Abstract
Methylation of cytosine nucleotides is governed by DNA methyltransferases (DNMTs) that establish de novo DNA methylation patterns in early embryonic development (e.g., DNMT3a and DNMT3b) or maintain those patterns on hemimethylated DNA in dividing cells (e.g., DNMT1). DNMTs continue to be expressed at high levels in mature neurons, however their impact on neuronal function and behavior are unclear. To address this issue we examined DNMT1 and DNMT3a expression following associative learning. We also generated forebrain specific conditional Dnmt1 or Dnmt3a knockout mice and characterized them in learning and memory paradigms as well as for alterations in long-term potentiation (LTP) and synaptic plasticity. Here, we report that experience in an associative learning task impacts expression of Dnmt3a, but not Dnmt1, in brain areas that mediate learning of this task. We also found that Dnmt3a knockout mice, and not Dnmt1 knockouts have synaptic alterations as well as learning deficits on several associative and episodic memory tasks. These findings indicate that the de novo DNA methylating enzyme DNMT3a in postmitotic neurons is necessary for normal memory formation and its function cannot be substituted by the maintenance DNA methylating enzyme DNMT1.
Keywords: methylation, behavior, long term potentiation, excitatory neurotransmission, fear conditioning
1. Introduction
DNA methylation is a mechanism underlying gene silencing that plays a critical role in establishing and maintaining cellular phenotypes throughout development (Guibert et al., 2009). Methylation of DNA is governed by a family of enzymes, the DNA methyltransferases (DNMTs), which catalyze the transfer of a methyl group to the 5’ position of cytosine nucleotides at CG dinucleotides using dietary sources of s-adenosyl-l-methoinine as the methyl donor. Distinct DNMT proteins establish de novo methylation patterns or act to maintain those patterns on unmethylated or hemimethylated DNA. Expression of the maintenance DNA methyltransferase DNMT1 and the de novo enzyme DNMT3a is high during development (Szyf et al., 1985) but DNMT mRNA and enzymatic activity are still observed at abundant levels in the adult CNS (Monk et al., 1987; Goto et al., 1994) suggesting the possibility that DNMTs and dynamic methylation states retain functional importance in the adult brain.
Methylation patterns have historically been considered static in postmitotic neurons, however recent work has suggested DNA demethylation at specific promoters may occur contributing to alterations in gene expression that impacts complex behavior and synaptic function (Martinowich et al., 2003; Weaver et al., 2004; Weaver et al., 2005; Weaver et al., 2006; Miller & Sweatt, 2007). In agreement with these findings, treatment with pharmacological DNMT inhibitor compounds has been shown to impair associative learning and inhibit the magnitude and maintenance of long term potentiation (LTP), widely considered a cellular correlate of learning, at hippocampal synapses (Levenson et al., 2006; Miller & Sweatt, 2007; Miller et al., 2008). While there is debate on the mechanism for how DNA demethylation occurs in postmitotic neurons, the potential role of this epigenetic process in cellular functions as well as in neuropsychiatric disorders (e.g., schizophrenia, drug addiction) has attracted a great deal of interest.
To examine the role of DNA methyltransferases in adult brain, mice lacking Dnmt1 and Dnmt3a have been generated. Constitutive Dnmt1 and Dnmt3a knockout mice are not viable, thus research on the role of DNMTs in adult brain function has utilized conditional knockout mice (Li et al., 1992; Okano et al., 1999; Fan et al., 2001; Golshani et al., 2005; Nguyen et al., 2007; Hutnick et al., 2009; Feng et al., 2010; LaPlant et al., 2010). In the current study we investigated the impact of a postnatal, forebrain-specific conditional knockout (CKO) of Dnmt1 or Dnmt3a on complex behavior and synaptic function. Our findings demonstrate clearly dissociable roles for Dnmt1 and Dnmt3a in associative learning tasks as well as in synaptic plasticity. We conclude that de novo and maintenance DNMTs cannot functionally compensate for each other in adult brain, and that DNMT3a is critical for normal adult behavior and synaptic function.
2. Materials and Methods
2.1. Dnmt1 and Dnmt3a conditional knockout mice
Floxed Dnmt1 and Dnmt3a lines and the CaMKIIα-Cre93 line were on a mixed 129/BALBC background backcrossed to a C57BL/6 line for > 10 generations. Male CaMKII-Cre93 mice were crossed with female floxed homozygous Dnmt1 or Dnmt3a mice, and resulting male Cre-floxed heterozygous Dnmt1 or Dnmt3a were crossed with female floxed homozygous Dnmt1 or Dnmt3a mice to generate conditional KOs (CKOs) of either Dnmt1 or Dnmt3a in forebrain. Littermates derived from these mating paradigms that were Cre- negative served as comparison control mice for the respective groups. Genomic DNA from tail samples was used for genotyping by PCR as previously described (Jackson-Grusby et al., 2001; Nguyen et al., 2007). Mice were tested in the following order in distinct cohorts for the behavioral tasks: cohort 1 – locomotor activity, rotarod, and cued fear conditioning; cohort 2 – spatial object recognition, novel object recognition, conditioned taste aversion, and contextual fear conditioning. Naïve mice were used for hippocampal slice electrophysiology. All experiments used 6–12 week old male mice maintained on a 12 hr light/dark cycle with ad libitum access to food and water except where noted. All experiments were scored by observers blind to the genotype of the mice. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center.
