329–1337. doi: 10.1089/ars.2006.8.1329. [DOI] [PubMed] [Google Scholar]
- 56.Rigas B, Williams JL. NO-donating NSAIDs and cancer: an overview with a note on whether NO is required for their action. Nitric Oxide. 2008;19:199–204. doi: 10.1016/j.niox.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feelisch M. The use of nitric oxide donors in pharmacological studies. Naunyn Schmiedebergs Arch Pharmacol. 1998;358:113–122. doi: 10.1007/pl00005231. [DOI] [PubMed] [Google Scholar]
- 58.Carpenter AW, Reighard KP, Saavedra JE, Schoenfisch MH. -Protected Diazeniumdiolate-Modified Silica Nanoparticles for Extended Nitric Oxide Release from Dental Composites. Biomater Sci. 2013;1:456–459. doi: 10.1039/C3BM00153A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoenfisch MH, Mowery KA, Rader MV, Baliga N, Wahr JA, et al. Improving the thromboresistivity of chemical sensors via nitric oxide release: fabrication and in vivo evaluation of NO-releasing oxygen-sensing catheters. Anal Chem. 2000;72:1119–1126. doi: 10.1021/ac991370c. [DOI] [PubMed] [Google Scholar]
- 60.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeda H, Akaike T, Yoshida M, Sato K, Noguchi Y. A new nitric oxide scavenger, imidazolineoxyl N-oxide derivative, and its effects in pathophysiology and microbiology. Curr Top Microbiol Immunol. 1995;196:37–50. doi: 10.1007/978-3-642-79130-7_5. [DOI] [PubMed] [Google Scholar]
- 62.Serafim RA, Primi MC, Trossini GH, Ferreira EI. Nitric oxide: state of the art in drug design. Curr Med Chem. 2012;19:386–405. doi: 10.2174/092986712803414321. [DOI] [PubMed] [Google Scholar]
- 63.Reynolds MM, Witzeling SD, Damodaran VB, Medeiros TN, Knodle RD, et al. Applications for nitric oxide in halting proliferation of tumor cells. Biochem Biophys Res Commun. 2013;431:647–651. doi: 10.1016/j.bbrc.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 64.Maciag AE, Nandurdikar RS, Hong SY, Chakrapani H, Diwan B, et al. Activation of the c-Jun N-terminal kinase/activating transcription factor 3 (ATF3) pathway characterizes effective arylated diazeniumdiolate-based nitric oxide-releasing anticancer prodrugs. J Med Chem. 2011;54:7751–7758. doi: 10.1021/jm2004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keefer LK, Nims RW, Davies KM, Wink DA. “NONOates” (1-substituted diazen-1-ium-1,2-diolates) as nitric oxide donors: convenient nitric oxide dosage forms. Methods Enzymol. 1996;268:281–293. doi: 10.1016/s0076-6879(96)68030-6. [DOI] [PubMed] [Google Scholar]
- 66.Siemens DR, Heaton JP, Adams MA, Kawakami J, Graham CH. Phase II study of nitric oxide donor for men with increasing prostate-specific antigen level after surgery or radiotherapy for prostate cancer. Urology. 2009;74:878–883. doi: 10.1016/j.urology.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Yasuda H, Nakayama K, Watanabe M, Suzuki S, Fuji H, et al. Nitroglycerin treatment may enhance chemosensitivity to docetaxel and carboplatin in patients with lung adenocarcinoma. Clin Cancer Res. 2006;12:6748–6757. doi: 10.1158/1078-0432.CCR-06-1124. [DOI] [PubMed] [Google Scholar]
- 68.Yasuda H, Yamaya M, Nakayama K, Sasaki T, Ebihara S, et al. Randomized phase II trial comparing nitroglycerin plus vinorelbine and cisplatin with vinorelbine and cisplatin alone in previously untreated stage IIIB/IV non-small-cell lung cancer. J Clin Oncol. 2006;24:688–694. doi: 10.1200/JCO.2005.04.0436. [DOI] [PubMed] [Google Scholar]
- 69.Chakrapani H, Showalter BM, Kong L, Keefer LK, Saavedra JE. V-PROLI/NO, a prodrug of the nitric oxide donor, PROLI/NO. Org Lett. 2007;9:3409–3412. doi: 10.1021/ol701419a. [DOI] [PubMed] [Google Scholar]
- 70. NO Donor. [Google Scholar]
