Abstract
The hallmark of endochondral bone development is the presence of cartilaginous templates, in which osteoblasts and stromal cells are generated to form mineralized matrix and support bone marrow hematopoiesis. However, the ultimate source of these mesenchymal cells and the relationship between bone progenitors in fetal life and those in later life are unknown. Fate-mapping studies revealed that cells expressing cre-recombinases driven by the collagen II (Col2) promoter/enhancer and their descendants contributed to, in addition to chondrocytes, early perichondrial precursors prior to Runx2 expression and, subsequently, to a majority of osteoblasts, Cxcl12 (chemokine (C-X-C motif) ligand 12)-abundant stromal cells and bone marrow stromal/mesenchymal progenitor cells in postnatal life. Lineage-tracing experiments using a tamoxifen-inducible creER system further revealed that early postnatal cells marked by Col2-creER, as well as Sox9-creER and aggrecan (Acan)-creER, progressively contributed to multiple mesenchymal lineages and continued to provide descendants for over a year. These cells are distinct from adult mesenchymal progenitors and thus provide opportunities for regulating the explosive growth that occurs uniquely in growing mammals.
Introduction
Most bones are formed through endochondral bone development, in which mesenchymal condensations first define the domain for the future bones, and then develop into the growth cartilage wherein chondrocytes appear and proliferate. Chondrocytes in the growth plate continue to proliferate well into adulthood in mice1. In the center of the developing cartilage mold, chondrocytes stop proliferating and become hypertrophic chondrocytes. These cells signal to induce the migration of mesenchymal cells into the marrow space; these cells then differentiate into osteoblasts that then form bone on top of the cartilaginous matrix. Perichondrial precursors expressing osterix (Osx) invade into the cartilage template along with blood vessels and eventually become both osteoblasts and stromal cells in the marrow space2. However, mesenchymal cells constituting earlier cells of the osteoblast lineage than Osx+ precursors in vivo have not been fully characterized. The transcription factor Sox9 is expressed in mesenchymal condensations, and osteochondroprogenitors are derived from Sox9+ cells, as Sox9-cre marks all chondrocytes and osteoblasts3. Sox9 binds to genes encoding major cartilaginous matrix proteins such as aggrecan (Acan) and type II collagen-α1 (Col2a1) and regulates their expression. How these early osteochondroprogenitors and their descendants relate to mesenchymal precursors in adult bone is unknown. In adult endochondral bones, the source of osteoblasts and stromal cells has been proposed to be mesenchymal stem cells (MSCs) or bone marrow stromal/mesenchymal progenitor cells (BMSCs), which are traditionally defined as cells capable of forming colonies in vitro (CFU-Fs: colony forming unit-fibroblasts) that can undergo multilineage differentiation in vitro and upon transplantation4. CFU-Fs are enriched among various adult marrow populations such as nestin (Nes)-GFP+ cells5, platelet-derived growth factor receptor-α (PDGFRα)+Sca1+CD45−Ter119− (PαS) cells6 and leptin receptor (LepR)/LepR-cre+ cells7 in mice and CD146+ pericytes in humans8. In this study, we sought to identify cells that robustly supply osteoblasts and stromal cells in the metaphyses of growing bones, and suggest the possibility that these cells might be the source of MSCs/BMSCs in adult bone marrow.
Results
Col2-cre/Col2-creER are expressed in early cells of the osteoblast lineage in fetal mice
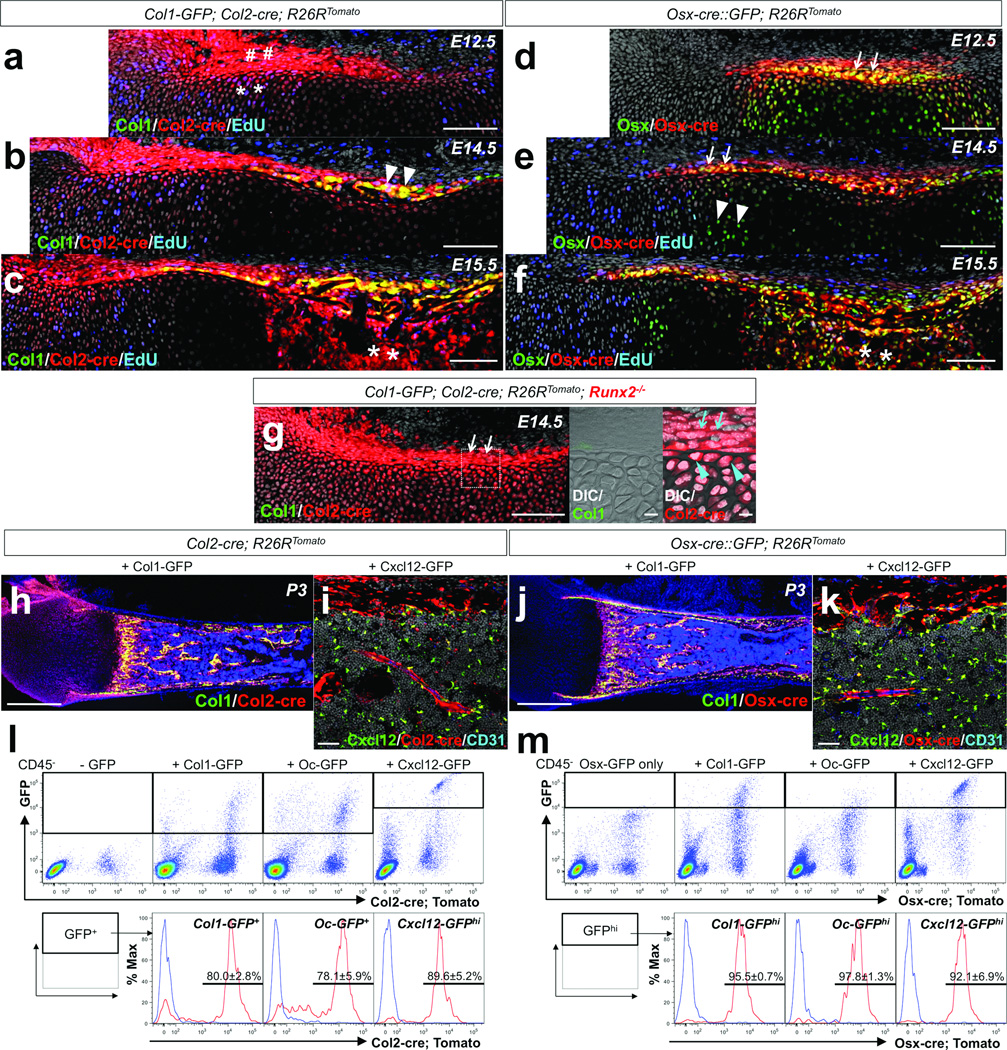
We chose to explore the hypothesis that cells defined by activities of the Sox9, aggrecan (Acan), and type II collagen (Col2) gene promoter/enhancers might encompass mesenchymal precursors of osteoblasts and stromal cells. Previous studies indicate that osteochondroprogenitors are marked by cre recombinases driven by the Col2a1 promoter9–12. First, we mapped cell fates using a Col2-cre; R26R-tdTomato reporter13, 14 in combination with Col1(2.3kb)-GFP15 as a readout of osteoblastic cells. In this system, cells expressing Col2-cre and their descendants become red, and if they concurrently express Col1-GFP, they become yellow. In addition, a thymidine analog, EdU, was administered shortly before analysis to evaluate cell proliferation. At embryonic day 12.5 (E12.5), red cells