2.2. Assessment of Dnmt1 and Dnmt3a expression by real-time PCR
Dnmt1, Dnmt3a, and Dnmt3b expression was examined in adult brain in order to verify the success of our conditional knockout strategy. Bilateral hippocampi, amygdala, prefrontal cortex, and cerebellum were dissected on an ice cold glass dish and placed on dry ice until storage at −80 °C. In separate experiments that assessed Dnmt1 and Dnmt3a gene expression in wild type mice after fear conditioning mice were sacrificed 30 minutes post-training. An untrained control group was sacrificed 30 minutes after exposure to the novel context or tone with no exposure to shock. In experiments that assessed gene expression after fear conditioning mice were sacrificed 30 minutes post-training. RNA was extracted from 4–6 animals/group using Trizol reagent and precipitated with isopropanol. After treatment with DNase I to remove residual contaminating genomic DNA, cDNA templates were synthesized using 250 ng RNA with Superscript III reverse transcriptase for 60 minutes at 50°C in the presence of random hexamers. Real-time PCR was performed in triplicate using optimized primers and the following protocol: 95°C for 10 minutes followed by 40 cycles of 95°C for 0.15 seconds and 60°C for 1 minute. Primer sequences used were, 5’-TGG AGA TTA AGC TCT GCC TGC TGT-3’ and 5’-TAG TCC TTG GTA GCA GCC TCC TCT TT-3’ for Dnmt1; 5’-AGT CCC TGC AAT GAC CTC TCC A-3’ and 5’-AAC TCA AAG AAG AGG CGG CCA-3’ for Dnmt3a; 5’-CTG AGA TCT CTG CTG ACA AAC-3’ and 5’-TGG TAC ATG GCC TTC CTA TAA-3’ for Dnmt3b; and the Gapdh (see Adachi et al., 2009). β-actin or GAPDH were chosen as housekeeping genes due to similar mRNA expression across all sample templates. Relative gene expression was calculated using the 2−ΔΔct method and expressed as fold-induction relative to control (Schmittgen & Livak, 2008).
2.3. Locomotor activity and rotarod testing
Locomotor activity was assessed for 2 hours in a fresh home cage using four photocell beams linked to computer acquisition software (San Diego Instruments). Motor coordination and motor learning were tested by 8 rotarod trials across 2 days (4 trials/day). Mice were placed on a rotating rod (IITC Life Science) which gradually increased speed over the 5-minute trial. Test sessions ended when the animals fell off the rod or after 5 minutes.
2.4. Context- and cue-dependent fear conditioning
Mice were individually tested in operant chambers (Med Associates) with electrifiable stainless steel grid floors and surrounded by a sound-attenuating external chamber. For the training phase of context-dependent fear conditioning, animals were habituated to the chamber [the conditioned stimulus (CS)] for 2 minutes, followed by 3 presentations of footshock [the unconditioned stimulus (US); 0.5 mA shock, 1 second duration with an inter-shock interval of 1 minute]. Ninety minutes after the training, short-term memory formation was assessed. The mice were placed in the same chambers and behavior was videotaped for 5 minutes. Freezing, defined as no movement except for respiration, was scored at 5 second intervals. For long-term memory formation, the mice were examined 24 hr after the training for a 5 minute time period. Extinction training was carried out for 5 days (i.e., re-exposure to context with no shock). For cue-dependent fear conditioning mice were trained via exposure to 3 tone-shock pairings (30 second white noise tone, 90 dB). Twenty-four hours after training animals underwent extinction training for 5 days in a novel context - a 3 minute baseline period followed by presentations of tone CS alone for 3 minutes. Cue-dependent FC was determined by subtracting baseline freezing from freezing observed during tone.
2.5. Footshock sensitivity and startle amplitude
At the conclusion of the behavioral experiments mice were exposed to increasing footshock intensity (range: 0.05–0.45 mA) to determine the threshold at which the animal responded by vocalization and by jumping. Startle was measured using SR-Lab Startle Response System (San Diego Instruments). Mice were placed into Plexiglass holders and allowed acclimation to the chamber and background white noise (70 dB) for 5 minutes. After acclimation, six startle stimuli (120 dB, 40 ms, white noise) were presented with an average interstimulus interval of 15 seconds (range 7–23 seconds).
2.6. Conditioned taste aversion learning
Mice were adapted to a restricted drinking schedule of two 15-minute drinking sessions per day for 7–10 days. On the training day mice were exposed to a 0.5% saccharin solution for 15 minutes during the morning drinking session rather than the usual presentation of water. Thirty minutes after the onset of saccharin intake animals were injected intraperitoneally (i.p.) with 0.14 M LiCl to induce malaise. A saccharin/water choice-test was given 48 hours after LiCl to determine genotypic effects on acquisition of the aversion. At the conclusion of the experiment animals were given a 0.04 % quinine/water choice test to assess taste sensitivity.