- 71.Favaloro JL, Kemp-Harper BK. The nitroxyl anion (HNO) is a potent dilator of rat coronary vasculature. Cardiovasc Res. 2007;73:587–596. doi: 10.1016/j.cardiores.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 72.Donzelli S, Espey MG, Flores-Santana W, Switzer CH, Yeh GC, et al. Generation of nitroxyl by heme protein-mediated peroxidation of hydroxylamine but not N-hydroxy-L-arginine. Free radical biology & medicine. 2008;45:578–584. doi: 10.1016/j.freeradbiomed.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukuto JM, Hobbs AJ, Ignarro LJ. Conversion of nitroxyl (HNO) to nitric oxide (NO) in biological systems: the role of physiological oxidants and relevance to the biological activity of HNO. Biochem Biophys Res Commun. 1993;196:707–713. doi: 10.1006/bbrc.1993.2307. [DOI] [PubMed] [Google Scholar]
- 74.Fukuto JM, Chiang K, Hszieh R, Wong P, Chaudhuri G. The pharmacological activity of nitroxyl: a potent vasodilator with activity similar to nitric oxide and/or endothelium-derived relaxing factor. J Pharmacol Exp Ther. 1992;263:546–551. [PubMed] [Google Scholar]
- 75.Pagliaro P, Mancardi D, Rastaldo R, Penna C, Gattullo D, et al. Nitroxyl affords thiol-sensitive myocardial protective effects akin to early preconditioning. Free radical biology & medicine. 2003;34:33–43. doi: 10.1016/s0891-5849(02)01179-6. [DOI] [PubMed] [Google Scholar]
- 76.Paolocci N, Katori T, Champion HC, St John ME, Miranda KM, et al. Positive inotropic and lusitropic effects of HNO/NO- in failing hearts: independence from beta-adrenergic signaling. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5537–5542. doi: 10.1073/pnas.0937302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paolocci N, Saavedra WF, Miranda KM, Martignani C, Isoda T, et al. Nitroxyl anion exerts redox-sensitive positive cardiac inotropy in vivo by calcitonin gene-related peptide signaling. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10463–10468. doi: 10.1073/pnas.181191198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miranda KM, Paolocci N, Katori T, Thomas DD, Ford E, et al. A biochemical rationale for the discrete behavior of nitroxyl and nitric oxide in the cardiovascular system. Proc Natl Acad Sci U S A. 2003;100:9196–9201. doi: 10.1073/pnas.1430507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paolocci N, Katori T, Champion HC, St John ME, Miranda KM, et al. Positive inotropic and lusitropic effects of HNO/NO- in failing hearts: independence from beta-adrenergic signaling. Proc Natl Acad Sci U S A. 2003;100:5537–5542. doi: 10.1073/pnas.0937302100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Norris AJ, Sartippour MR, Lu M, Park T, Rao JY, et al. Nitroxyl inhibits breast tumor growth and angiogenesis. Int J Cancer. 2008;122:1905–1910. doi: 10.1002/ijc.23305. [DOI] [PubMed] [Google Scholar]
- 81.Keefer LK. Nitric oxide (NO)- and nitroxyl (HNO)-generating diazeniumdiolates (NONOates): emerging commercial opportunities. Curr Top Med Chem. 2005;5:625–636. doi: 10.2174/1568026054679380. [DOI] [PubMed] [Google Scholar]
- 82.Fitzhugh AL, Keefer LK. Diazeniumdiolates: pro- and antioxidant applications of the “NONOates”. Free Radic Biol Med. 2000;28:1463–1469. doi: 10.1016/s0891-5849(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 83.Andrei D, Salmon DJ, Donzelli S, Wahab A, Klose JR, et al. Dual mechanisms of HNO generation by a nitroxyl prodrug of the diazeniumdiolate (NONOate) class. J Am Chem Soc. 2010;132:16526–16532. doi: 10.1021/ja106552p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miranda KM, Katori T, Torres de Holding CL, Thomas L, Ridnour LA, et al. Comparison of the NO and HNO donating properties of diazeniumdiolates: primary amine adducts release HNO in Vivo. J Med Chem. 2005;48:8220–8228. doi: 10.1021/jm050151i. [DOI] [PubMed] [Google Scholar]