were observed in the growth cartilage (Figure 1a, asterisks) and perichondrium (Figure 1a, sharps), where many of these cells were proliferating (See also Supplementary Figure 1a for an earlier day). At E14.5, almost all Col1-GFP+ cells appearing in the osteogenic perichondrium were yellow (Figure 1b, arrowheads); thus, these perichondrial preosteoblasts were derived from Col2-cre+ cells. Active cell proliferation was observed in the perichondrium, but not in its adjacent hypertrophic chondrocytes (Figure 1b). At E15.5, Col2-cre-derived cells proliferated within the marrow space, as the nascent primary ossification center was occupied by red cells (Figure 1c, asterisks).
Figure 1. Fate mapping of Col2-cre+ and Osx-cre+ cells during endochondral ossification.
a–c. Fate mapping of Col2-cre+ cells was performed using Col1-GFP; Col2-cre; R26RTomato mice, with EdU administration prior to analysis. (a) Embryonic day 12.5 (E12.5); sharps: Tomato+ perichondrial cells, asterisks: Tomato+ chondrocytes. (b) E14.5; arrowheads: Col1+Tomato+ perichondrial cells. (c) E15.5; asterisks: Tomato+ cells in primary ossification center. Green: EGFP, red: tdTomato, blue: Alexa647, gray: DAPI. Scale bars: 100µm.
d–f. Fate mapping of Osx-cre+ cells was performed using Osx-cre∷GFP; R26RTomato mice, with EdU administration prior to analysis (e,f). (d) E12.5; arrows: Osx+ perichondrium. (e) E14.5; arrows: Osx+Col1− perichondrium, arrowheads: Osx+ prehypertrophic chondrocytes. (f) E15.5; asterisks: Osx+ cells in primary ossification center. Green: EGFP, red: tdTomato, blue: Alexa647, gray: DAPI. Scale bars: 100µm.
g. Fate mapping of Col2-cre+ cells in Runx2-deficient bone anlage at E14.5 was performed using Col1-GFP; Col2-cre; R26RTomato; Runx2−/− mice. Middle and right panels: dotted area of left panel revealing perichondrium. Arrows: Tomato+ perichondrial cells, arrowheads: Tomato+ chondrocytes beneath perichondrium. Green: EGFP, red: tdTomato, gray: DAPI and DIC (differential interference contrast). Scale bars: 100µm (left panel) and 10µm (center and right panels).
h,j. Comparative fate mapping in postnatal day 3 (P3) bones was performed using Col1-GFP; Col2-cre; R26RTomato (h) and Col1-GFP; Osx-cre; R26RTomato (j) mice. Green: EGFP, red: tdTomato, blue: DAPI. Scale bars: 500µm.
i,k. Comparative fate mapping in P3 bones was performed using Cxcl12-GFP; Col2-cre; R26RTomato (i) and Cxcl12-GFP; Osx-cre; R26RTomato (k) mice. Shown are diaphyseal endocortices and bone marrows, stained for CD31. Green: EGFP, red: tdTomato, blue: Alexa633, gray: DAPI. Scale bars: 50µm.
l,m. Flow cytometry analysis was performed using dissociated bone cells harvested at P3. CD45− fraction was gated for GFP. (l) Representative dot plots of cells from Col2-cre; R26RTomato mice that also carry Col1-GFP (left middle panel), Oc-GFP (right middle panel) or Cxcl12-GFP (rightmost panel). Leftmost panel: no GFP control. Lower panels: histograms of the GFP+ fraction developed for Tomato; blue lines: GFP+Tomato control cells. (m) Representative dot plots of cells from Osx-cre∷GFP; R26RTomato mice that also carry Col1-GFP (left middle panel), Oc-GFP (right middle panel) or Cxcl12-GFP (rightmost panel). Leftmost panel: Osx-GFP only control. Lower panels: histograms of the GFPhigh fraction developed for Tomato; blue lines: GFPhighTomato control cells. Upper panels; X-axis: tdTomato, Y-axis: GFP. Lower panels; X-axis: tdTomato. n=3 mice per group. All data are represented as mean ± SD.
Second, we mapped cell fates using an Osx-cre∷GFP; R26R-tdTomato reporter16. In this system, cells expressing Osx-cre∷GFP become green in the nucleus, and these cells and their descendants become red. At E12.5, Osx+ yellow cells (expressing Osx-cre∷GFP and tdTomato) were observed in the growth cartilage and perichondrium, in a domain more restricted than that of Col2-cre targeted cells (Figure 1d, arrows). At E14.5, Osx+ yellow cells dominated the inner part of the perichondrium in a domain broader than Col1+ cells seen in Fig.1b (Figure 1e, arrows), with some of them in proliferation (Supplementary Figure 1f,g). Osx+ prehypertrophic chondrocytes appearing green were not proliferating (Figure 1e, arrowheads). At E15.5, mesenchymal cells appearing in the primary ossification center were largely yellow (Figure 1f, asterisks), and therefore expressing Osx. These comparative fate mapping analyses suggest that one fate of Col2+ cells may be to become Osx+ cells in the perichondrium and the marrow space.