2.7. Spatial and novel object recognition tasks
For spatial object recognition, on day 1 mice were habituated for 10 minutes to an open field chamber (W × L, 39 × 39 cm) decorated with distinct visual stimuli (e.g., solid vertical lines, large star shape). Twenty-four hours later mice were returned to the open field. Mice were given a 10-minute to explore 3 identical objects (e.g., metal tubing) spaced ~8 cm from the walls of the open field. Twenty-four hours later (day 3) the location of 1 object was changed and mice were tested for 6 minutes to determine the ratio of time spent exploring the moved (“spatial”) object to time spent exploring the other 2 objects. The novel object recognition protocol was similar to the spatial object recognition paradigm, however on the test day one object was replaced with a novel object and the ratio of time spent with novel vs. familiar objects was quantified. Animals that did not explore all objects during training and test sessions were excluded. A separate group of animals (n = 5–6/group) were used to determine potential object exploration bias (i.e., whether the animals initially prefer exploring one object versus another). There were no differences in amount of time spent exploring the different objects used for the experiments (not shown). Spatial and novel object recognition tests and training were carried out in dim lighting.
2.8. Hippocampal slice recordings
Hippocampal slices were prepared as described previously (Morris et al., 2013). Input-output relationship was determined by providing an ascending series of stimulus input intensities (range 40 – ~240 µA) until the maximum field excitatory postsynaptic potential (fEPSP) response was determined. An input stimulus intensity that induced 40–50% of the maximum response was used for measuring paired-pulse ratio (PPR) and theta burst-induced LTP. An input intensity that induced ~75% of the maximum response was used for high frequency stimulation (HFS)–induced LTP. PPR was induced by giving 2 pulses at decreasing interpulse intervals (400, 200, 100, 50, 30, and 20 ms) and analyzed by dividing the fEPSP slope of pulse 2 by pulse 1. Following 20 minutes of stable baseline fEPSP slope, LTP was induced by theta burst (3 trains with 3 100 Hz bursts/train. Each burst consisted of 5 pulses with an interburst interval of 200 ms and intertrain interval of 10 s), or HFS (4 trains of 100 pulses/train at 100 Hz, inter-train interval of 20 seconds).
2.9. Statistical analysis
Data were analyzed by Student’s t-tests, two-way ANOVA with repeated measures, or one-way ANOVA, with post-hoc Fischer’s LSD tests conducted following significant interaction effects. Input-output slopes from hippocampal field recordings were fit by linear regression and statistical significance of slope differences was determined by t-test. Slopes that did not achieve a fit of r2 > 0.80 were discarded. Data are presented as mean ± SEM. All analyses were two-tailed with p-value ≤ 0.05 considered statistically significant.
3. Results
3.1 Fear conditioning increases Dnmt3a expression in brain
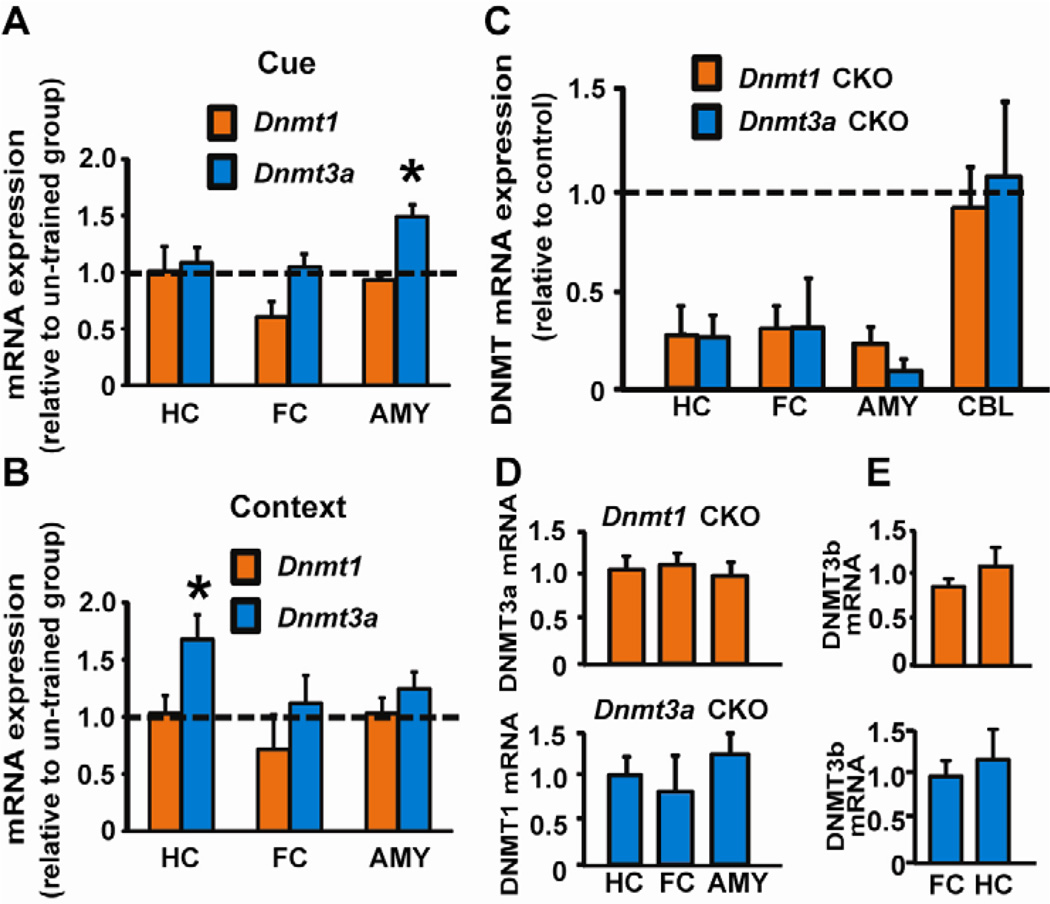
We examined Dnmt1 and Dnmt3a expression in C57BL/6 mice following training in a fear conditioning task, using quantitative PCR (qPCR) on tissue from brain regions important for associative fear learning. Dnmt1 mRNA was unchanged in hippocampus, frontal cortex or amygdala following training in cued or contextual fear conditioning (Fig. 1A, B). In contrast, Dnmt3a mRNA was rapidly (30 minutes) increased in the amygdala, a brain region necessary for associative fear learning, following training in cued fear conditioning (Fig. 1A, B). At this same time point following training in contextual fear conditioning, Dnmt3a was significantly increased in the hippocampus, which is required for contextual, not cued, fear learning (Kim & Fanselow, 1992; Kim & Jung, 2006).