- 85.Maragos CM, Morley D, Wink DA, Dunams TM, Saavedra JE, et al. Complexes of .NO with nucleophiles as agents for the controlled biological release of nitric oxide. Vasorelaxant effects. J Med Chem. 1991;34:3242–3247. doi: 10.1021/jm00115a013. [DOI] [PubMed] [Google Scholar]
- 86.Salmon DJ, Torres de Holding CL, Thomas L, Peterson KV, Goodman GP, et al. HNO and NO release from a primary amine-based diazeniumdiolate as a function of pH. Inorg Chem. 2011;50:3262–3270. doi: 10.1021/ic101736e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Basudhar D, Bharadwaj G, Cheng RY, Jain S, Shi S, et al. Synthesis and chemical and biological comparison of nitroxyl- and nitric oxide-releasing diazeniumdiolate-based aspirin derivatives. J Med Chem. 2013;56:7804–7820. doi: 10.1021/jm400196q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shen J, Ambrosone CB, Zhao H. Novel genetic variants in microRNA genes and familial breast cancer. Int J Cancer. 2009;124:1178–1182. doi: 10.1002/ijc.24008. [DOI] [PubMed] [Google Scholar]
- 89.Mar-Aguilar F, Luna-Aguirre CM, Moreno-Rocha JC, Araiza-Chavez J, Trevino V, et al. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac J Clin Oncol. 2013;9:53–59. doi: 10.1111/j.1743-7563.2012.01548.x. [DOI] [PubMed] [Google Scholar]
- 90.Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175–2181. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 91.Costa PM, Cardoso AL, Nobrega C, Pereira de Almeida LF, Bruce JN, et al. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum Mol Genet. 2013;22:904–918. doi: 10.1093/hmg/dds496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan-nan B, Zhao-yan Y, Li-xi L, jiang Y, Qing-jie X, et al. MicroRNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem Biophys Res Commun. 2014;443:802–807. doi: 10.1016/j.bbrc.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 93.Okayama H, Saito M, Oue N, Weiss JM, Stauffer J, et al. NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. Int J Cancer. 2013;132:9–18. doi: 10.1002/ijc.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olivieri F, Rippo MR, Monsurro V, Salvioli S, Capri M, et al. MicroRNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res Rev. 2013;12:1056–1068. doi: 10.1016/j.arr.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 95.Pan X, Wang ZX, Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther. 2010;10:1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 96.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma Y, Xia H, Liu Y, Li M. Silencing miR-21 Sensitizes Non-Small Cell Lung Cancer A549 Cells to Ionizing Radiation through Inhibition of PI3K/Akt. Biomed Res Int. 2014;2014:617868. doi: 10.1155/2014/617868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gwak HS, Kim TH, Jo GH, Kim YJ, Kwak HJ, et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012;7:e47449. doi: 10.1371/journal.pone.0047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corcoran C, Friel AM, Duffy MJ, Crown J, O'Driscoll L. Intracellular and extracellular microRNAs in breast cancer. Clin Chem. 2011;57:18–32. doi: 10.1373/clinchem.2010.150730. [DOI] [PubMed] [Google Scholar]