Runx2 is a crucial transcription factor in osteoblastic differentiation genetically upstream of Osx17. To understand whether Col2+ cells require Runx2 expression, as, for example, nestin-GFP+ perichondrial cells do18, we analyzed mice carrying Runx2-null alleles19 in addition to the Col2-cre; R26R-tomato; Col1-GFP transgenes. In these mice, no Col1-GFP+ cell appeared in the perichondrium at E14.5. However, despite the absence of Runx2, Col2-cre-derived red cells were still present in the perichondrium (Figure 1g, arrows) and its underlying growth cartilage (Figure 1g, right panel, arrowheads). Therefore, Runx2 is dispensable for generating these Col2-cre-derived progenitor cells, although it is required for their further differentiation into osteoblasts. This places Col2+ cells in the perichondrium upstream of Runx2 expression in the lineage.
We next examined the distribution of Col2-cre targeted red cells at postnatal day 3 (P3), when bone marrow hematopoiesis had been established. Col2-cre targeted red cells contributed not only to chondrocytes and perichondrial cells in the growth cartilage, but also to Col1-GFP+ osteoblasts, osteocytes and Cxcl12high stromal cells20, 21 in the marrow space (Figure 1h,i, see also Supplementary Figure 1b,c). Osx-cre targeted red cells contributed to all these cell types22, 23 (Figure 1j,k, see also Supplementary Figure 1d,e). Flow cytometry analysis of dissociated bone cells revealed that Osx-cre targeted cells contributed to essentially all osteoblasts (95.5±0.7% of Col1-GFPhigh and 97.8±1.3% of Oc-GFPhigh15) and Cxcl12-expressing stromal cells (92.1±6.9% of Cxcl12-GFPhigh, n=3 per group, data represented as mean ± SD) 24 (Figure 1m). Col2-cre targeted cells also contributed to a great majority of osteoblasts (80.0±2.8% of Col1-GFP+ and 78.1±5.9% of Oc-GFP+) and Cxcl12-expressing stromal cells (89.6±5.2% of Cxcl12-GFPhigh, n=3 per group, data represented as mean ± SD) (Figure 1l). Therefore, most osteoblasts and Cxcl12-expressing stromal cells in endochondral bones are derived from mesenchymal progenitors/precursors that express Col2-cre and Osx-cre at some point in their development.
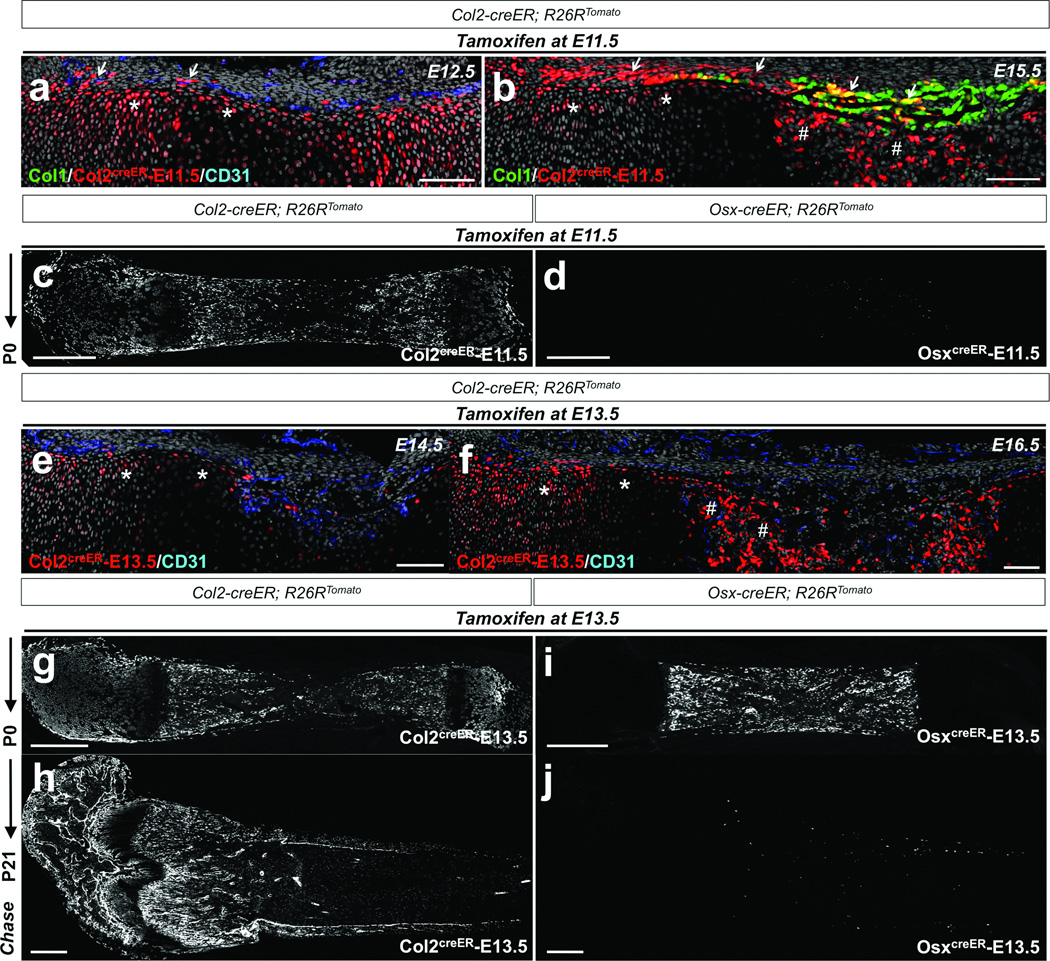
To further clarify the relationships between Col2+ cells and Osx+ cells within the mesenchymal lineage, we took advantage of tamoxifen-inducible creER recombinases (Col2-creER25 or Osx-creER2) and performed pulse-chase experiments with a single tamoxifen injection. An E11.5 pulse to Col2-creER mice marked perichondrial cells and chondrocytes at E12.5 (Figure 2a), and their descendants (Col2creER-E11.5) contributed to the perichondrium and the primary ossification center at E15.5 (Figure 2b) and yielded a number of Tomato+ cells throughout the bone at P0 (Figure 2c) and P21 (Supplementary Figure 2a). In contrast, an E11.5 pulse to Osx-creER mice did not give rise to descendants at P0 (Figure 2d), suggesting that Col2-creER+ cells appeared earlier than Osx-creER+ cells during early skeletal development. An E13.5 pulse to Col2-creER mice marked chondrocytes beneath the perichondrium, as well as perichondrial cells, at E14.5 (Figure 2e), and their descendants (Col2creER-E13.5) contributed to the primary ossification center at E16.5 (Figure 2f). Col2creER-E13.5 cells continued to yield Tomato+ cells robustly in the growth cartilage, the perichondrium and the bone at P0 (Figure 2g) and including the