Fig. 1.
Dnmt1 and Dnmt3a mRNA expression in mouse brain. (A) After training in cued fear conditioning Dnmt3a expression was increased in the amygdala (AMY) of C57BL/6 mice relative to un-trained mice [t(8) = 3.219, p = 0.012]. A trend (p = 0.062) for reduced Dnmt1 expression was evident in frontal cortex (FC). (B) Training in contextual fear conditioning increased Dnmt3a expression in hippocampus (HC) [t(10) = 2.30, p = 0.049] with a trend in AMY (p = 0.16); Dnmt1 expression was unchanged in HC, FC, and AMY. (C) Consistent with forebrain-specific deletion, Dnmt1 or Dnmt3a mRNA was reduced in 6-week old CKOs in HC, AMY, and FC, with no change in cerebellum (CBL). (D) No compensation in Dnmt3a expression when Dnmt1 was knocked down (top panel), and vice versa (bottom panel). (E) No changes in Dnmt3b expression in Dnmt1 and Dnmt3a CKO mice (top and bottom panels, respectively) in comparison to control animals. *p < 0.05 compared with un-trained CTL (n= 4–6/group in all groups).
3.2 Dnmt3a CKO mice exhibit associative learning deficits
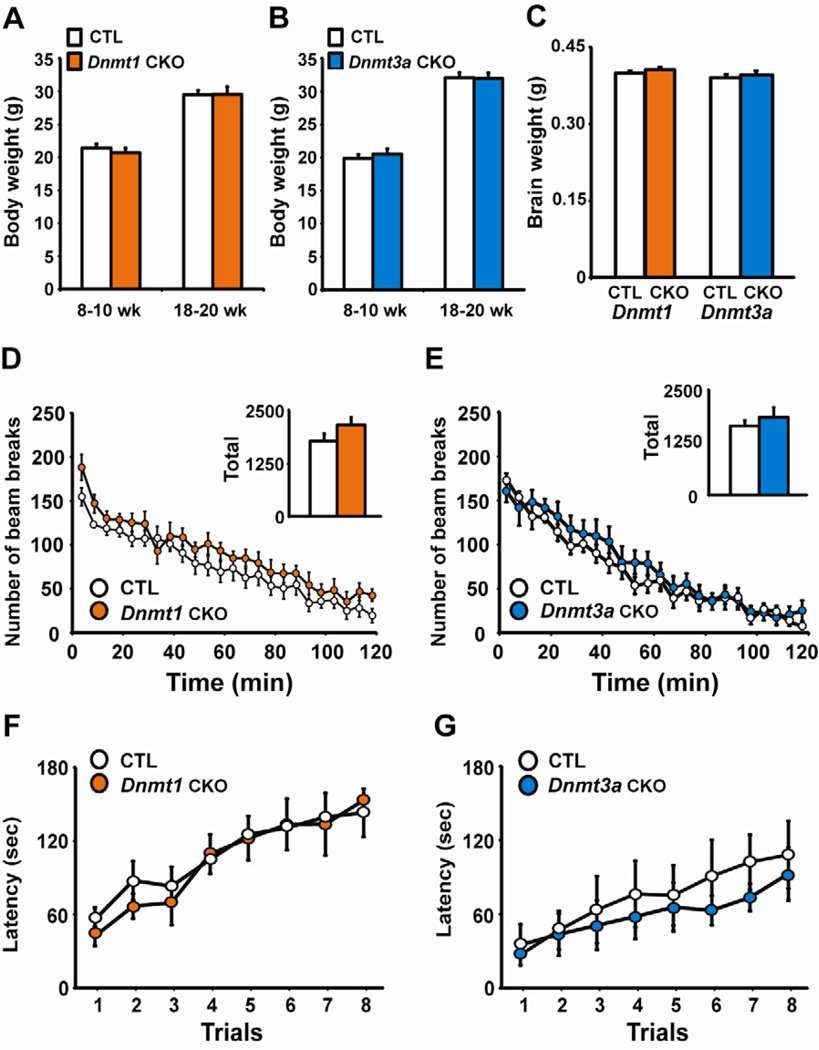
Given the dissociable effects of fear conditioning on Dnmt expression we hypothesized that DNMT1 and DNMT3a are uniquely involved in learning this task, which requires animals to associate a previously innocuous conditioned stimulus (CS) with a salient unconditioned stimulus (US). To test this hypothesis conditional Dnmt1 or Dnmt3a knockout mice were generated using the calcium-calmodulin-dependent protein kinase II promoter (CaMKIIα) to drive expression of Cre recombinase (CaMKIIα –Cre93 line) to delete Dnmt1 or Dnmt3a in forebrain neurons starting at approximately two weeks of age (Chen et al., 2001). This conditional knockout (CKO) strategy reduced Dnmt1 or Dnmt3a mRNA expression ~70–85% in adult forebrain regions including the hippocampus, frontal cortex and amygdala without affecting expression in the cerebellum or compensating other Dnmt gene expression (Fig. 1C, D, E). The Dnmt1 or Dnmt3a knockout mice also had indistinguishable body weight, brain weight, locomotor activity, and motor learning relative to littermate control (CTL) mice (Fig. 2A–G).