- 100.Basudhar D, Bharadwaj G, Cheng RY, Jain S, Shi S, et al. Synthesis and chemical and biological comparison of nitroxyl- and nitric oxide-releasing diazeniumdiolate-based aspirin derivatives. Journal of medicinal chemistry. 2013;56:7804–7820. doi: 10.1021/jm400196q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pupo E, Pla AF, Avanzato D, Moccia F, Cruz JE, et al. Hydrogen sulfide promotes calcium signals and migration in tumor-derived endothelial cells. Free Radic Biol Med. 2011;51:1765–1773. doi: 10.1016/j.freeradbiomed.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 102.Lv M, Li Y, Ji MH, Zhuang M, Tang JH. Inhibition of invasion and epithelial-mesenchymal transition of human breast cancer cells by hydrogen sulfide through decreased phospho-p38 expression. Mol Med Rep. 2014;10:341–346. doi: 10.3892/mmr.2014.2161. [DOI] [PubMed] [Google Scholar]
- 103.Scieglinska D, Piglowski W, Chekan M, Mazurek A, Krawczyk Z. Differential expression of HSPA1 and HSPA2 proteins in human tissues; tissue microarray-based immunohistochemical study. Histochem Cell Biol. 2011;135:337–350. doi: 10.1007/s00418-011-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaur P, Nagaraja GM, Zheng H, Gizachew D, Galukande M, et al. A mouse model for triple-negative breast cancer tumor-initiating cells (TNBC-TICs) exhibits similar aggressive phenotype to the human disease. BMC Cancer. 2012;12:120. doi: 10.1186/1471-2407-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Souza AP, Albuquerque C, Torronteguy C, Frasson A, Maito F, et al. HspBP1 levels are elevated in breast tumor tissue and inversely related to tumor aggressiveness. Cell Stress Chaperones. 2009;14:301–310. doi: 10.1007/s12192-008-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen D, Bhat-Nakshatri P, Goswami C, Badve S, Nakshatri H. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res. 2013;73:5821–5833. doi: 10.1158/0008-5472.CAN-13-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tabuchi Y, Takasaki I, Wada S, Zhao QL, Hori T, et al. Genes and genetic networks responsive to mild hyperthermia in human lymphoma U937 cells. Int J Hyperthermia. 2008;24:613–622. doi: 10.1080/02656730802140777. [DOI] [PubMed] [Google Scholar]
- 108.Hamamura RS, Ohyashiki JH, Kurashina R, Kobayashi C, Zhang Y, et al. Induction of heme oxygenase-1 by cobalt protoporphyrin enhances the antitumour effect of bortezomib in adult T-cell leukaemia cells. Br J Cancer. 2007;97:1099–1105. doi: 10.1038/sj.bjc.6604003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Budina-Kolomets A, Balaburski GM, Bondar A, Beeharry N, Yen T, et al. Comparison of the activity of three different HSP70 inhibitors on apoptosis, cell cycle arrest, autophagy inhibition, and HSP90 inhibition. Cancer Biol Ther. 2014;15:194–199. doi: 10.4161/cbt.26720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakatsu N, Yoshida Y, Yamazaki K, Nakamura T, Dan S, et al. Chemosensitivity profile of cancer cell lines and identification of genes determining chemosensitivity by an integrated bioinformatical approach using cDNA arrays. Mol Cancer Ther. 2005;4:399–412. doi: 10.1158/1535-7163.MCT-04-0234. [DOI] [PubMed] [Google Scholar]
- 111.Schulze PC, Liu H, Choe E, Yoshioka J, Shalev A, et al. Nitric oxide-dependent suppression of thioredoxin-interacting protein expression enhances thioredoxin activity. Arterioscler Thromb Vasc Biol. 2006;26:2666–2672. doi: 10.1161/01.ATV.0000248914.21018.f1. [DOI] [PubMed] [Google Scholar]
- 112.Yamaguchi F, Hirata Y, Akram H, Kamitori K, Dong Y, et al. FOXO/TXNIP pathway is involved in the suppression of hepatocellular carcinoma growth by glutamate antagonist MK-801. BMC Cancer. 2013;13:468. doi: 10.1186/1471-2407-13-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamaguchi F, Takata M, Kamitori K, Nonaka M, Dong Y, et al. Rare sugar D-allose induces specific up-regulation of TXNIP and subsequent G1 cell cycle arrest in hepatocellular carcinoma cells by stabilization of p27kip1. Int J Oncol. 2008;32:377–385. [PubMed] [Google Scholar]