secondary ossification center in the epiphyseal region at P21 (Figure 2h). Osx-creER+ cells at E13.5 proliferate in the primary ossification center at E16.52 but do not persist in the perichondrium18. Their descendants (OsxcreER-E13.5) appeared as osteoblasts and stromal cells among cells derived from the primary ossification center, but not those of the secondary ossification center at P0 (Figure 2i), and then gradually disappeared from the metaphysis by P21 (Figure 2j and Supplementary Figure 2b). These data underscore the transient nature of embryonic Osx-creER+ cells, supporting the notion that these cells are replenished by their precursors, probably derived from Col2-creER cells during early bone development.
Figure 2. Col2-creERmarks cells earlier than Osx-creER+ cells in the osteoblast lineage in fetal mice.
a,b. Pregnant mice received 1mg tamoxifen at E11.5, and Col1-GFP; Col2-creER; R26RTomato mice were chased for 24 hours (a; E12.5) or 4 days (b; E15.5). Shown are distal halves of growth cartilages. Perichondrium is on top. Femur sections were stained for nuclei and CD31. Arrows: Tomato+ perichondrial cells, asterisks: Tomato+ chondrocytes, sharps: Tomato+ cells in primary ossification center. Green: EGFP, red: tdTomato, blue: Alexa633, gray: DAPI.
c.d. Lineage-tracing of embryonic Col2-creER+ and Osx-creER+ cells was performed by injecting 1mg tamoxifen into pregnant mice at E11.5, and Col2-creER; R26RTomato (c) or Osx-creER; R26RTomato (d) mice were chased until P0. Shown are femur sections with the distal side on the left. Col2creER-E11.5 represents descendants of Col2-creER+ cells at E11.5. White indicates tdTomato.
e,f. Pregnant mice received 1mg tamoxifen at E13.5, and Col2-creER; R26RTomato mice were chased for 24 hours (e; E14.5) or 3 days (f; E16.5). Femur sections were stained for nuclei and CD31. Asterisks: Tomato+ chondrocytes and sharps: Tomato+ cells in primary ossification center. Red: tdTomato, blue: Alexa633, gray: DAPI.
g–j. Lineage-tracing of embryonic Col2-creER+ and Osx-creER+ cells was performed by injecting 1mg tamoxifen into pregnant mice at E13.5, and Col2-creER; R26RTomato (g,h) or Osx-creER; R26RTomato (i,j) mice were chased until indicated postnatal day (P0 or P21). Shown are femur sections with the distal side on the left. Col2creER-E13.5 represents descendants of Col2-creER+ cells at E13.5. White indicates tdTomato. Scale bars: 100µm (a,b,e,f) or 500µm (c,d,g–j).
Col2-creER-expressing cells at an early postnatal stage generate multiple mesenchymal lineages
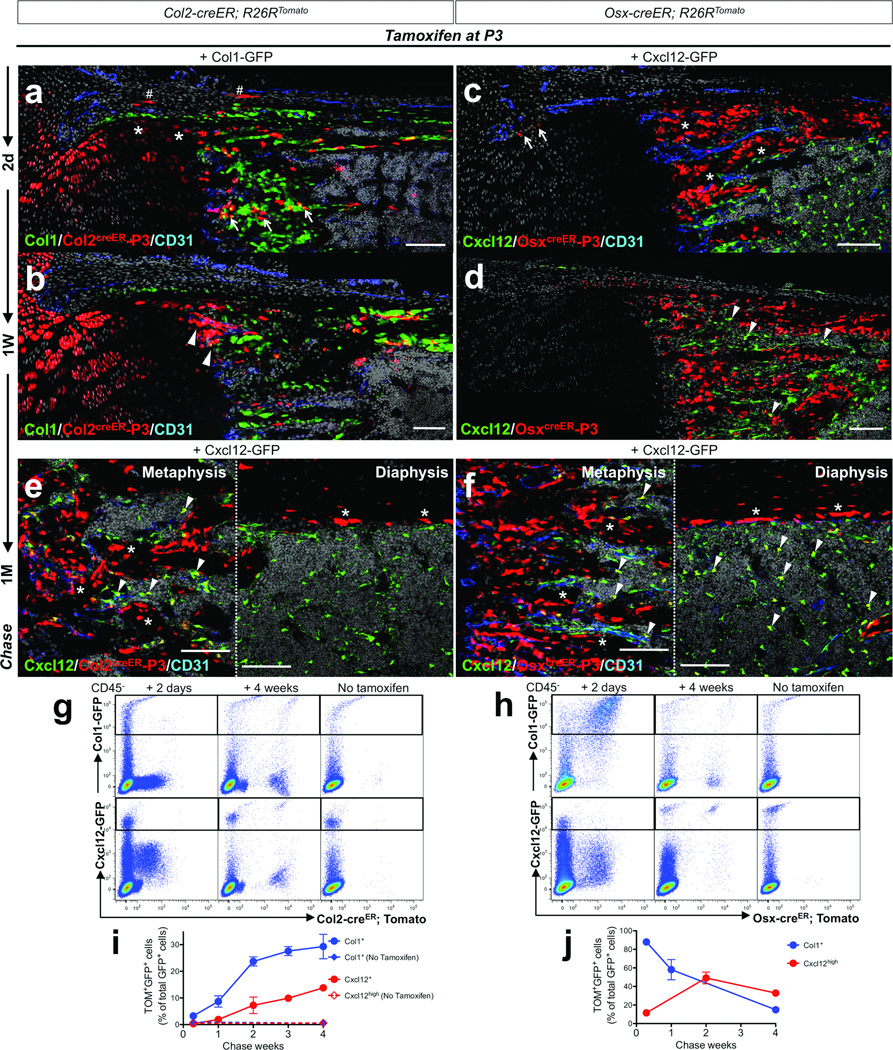
Because cells expressing Col2-creER in fetal life appear to be early cells in the osteoblast lineage, we next asked whether this transgene is also active in postnatal life. For this purpose, Col2-creER; R26R-tomato25 mice were pulsed with a low-dose tamoxifen injection at P3. A small number of Tomato+ cells were detected at P5 and P10 in the absence of tamoxifen administration (Supplementary Figure 2d,e). Two days after a tamoxifen pulse at P3 (Col2creER-P3), chondrocytes in the growth plate and beneath the perichondrium, as well as cells in the perichondrium and metaphyseal spongiosa, were labeled by Col2-creER (Figure 4a). After a week, a group of Col2creER-P3 cells appeared directly under the growth plate in the primary spongiosa and beneath the unlabeled cells of the perichondrium with further contiguity to the endocortical