Fig. 2.
Normal body and brain weights and motor behavior in Dnmt1 and Dnmt3a CKO mice. (A–C) Dnmt1 or Dnmt3a CKO did not adversely affect body weight or brain weight measured at 10 weeks, n=10–12/group. (D–E) Locomotor activity was normal in Dnmt1 (n=8) and Dnmt3a (n=10) CKO mice relative to control littermates (CTL, n=8, 10). Inset displays total locomotion over the 2-hour test. (F–G) Dnmt1 (n=8) and Dnmt3a (n=10) CKOs had normal motor coordination and normal motor learning relative to CTL (n=8, 10) as demonstrated by performance on the rotarod task.
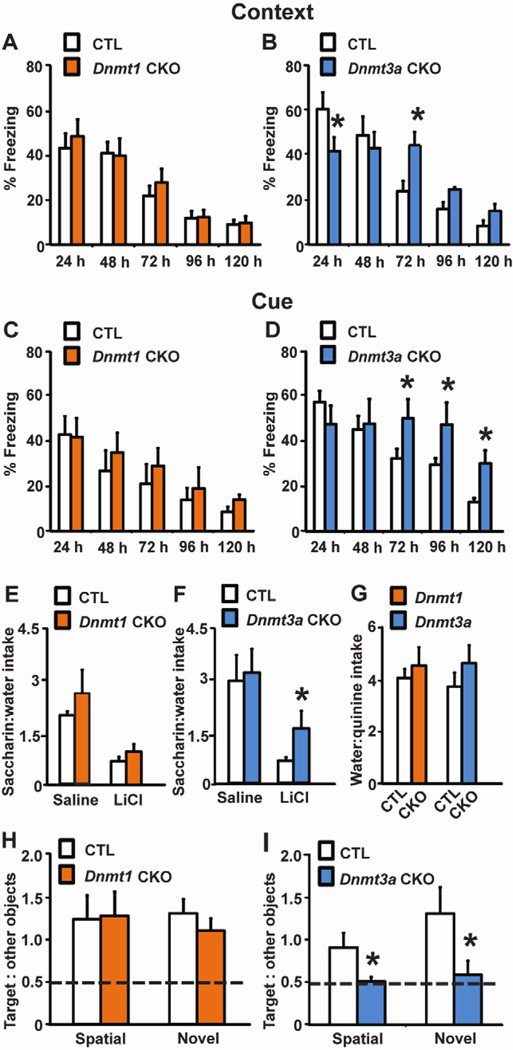
Dnmt1 CKO mice performed similarly to CTL in contextual and cued fear conditioning (Fig. 3A, C). By contrast, Dnmt3a CKO mice had impaired memory formation 24 hours post-training along with abnormal extinction 72 hours after training in contextual fear conditioning (Fig. 3B). In a cued fear learning task persistent deficits in the ability to extinguish the tone-footshock association manifest 3 days post-training and persisted for the remainder of testing (Fig. 3D), however long term memory tested 24 hours after training did not differ from CTL mice. Dnmt3a CKOs had normal nociception, startle responses and baseline freezing (not shown) ruling out differential pain sensitivity, audition, or freezing in a novel environment as confounding factors in our study. A separate cohort of mice was used to test short-term fear learning. Dnmt1 and Dnmt3a CKO mice did not exhibit any differences compared with CTL mice in context fear conditioning tested 90 minutes after training (data not shown). To further investigate a specific role for DNMT3a in associative learning, we examined CKO mice in a conditioned taste aversion (CTA) paradigm, in which a novel taste CS is paired with an illness inducing unconditioned stimulus and results in subsequent avoidance of the novel taste. Again Dnmt1 CKO mice learned normally, while Dnmt3a CKOs displayed less of an aversion to 0.5% saccharin solution compared with CTL mice 48 hours after this novel taste CS was paired with 0.14M LiCl, a US which induces malaise (Fig. 3E–G). CTA learning relies on brain circuits distinct from those involved in fear learning (Yamamoto et al., 1994), suggesting a general involvement of DNMT3a in associative learning.
Fig. 3.