- 114.Kim S, Kim JY, Kim do H, Jung WH, Koo JS. Analysis of phyllodes tumor recurrence according to the histologic grade. Breast Cancer Res Treat. 2013;141:353–363. doi: 10.1007/s10549-013-2684-x. [DOI] [PubMed] [Google Scholar]
- 115. DDIT3. [Google Scholar]
- 116.Oyadomari S, Takeda K, Takiguchi M, Gotoh T, Matsumoto M, et al. Nitric oxide-induced apoptosis in pancreatic beta cells is mediated by the endoplasmic reticulum stress pathway. Proc Natl Acad Sci U S A. 2001;98:10845–10850. doi: 10.1073/pnas.191207498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kawahara K, Oyadomari S, Gotoh T, Kohsaka S, Nakayama H, et al. Induction of CHOP and apoptosis by nitric oxide in p53-deficient microglial cells. FEBS Lett. 2001;506:135–139. doi: 10.1016/s0014-5793(01)02898-8. [DOI] [PubMed] [Google Scholar]
- 118.Gotoh T, Oyadomari S, Mori K, Mori M. Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP. J Biol Chem. 2002;277:12343–12350. doi: 10.1074/jbc.M107988200. [DOI] [PubMed] [Google Scholar]
- 119.Barton M, Traupe T, Haudenschild CC. Endothelin, hypercholesterolemia and atherosclerosis. Coron Artery Dis. 2003;14:477–490. doi: 10.1097/00019501-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 120.Piacentini L, Gray M, Honbo NY, Chentoufi J, Bergman M, et al. Endothelin-1 stimulates cardiac fibroblast proliferation through activation of protein kinase C. J Mol Cell Cardiol. 2000;32:565–576. doi: 10.1006/jmcc.2000.1109. [DOI] [PubMed] [Google Scholar]
- 121.Tsukahara H, Sekine K, Miura M, Todoroki Y, Ohshima Y, et al. Vasoactive and natriuretic mediators in umbilical cord blood: a report of our observation and review of the literature. Early Hum Dev. 2002;69:57–64. doi: 10.1016/s0378-3782(02)00038-5. [DOI] [PubMed] [Google Scholar]
- 122.Larson SR, Zhang X, Dumpit R, Coleman I, Lakely B, et al. Characterization of osteoblastic and osteolytic proteins in prostate cancer bone metastases. Prostate. 2013;73:932–940. doi: 10.1002/pros.22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bruneau BG, de Bold AJ. Selective changes in natriuretic peptide and early response gene expression in isolated rat atria following stimulation by stretch or endothelin-1. Cardiovasc Res. 1994;28:1519–1525. doi: 10.1093/cvr/28.10.1519. [DOI] [PubMed] [Google Scholar]
- 124.Lee JG, Zheng R, McCafferty-Cepero JM, Burnstein KL, Nanus DM, et al. Endothelin-1 enhances the expression of the androgen receptor via activation of the c-myc pathway in prostate cancer cells. Mol Carcinog. 2009;48:141–149. doi: 10.1002/mc.20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rauh A, Windischhofer W, Kovacevic A, DeVaney T, Huber E, et al. Endothelin (ET)-1 and ET-3 promote expression of c-fos and c-jun in human choriocarcinoma via ET(B) receptor-mediated G(i)- and G(q)-pathways and MAP kinase activation. Br J Pharmacol. 2008;154:13–24. doi: 10.1038/bjp.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miyauchi T, Masaki T. Pathophysiology of endothelin in the cardiovascular system. Annu Rev Physiol. 1999;61:391–415. doi: 10.1146/annurev.physiol.61.1.391. [DOI] [PubMed] [Google Scholar]
- 127.Dessauer CW, Nguyen BT. Relaxin stimulates multiple signaling pathways: activation of cAMP, PI3K, and PKCzeta in THP-1 cells. Ann N Y Acad Sci. 2005;1041:272–279. doi: 10.1196/annals.1282.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Besier S, Ludwig A, Brade V, Wichelhaus TA. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol Microbiol. 2003;47:463–469. doi: 10.1046/j.1365-2958.2003.03307.x. [DOI] [PubMed] [Google Scholar]
- 130.Friebe A, Koesling D. Regulation of nitric oxide-sensitive guanylyl cyclase. Circ Res. 2003;93:96–105. doi: 10.1161/01.RES.0000082524.34487.31. [DOI] [PubMed] [Google Scholar]