surface (Figure 3a, arrowheads, see also Figure 4b). After a month, Col2creER-P3 cells became osteoblasts and stromal cells in the metaphysis and epiphysis, in addition to chondrocytes in the growth plate and articular cartilage (Figure 3b, see also Figure 4e). During an extended chase period over a year after the pulse, Col2creER-P3 cells continued to yield chondrocytes, osteoblasts and stromal cells in the metaphysis, and also became adipocytes in the metaphyseal bone marrow (Figure 3c,d and Supplementary Figure 2f). Of note, Col2-creER does not mark marrow cells in the middle of the diaphysis at P3, even though Col2-cre marks all stromal and osteoblastic regions of bone. Presumably, this contrast results from the Col2 promoter driving the expression of creER no longer being active in marrow stromal cells in the diaphysis at P3.
Figure 4. Col2-creER+ cells generate multiple mesenchymal lineages in postnatal growing bones.
a,b. Col1-GFP; Col2-creER; R26RTomato mice received 0.1mg tamoxifen at P3 and were chased for 48 hours (a) or a week (b). Shown are distal halves of growth cartilages at the junction of growth plate and primary spongiosa. Perichondrium is on top. Femur sections were stained for nuclei and CD31. Asterisks: Tomato+ chondrocytes, sharps: Tomato+ perichondrial cells, arrows: Tomato+ cells in primary spongiosa, arrowheads: a group of Tomato+ cells directly under the growth plate and beneath the perichondrium. c,d. Cxcl12-GFP; Osx-creER; R26RTomato mice received 0.1mg tamoxifen at P3 and were chased for 48 hours (c) or a week (d). Shown are distal halves of growth cartilages at the junction of growth plate and primary spongiosa. Perichondrium is on top. Femur sections were stained for nuclei and CD31. Arrows: Tomato+ perichondrial cells, asterisks: Tomato+ osteoblasts, arrowheads: GFP+Tomato+ stromal cells. e,f. Cxcl12-GFP; Col2-creER; R26RTomato (e) or Cxcl12-GFP; Osx-creER; R26RTomato (f) mice received 0.1mg tamoxifen at P3 and were chased for a month. Shown are metaphyseal spongiosa (left panel) and diaphyseal endocortex (right panel). Femur sections were stained for nuclei and CD31. Asterisks: Tomato+ osteoblasts, arrowheads: GFP+Tomato+ stromal cells. For a–f, Green: EGFP, red: tdTomato, blue: Alexa633, gray: DAPI. Scale bars: 100µm.
g,h. Flow cytometry analysis was performed using dissociated bone cells harvested from mice that received 0.1mg tamoxifen at P3 (except no tamoxifen controls) and were chased for indicated periods. CD45− fraction was gated for GFP. (g) Representative dot plots of cells from Col1-GFP; Col2-creER; R26RTomato (upper panels) and Cxcl12-GFP; Col2-creER; R26RTomato (lower panels) mice. (h) Representative dot plots of cells from Col1-GFP; Osx-creER; R26RTomato (upper panels) and Cxcl12-GFP; Osx-creER; R26RTomato (lower panels) mice. Left panels: 2 days chase, middle panels: 4 weeks chase, right panels: no tamoxifen controls. X-axis represents tdTomato, and Y-axis represents GFP.
i,j. Percentage of Col2creER-P3 cells (o) or OsxcreER-P3 cells (p) among Col1-GFP+ (blue lines) osteoblasts or Cxcl12-GFP+ (red lines) stromal cells during the chase. X-axis represents the duration of the chase, and Y-axis represents the percentage of Tomato+GFP+ cells among total GFP+ cells. Dotted lines represent no tamoxifen controls. n=3–6 mice per group (see Methods for precise n values). All data are represented as mean ± SD.