Dnmt3a CKOs have learning and memory deficits. (A–B) In context-dependent fear conditioning Dnmt1 (n=10) CKOs have normal learning (CTL, n=12), while Dnmt3a (n=10) CKOs exhibited deficits in long-term memory 24 hours after training [t(16) = 2.158, p = 0.047] and extinction 72 hours post-training [t(16) = 2.633, p = 0.018] compared to CTL (n=8). (C–D) in cue-dependent fear conditioning, Dnmt1 (n=12) CKOs learned normally, while Dnmt3a (n=8) CKOs had impaired extinction relative to CTL (n=12, 8) [72 hours post-training: t(14) = 2.140, p = 0.05; 96 hours : t(14) = 2.166, p =0.049; 120 hours : t(14) = 3.011, p = 0.009]. (E–G) Saline–treated Dnmt1 (n=11) and Dnmt3a (n=8) CKOs had normal 0.5% saccharin preference compared to CTL (n=11, 10). Forty-eight hours after training LiCl-treated Dnmt1 (n=11) CKOs showed normal acquisition of an induced taste aversion to saccharin, however Dnmt3a (n=9) CKOs exhibited deficits in memory as evidenced by increased saccharin:water intake ratios relative to LiCl-treated CTL mice (t(14) = 2.167, p = 0.048). (G) Deficits in CTA learning could not be explained by genotypic differences in taste sensitivity as both CKOs preferred water to 0.04% quinine equally to CTL. (H) Dnmt1 (n=8) CKOs had normal spatial or novel object recognition relative to CTL (n=11). (I) Dnmt3a CKOs had severe deficits and performed near chance level (dashed line) for both spatial object recognition and novel object recognition [spatial, t(18) = 2.226, p = 0.039; novel, t(14) = 2.275, p = 0.038]; Dnmt3a CKO, spatial object recognition n=10, novel object recognition n= 8; CTL, spatial object recognition n=10, novel object recognition n= 8). *p < 0.05 vs. CTL
To determine if the effects of Dnmt3a CKO were specific for associative learning, we tested spatial and novel object recognition, in which normal mice spend more time exploring objects that have been displaced from their original spatial location or from novel objects. Dnmt3a CKO mice failed to form memories for spatial object location or for the nature of the object itself, as demonstrated by no preference for relocated or novel objects, whereas Dnmt1 CKO mice displayed no deficits (Fig. 3H, I).
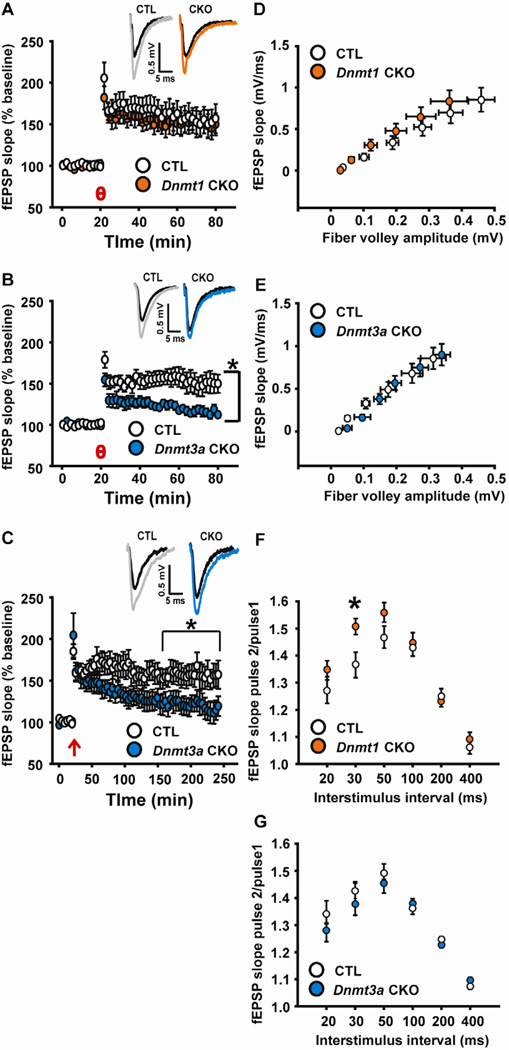
3.3 Long-term potentiation in hippocampal CA1 is deficient in Dnmt3a CKO mice
Long-term potentiation (LTP), a putative cellular mechanism underlying learning, was examined using theta burst stimulation (TBS)- a physiologically relevant stimulus that mimics in vivo hippocampal firing during learning (Otto et al., 1991). Field recordings from hippocampal slices from Dnmt1 CKO mice revealed normal TBS-induced LTP in the CA1 subregion of HC (Fig. 4A). In contrast, Dnmt3a CKO slices had impairments in the induction and maintenance of LTP compared to slices from CTL mice (Fig. 4B). We next used 4-train high frequency stimulation (HFS), a robust stimulus that, unlike the theta burst protocol, produces long-lasting LTP. We found that although the LTP magnitude was not significantly different from CTL for ~40 minutes after HFS, deficits in LTP magnitude subsequently manifest in Dnmt3a CKO slices and LTP was not maintained (Fig. 4C). Basal synaptic function as well as short-term plasticity appeared largely normal in Dnmt1 and Dnmt3a CKOs as demonstrated by input-output relationships and paired pulse ratio (PPR) (Fig. 4D–G), suggesting the deficits were specific for long-term synaptic plasticity. Dnmt1 CKO exhibited enhanced PPR relative to CTL at one of six inter-stimulus intervals tested, suggesting the possibility of mild neurotransmitter release deficits that do not significantly impact the learning and memory behaviors we assessed (Fig. 4F).
Fig. 4.