- 131.Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci U S A. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 133.Goldsmith ZG, Dhanasekaran DN. G protein regulation of MAPK networks. Oncogene. 2007;26:3122–3142. doi: 10.1038/sj.onc.1210407. [DOI] [PubMed] [Google Scholar]
- 134.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 135.Menard S, Tagliabue E, Campiglio M, Pupa SM. Role of HER2 gene overexpression in breast carcinoma. J Cell Physiol. 2000;182:150–162. doi: 10.1002/(SICI)1097-4652(200002)182:2<150::AID-JCP3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 136.Prenzel N, Fischer OM, Streit S, Hart S, Ullrich A. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer. 2001;8:11–31. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 137.Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 138.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 139.Buchholz TA, Wazer DE. Molecular biology and genetics of breast cancer development: a clinical perspective. Semin Radiat Oncol. 2002;12:285–295. doi: 10.1053/srao.2002.35248. [DOI] [PubMed] [Google Scholar]
- 140.Pusztai L, Cristofanilli M, Paik S. New generation of molecular prognostic and predictive tests for breast cancer. Semin Oncol. 2007;34:S10–S16. doi: 10.1053/j.seminoncol.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 141.Twelves C, Vahdat LT, Cortes J. Novel treatment options in the management of metastatic breast cancer. Clin Adv Hematol Oncol. 2011;9:1–16. [PubMed] [Google Scholar]
- 142.Huang Z, Zeng ZY, Peng HW, Xia LP, Hou JH. [Inducible nitric oxide synthase and basic fibroblast growth factor expression as prognostic factors in supraglottic squamous cell carcinoma] Ai Zheng. 2003;22:978–981. [PubMed] [Google Scholar]
- 143.Hara A, Okayasu I. Cyclooxygenase-2 and inducible nitric oxide synthase expression in human astrocytic gliomas: correlation with angiogenesis and prognostic significance. Acta Neuropathol. 2004;108:43–48. doi: 10.1007/s00401-004-0860-0. [DOI] [PubMed] [Google Scholar]
- 144.Qiu LQ, Sinniah R, Hsu SI. Role of differential and cell type-specific expression of cell cycle regulatory proteins in mediating progressive glomerular injury in human IgA nephropathy. Lab Invest. 2004;84:1112–1125. doi: 10.1038/labinvest.3700144. [DOI] [PubMed] [Google Scholar]
- 145.Li LG, Xu HM. Inducible nitric oxide synthase, nitrotyrosine and apoptosis in gastric adenocarcinomas and their correlation with a poor survival. World J Gastroenterol. 2005;11:2539–2544. doi: 10.3748/wjg.v11.i17.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pinlaor S, Sripa B, Ma N, Hiraku Y, Yongvanit P, et al. Nitrative and oxidative DNA damage in intrahepatic cholangiocarcinoma patients in relation to tumor invasion. World J Gastroenterol. 2005;11:4644–4649. doi: 10.3748/wjg.v11.i30.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Switzer CH, Ridnour LA, Cheng R, Heinecke J, Burke A, et al. S-Nitrosation Mediates Multiple Pathways That Lead to Tumor Progression in Estrogen Receptor-Negative Breast Cancer. For Immunopathol Dis Therap. 2012;3:117–124. doi: 10.1615/ForumImmunDisTher.2012006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang W, He XJ, Ma YY, Wang HJ, Xia YJ, et al. Inducible nitric oxide synthase expression correlates with angiogenesis, lymphangiogenesis, and poor prognosis in gastric cancer patients. Hum Pathol. 2011;42:1275–1282. doi: 10.1016/j.humpath.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 149.Lee JM, Kim IS, Kim H, Lee JS, Kim K, et al. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol Cell. 2010;37:183–195. doi: 10.1016/j.molcel.2009.12.022. [DOI] [PubMed] [Google Scholar]