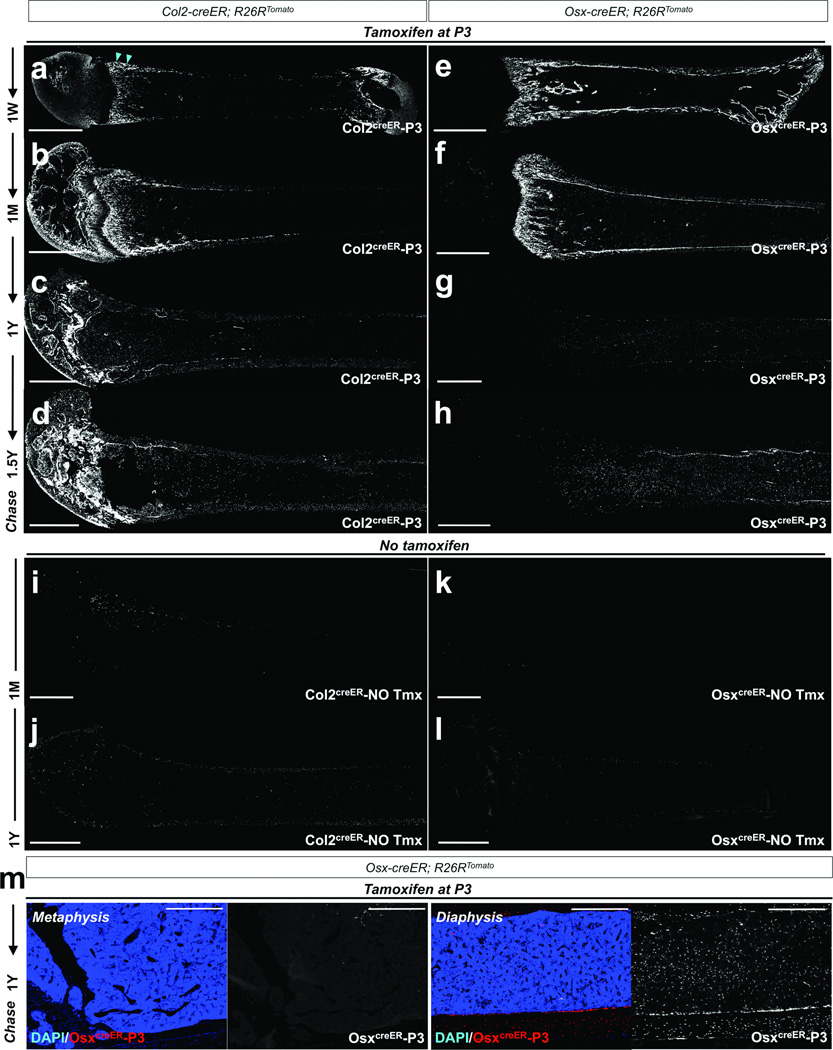
Figure 3. Col2-creER+ cells encompass early cells of the osteoblast lineage in postnatal mice.
a–h. Lineage-tracing of postnatal Col2-creER+ and Osx-creER+ cells was performed by injecting 0.1mg tamoxifen into P3 mice. Col2-creER; R26RTomato (a–d) or Osx-creER; R26RTomato (e–h) mice were chased for a week (a,e), a month (b,f), a year (c,g) and 18 months (d,h). Shown are femur sections with the distal side on the left. Col2creER-P3 represents descendants of Col2-creER+ cells at P3. Arrowheads in (g) point to Col2creER-P3 cells under the growth plate and beneath the perichondrium. White indicates tdTomato..
i–l. No tamoxifen controls of Col2-creER; R26RTomato (i,j) or Osx-creER; R26RTomato (k,l) mice at 1 month of age (i,k) or 1 year of age (j,l) are shown. Shown are femur sections with the distal side on the left. White indicates tdTomato.
m. Osx-creER; R26RTomato mice received 0.1mg tamoxifen at P3 and were chased for a year. Shown are higher magnification views of Figure 3g. OsxcreER-P3 represents descendants of Osx-creER+ cells at P3. Left and right two panels show the magnified views of the metaphysis and diaphysis, respectively. Subpanels on the right show a single color view of tdTomato. Blue indicates DAPI and red and white indicate tdTomato. Scale bars: 1mm (a–l) or 500µm (m).
The contrast between the cells marked by Col2-creER and Osx-creER transgenes is instructive. Shortly after the pulse, a majority of osteoblasts on the bone surface and cells in the perichondrium were marked by Osx-creER (Figure 4c). After a week of chase, OsxcreER-P3 cells continued to be present on the bone surface and in the marrow space (Figure 3e, see also Figure 4d). After a month, OsxcreER-P3 cells became osteoblasts and stromal cell in the metaphysis and diaphysis, but not in the epiphysis (Figure 3f, see also Figure 4f). In contrast to the persistence of Col2creER-P3 cells in the one year chase, OsxcreER-P3 cells gradually disappeared from the metaphysis and became increasingly present as stromal cells in the diaphyseal bone marrow and osteoblasts on the endocortical surface (Figure 3g,h,m and Supplementary Figure 2c). The Osx-creER; R26R-tomato system had little to no tamoxifen-independent activities (Figure 3k,l). Thus, Osx-creER+ cells at P3 generate cells in the metaphysis only transiently, but persist in the marrow stroma for a prolonged period.