Dissociable effects of Dnmt1 and Dnmt3a CKO on LTP in hippocampal CA1. (A–B) Hippocampal slices prepared from Dnmt1 CKO mice (n=10) had normal theta burst-induced LTP relative to CTL (n=9), however Dnmt3a CKO slices had deficits in LTP following theta burst (time × genotype interaction F(159, 3517) = 4.006, p < 0.001, 90.0 % of points after theta burst p ≤ 0.05). (C) Dnmt3a CKO mice were also impaired following high-frequency stimulation (time × genotype interaction F(427, 6213) = 3.392, p < 0.001, 68.77% of points after high frequency stimulation p ≤ 0.05; 100% of points significant 157 minutes after LTP induction until end of recording) relative to CTL (Dnmt3a CKO, theta burst n=10, HFS n=6; CTL, theta burst n=12, HFS n=7). θ = theta burst; ↑= high frequency stimulation. Insets (A–C) - Representative traces from CTL and Dnmt1 or Dnmt3a CKO slices 30 (A, B) or 120 minutes (C) after LTP induction. Black traces = baseline, gray (CTL), or colored traces are after LTP induction. (D, E) Input-output relationships were normal in both Dnmt1 (n=11) and Dnmt3a (n=6) CKOs relative to control littermate (CTL, n=9, 8) hippocampal slices. (F, G) Dnmt1 (n=10) CKOs exhibited significantly enhanced paired pulse ratio at 30 ms interstimulus interval (t(15) = 2.346, p = 0.033), suggesting decreased neurotransmitter release probability compared with CTL (n=7). Dnmt3a (n=6) CKO mice had paired pulse ratios that did not differ from CTL (n=8). *p ≤ 0.05 vs. CTL.
4. Discussion
In the current study we report a critical role for DNMT3a in complex behavior and synaptic function in the adult CNS. Our studies demonstrate that experience in an associative fear learning task impacts expression of Dnmt3a, but not Dnmt1, in brain areas that mediate learning of this task. To further explore this finding, we examined conditional Dnmt3 KO mice and observed impairments in learning and memory as assessed by several tasks that assay distinct forms of learning and memory. Dnmt3a CKO mice had impairments in spatial displaced object or novel object recognition. Associative memory formation and extinction of previously learned associations were also deficient in Dnmt3a CKOs as evidenced by impaired performance relative to CTL mice in both contextual and cued fear conditioning and a CTA paradigm. Conversely, in all cases of learning and memory task performance, we found no phenotypes in Dnmt1 CKO mice.
The hippocampal electrophysiological phenotypes promoted by Dnmt3a or Dnmt1 CKO were also dissociable. The impact of Dnmt1 CKO on electrophysiological properties in the CA1 subregion of hippocampal slices was relatively mild as we found only an increase in PPF at a short (30 ms) interpulse interval and no changes in input-output functions. Furthermore, LTP was normal in Dnmt1 CKOs, in contrast to Dnmt3a CKOs that were impaired using a theta burst stimulus or more robust high frequency stimulation. These data are consistent with the behavioral results in which we found no impact of Dnmt1 CKO on learning whereas we observed learning deficits in the Dnmt3a CKOs.
The present study provides evidence for dissociable roles for DNMT1 and DNMT3a in associative learning, extinction learning, and synaptic plasticity. These data are consistent with previous work showing DNMT3a manipulation in forebrain can induce robust behavioral phenotypes (LaPlant et al., 2010; Oliveira et al., 2012). Experience in a fear learning task or chronic treatment with drugs of abuse has been shown to alter the expression of Dnmt3a, and not Dnmt1, in the forebrain, an effect consistent with a distinct role for these enzymes in adult behavior (Miller & Sweatt, 2007; LaPlant et al., 2010). Furthermore, a recent study found decreased Dnmt3a expression in the brains of aged mice that correlated with poor performance on tests of memory, while restoration of the level of Dnmt3a2, one of two transcripts from the Dnmt3a gene, improved memory in fear conditioning and spatial object recognition (Oliveira et al., 2012).
Our findings are seemingly in contrast to a recent study that reported deletion of Dnmt1 or Dnmt3a in forebrain, using the same CaMKIIα-Cre93 line, did not impact learning although a concurrent knockout of both Dnmt1 and Dnmt3a, not tested in our study, produced learning and memory as well as synaptic plasticity deficits (Feng et al., 2010). In the previous study, learning and memory was assessed by the Morris water maze and by contextual fear conditioning 24 hours after training. In the current study, we sought to examine the conditional Dnmt1 and Dnmt3a CKOs in several other learning and memory paradigms. In the fear conditioning paradigm, we observed a significant deficit in context dependent learning of the Dnmt3a CKO mice at 24 hours after training as well as in rates of extinction. The Dnmt3a CKO mice in cue dependent fear conditioning had significant differences in extinction of the cue starting at 72 hours after training that persisted for at least 120 hours. We also examined the individual Dnmt1 and Dnmt3a CKOs in novel and spatial objection recognition as well as conditioned taste aversion where we noticed specific deficits in learning associated with the loss of Dnmt3a and not Dnmt1. These data suggest that DNMT3a is involved in specific types of learning and memory distinct from DNMT1. The reason for contrasting behavioral results between our current study and Feng et al. (2010) are not clear, however it is possible that gender and/ or age differences between mice used in their experiments and ours may have contributed. Our study used only male mice between 6–12 weeks of age, whereas their study included both males and females and the age of the mice was not reported. We also examined our individual Dnmt1 and Dnmt3a CKOs for alterations in LTP as well as paired pulse facilitation. Consistent with our results the study by Feng reported no deficits in LTP in the single Dnmt1 CKO mice using a tetanic stimulation and also did not find alterations in prepulse facilitation or in analysis of input-output curves. In Dnmt3a CKO slices we observed significant deficits in the induction and maintenance of LTP in the Dnmt3a CKOs using theta burst stimulation and a high frequency stimulation, however we did not observe any changes in paired pulse facilitation or in the analysis of input-output curves. The Feng et al study stated that the single knockout mice showed no deficit in LTP, however data was not shown regarding the analysis of LTP in the Dnmt3a single knockout mice leaving it difficult to make a direct comparison between the two studies. The consistent findings that loss of DNMT1 or DNMT3a do not impact paired pulse facilitation or basal synaptic transmission as assessed by input-output curves suggests that DNMT3a impacts specific aspects of synaptic plasticity and not global aspects.
The mechanisms responsible for increased Dnmt3a expression following learning in the fear-conditioning paradigm are unknown. However, taken together with our findings that Dnmt3a CKO mice have learning and synaptic deficits, it is possible that the upregulation of Dnmt3a triggers de novo DNA methylation on genes involved in these processes while the deficits observed in the Dnmt3a KO mice are the result of the loss of this mechanism. This hypothesis would be in agreement with recent in vitro work demonstrating that NMDA receptor mediated synaptic activity drives DNA demethylation in hippocampal neurons (Nelson et al., 2008). Collectively, these data suggest an important and critical role for transcriptional repression in learning and synaptic function (Monteggia & Kavalali, 2009).
Our findings that fear conditioning regulated Dnmt3a expression and that loss of Dnmt3a resulted in learning and synaptic deficits support the growing literature for an importance of epigenetic mechanisms in the adult brain. The gene targets that may be ultimately impacted in learning and memory by Dnmt3a are currently unknown. Previous work has demonstrated that experience in appetitive or aversive learning tasks impacts the methylation of various genes with known involvement in memory formation including BDNF, protein phosphatase- 1, reelin, calcineurin, and the immediate early genes Erg1 and c-fos (Miller & Sweatt, 2007; Lubin et al., 2008; Miller et al., 2010; Mizuno et al., 2012; Day et al., 2013). The further identification and characterization of putative targets will be important in deciphering the role of DNA methylation changes in learning and memory, as little is known about this process in the adult central nervous system. Taken together, our data suggest that de novo and maintenance DNMTs play functionally distinct roles in the adult brain in learning and memory.
Acknowledgements
This work was supported by National Institute of Health grant MH081060 (LMM) and a NARSAD Independent Investigator Award (MJM). We thank Dr. Rudolf Jaensich for generously providing the floxed DNMT1, DNMT3a, and CaMKII-Cre93 mice. The authors would also like to thank Elizabeth Gordon, Melissa Maghoub, and Aroon Karra for assistance with breeding and genotyping of the mice. The authors would like to acknowledge Dr. Ege T. Kavalali as well as members of the Monteggia laboratory for discussions and comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi M, Autry AE, Covington HE, Monteggia LM. MeCP2-mediated transcription repression in the basolateral amygdala may underlie heightened anxiety in a mouse model of Rett syndrome. J Neurosci. 2009;29:4218–4227. doi: 10.1523/JNEUROSCI.4225-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD. DNA methylation regulates associative reward learning. Nat Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, Trumpp A, Poon C, Wilson CB, Jaenisch R. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical longterm potentiation. Thalamus & related systems. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibert S, Forne T, Weber M. Dynamic regulation of DNA methylation during mammalian development. Epigenomics. 2009;1:81–98. doi: 10.2217/epi.09.5. [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Human molecular genetics. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science. 1992;256:675–677. doi: 10.1126/science.1585183. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Jung MW. Neural circuits and mechanisms involved in Pavlovian fear conditioning: a critical review. Neurosci Biobehav Rev. 2006;30:188–202. doi: 10.1016/j.neubiorev.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiology of learning and memory. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes Brain Behav. 2012;11:651–659. doi: 10.1111/j.1601-183X.2012.00805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Mahgoub M, Na ES, Pranav H, Monteggia LM. Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J Neurosci. 2013;33:6401–6411. doi: 10.1523/JNEUROSCI.1001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci. 2012 doi: 10.1038/nn.3151. [DOI] [PubMed] [Google Scholar]
- Otto T, Eichenbaum H, Wiener SI, Wible CG. Learning-related patterns of CA1 spike trains parallel stimulation parameters optimal for inducing hippocampal long-term potentiation. Hippocampus. 1991;1:181–192. doi: 10.1002/hipo.450010206. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Szyf M, Kaplan F, Mann V, Giloh H, Kedar E, Razin A. Cell cycle-dependent regulation of eukaryotic DNA methylase level. The Journal of biological chemistry. 1985;260:8653–8656. [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shimura T, Sako N, Yasoshima Y, Sakai N. Neural substrates for conditioned taste aversion in the rat. Behav Brain Res. 1994;65:123–137. doi: 10.1016/0166-4328(94)90097-3. [DOI] [PubMed] [Google Scholar]