To assess quantitatively how cells marked by Col2-creER or Osx-creER contribute differentially to defined cell types over time, we performed flow cytometry analysis of cells isolated from enzymatically-digested bones from triple transgenic mice (Col1-GFP or Cxcl12-GFP, Col2-creER or Osx-creER and R26R-tomato) with a single tamoxifen injection at P3. Shortly after the pulse, only a small fraction of cells marked by Col2-creER expressed these GFP reporters (48 hours after tamoxifen injection, Col1-GFP+; 3.2±1.2% [n=4], Cxcl12-GFPhigh, 0.3±0.1% [n=3], data represented as mean ± SD), suggesting that Col2-creER+ cells were largely distinct from osteoblasts or Cxcl12-expressing stromal cells (Figure 4g, left panels). The fraction of Col2creER-P3 cells among osteoblasts and Cxcl12-expressing stromal cells increased progressively as the chase period extended (Col1-GFP+; from 8.7±2.1% [n=3], 23.7±1.7% [n=5], 27.6±1.7% [n=3] to 29.3±4.6% [n=6], Cxcl12-GFPhigh; from 1.9±0.3% [n=4], 7.3±3.1% [n=4], 9.9±1.2% [n=4] to 13.8±0.5% [n=4], for the first, second, third and fourth week, respectively, Figure 4g, middle panels, and Figure 4i, blue and red lines, data represented as mean ± SD). No increase of Tomato+ cells was observed without tamoxifen injection (Figure 4g, right panels). In contrast, 87.9±1.4% [n=4] and 11.6±3.5% [n=3] of cells marked by Osx-creER shortly after the pulse were Col1-GFP+ osteoblasts and Cxcl12-expressing stromal cells, respectively, (Figure 4h, left panels, data represented as mean ± SD). The fraction of OsxcreER-P3 cells decreased among osteoblasts (58.1±10.9% [n=7] and 14.9±0.1% [n=3] for the first and fourth week, respectively, Figure 4h, upper middle panel, and Figure 4j, blue line), and increased among Cxcl12-expressing stromal cells (49.2±6.3% [n=3] and 32.9±1.1% [n=4] for the second and fourth week, respectively, Figure 4h, lower middle panel, and Figure 4j, red line, data represented as mean ± SD). These studies reinforce the results from the histological findings that Col2-creER-marked cells robustly contribute to osteoblasts and stromal cells, while Osx-creER-marked cells transiently become osteoblasts and persist longer in bone marrow as stromal cells.
Relationship of growth skeletal progenitor cells and adult bone marrow stromal/mesenchymal progenitor cells
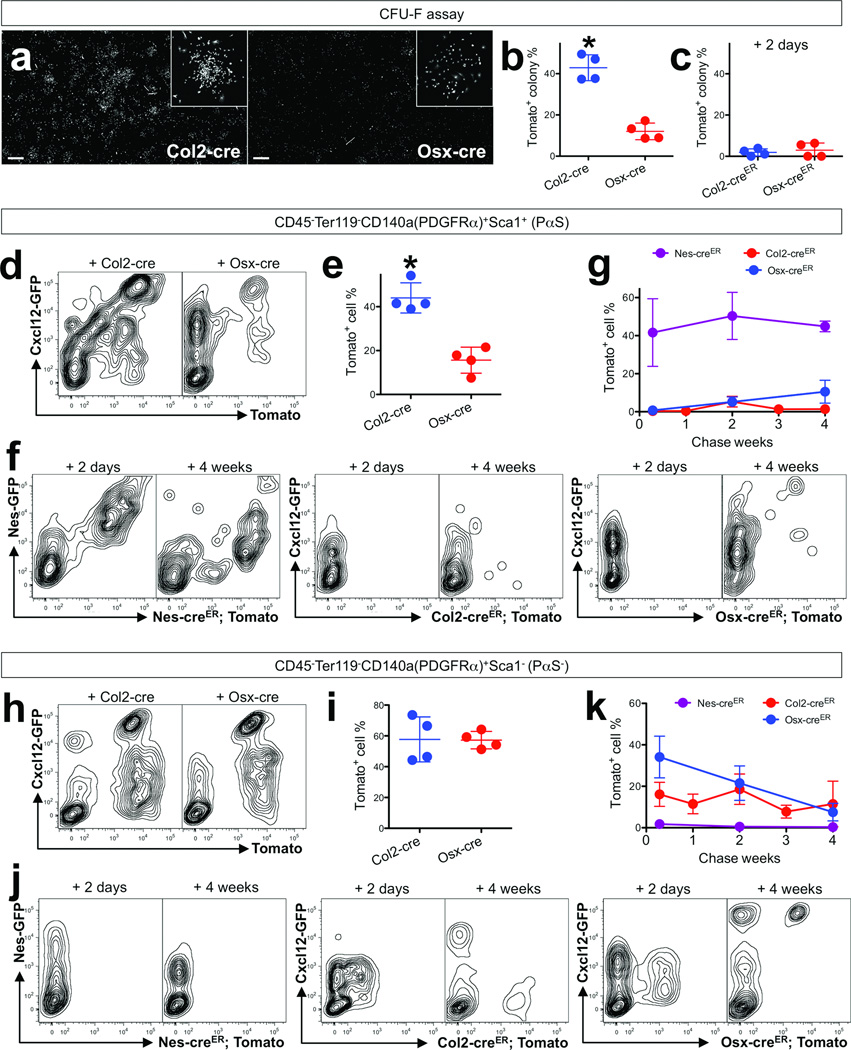
In order to determine the relationship of the growth skeletal stem/progenitor cells that we have defined here with the more traditionally defined BMSCs, we conducted colony-forming unit fibroblast (CFU-F) assays using unfractionated bone marrow cells isolated from P5 mice. Quantification of Tomato+ colonies revealed that Col2-cre targeted a higher proportion of CFU-Fs than Osx-cre did (Tomato+ CFU-F: Col2-cre; 42.9±6.3%, Osx-cre; 12.0±4.0%, Figure 5a,b, see also Supplementary Figure 3a, n=4 per group, data represented as mean ± SD), indicating that a larger fraction of BMSCs were derived from Col2-cre+ cells than from Osx-cre+ cell during development. To understand if BMSCs themselves express Col2 or Osx, CFU-F assays were performed using bone marrow cells harvested from Col2-creER or Osx-creER; R26R-tomato mice 2 days after the pulse. Only a small proportion of CFU-Fs were Tomato+ using this protocol (Tomato+ CFU-F: Col2-creER; 1.9±1.7%, Osx-creER; 3.0±3.5%, Figure 5c, see also Supplementary Figure 3b, n=4 per group, data represented as mean ± SD), suggesting that the majority of BMSCs were rather descendants of cells expressing Col2-cre. Second, we conducted flow cytometry analysis of a PαS fraction using dissociated bone cells harvested from these mice at P5 (Supplementary Figure 3c). A PαS fraction represents <1% of the CD45−Ter119− fraction and is enriched for CFU-Fs6. Col2-cre targeted a higher proportion of PαS cells than Osx-cre did (Tomato+ PαS cells: Col2-cre; 44.0±6.9%, Osx-cre; 15.6±6.9%, Figure 5d,e, n=4 per group, data represented as mean ± SD), as anticipated from the fraction of CFU-Fs marked by these cre lines. A fraction of PαS cells was Cxcl12-GFPhigh, and a small percentage of Cxcl12-GFPhigh cells was positive for Sca1; these fractions appeared to decrease with age (Supplementary Figure 3d,e). A great portion of Cxcl12-GFPhigh cells was PDGFRα+Sca1− cells (Supplementary Figure 3f). Nes-GFP+ cells include all CFU-Fs5 and are descended from Col2-creER+ cells during bone development18. Analysis of Nes-GFP; Col2-cre; R26R-tomato bone cells revealed that 46.7±5.1% of Nes-GFP+ cells were targeted by Col2-cre (Supplementary Figure 4a, n=5, data represented as mean ± SD). We further interrogated the PαS fraction using Nes-GFP; Nes-creER; R26R-tomato mice and a single tamoxifen injection at P3. A high fraction of PαS cells was marked by Nes-creER and their descendants throughout the entire first month of chase (48 hours after the pulse, 41.6±17.8% [n=5], then 50.4±12.4% [n=3] and 44.9±2.8% [n=4] for the second and fourth week of chase, respectively, Fig.3f,g, data represented as mean ± SD). Nes-creER-marked cells contributed to Cxcl12-GFPhigh cells transiently (50.1±4.2% [n=3], 11.6±2.3% [n=4] then 6.0±0.8% [n=3], for the second day, the first and fourth week of chase, respectively, data represented as mean ± SD, Supplementary Figure 4c), while their contribution to endothelial cells is predominant and persistent18. Virtually no PαS cell was marked by Col2-creER or Osx-creER shortly after the pulse (0.3±0.4% [n=4] and 0.7±0.3% [n=6], respectively, Figure 5f,g, data represented as mean ± SD). During the chase, the fraction of PαS cells derived from OsxcreER-P3 cells gradually increased (5.2±2.6% [n=5] and 10.5±6.0% [n=8] for the second and fourth week, respectively), while those derived from Col2creER-P3 cells only temporarily increased (0.4±0.3% [n=5], 5.3±2.8% [n=7], 1.3±1.6% [n=5] and 1.4±1.3% [n=9] for the first to fourth week, respectively, data represented as mean ± SD) (Figure 5f,g). Similarly, an E13.5 pulse to Col2-creER mice marked virtually no PαS after two weeks of chase at P7 (Supplementary Figure 4b). Therefore, while postnatal Col2-creER+ cells robustly contributed to Col1-GFP+ osteoblasts and Cxcl12high stromal cells during bone growth, their contribution to a PαS fraction appears to be minimal. Since Col2-cre+ cells do contribute substantially to the PαS population, it is possible that they do so at a time that we have not tested with Col2-creER mice. A PDGFRα+Sca1− fraction encompasses most BMSCs7. Col2-cre and Osx-cre also targeted a similar proportion of PDGFRα+Sca1− cells (Tomato+ PDGFRα+Sca1− cells: Col2-cre; 57.7±14.6%, Osx-cre; 57.2±5.7%, Figure 5h,i, n=4 per group, data represented as mean ± SD). A sizable fraction of PDGFRα+Sca1− cells were marked by Col2-creER or Osx-creER shortly after the pulse (16.1±5.8% [n=4] and 34.1±10.0% [n=6], respectively), while only marginally marked by Nes-creER (1.8±1.3% [n=5], Figure 5j,k, data represented as mean ± SD).
Figure 5. Relationship of growth skeletal progenitor cells and adult bone marrow stromal/mesenchymal progenitor cells.
a. Colony-forming unit fibroblast (CFU-F) assay was performed using unfractionated P5 bone marrow cells plated at a clonal density (106 cells per 9.6cm2) and cultured for 10 days. Cells were harvested from Col2-cre; R26RTomato (a, left panel), Osx-cre; R26RTomato (a, left panel) mice, Col2-creER; R26RTomato or Osx-creER; R26RTomato mice that received 0.1mg tamoxifen at P3. White indicates tdTomato. Insets highlight examples of Tomato+ colonies. Scale bars: 2mm.
b,c. Percentage of Tomato+ colonies among total colonies (>50 cells) was enumerated. P5 bone marrow cells from Col2-cre; R26RTomato or Osx-cre; R26RTomato mice (b), and Col2-creER; R26RTomato or Osx-creER; R26RTomato mice that received 0.1mg tamoxifen at P3 (c) were subjected to CFU-F assay. n=4 mice per group, *p<0.05. All data are represented as mean ± SD.
d,e,h,i. Flow cytometry analysis was performed using dissociated bone cells stained for CD45, Ter119, PDGFRα (CD140a) and Sca1. (d) Cells were harvested from Cxcl12-GFP; Col2-cre; R26RTomato (left panel) or Cxcl12-GFP; Osx-cre; R26RTomato (right panel) mice at P14. Representative contour plots of CD45−Ter119−CD140a+Sca1+ (PαS) (d) or CD45−Ter119−CD140a+Sca1− (PαS−) (h) fraction of the indicated genotype. X-axis represents tdTomato, and Y-axis represents Cxcl12-GFP. (e,i) Percentage of Tomato+ PαS cells (e) and Tomato+ PαS− cells among total PαS cells. n=4 mice per group. *p<0.05. All data are represented as mean ± SD.
f,j. Flow cytometry analysis was performed using dissociated bone cells stained for CD45, Ter119, PDGFRα (CD140a) and Sca1. Representative contour plots of CD45−Ter119−CD140a+Sca1+ (PαS) (f) or CD45−Ter119−CD140a+Sca1− (PαS−) (j) fraction of Nes-GFP; Nes-creER; R26RTomato (left panels), Cxcl12-GFP; Col2-creER; R26RTomato (middle panels) or Cxcl12-GFP; Osx-creER; R26RTomato (right panels) mice that received 0.1mg tamoxifen at P3. Left subpanels show 2 days of chase and right subpanels show 4 weeks of chase. X-axis represents tdTomato, and Y-axis represents GFP.
g,k. Percentage of NescreER-P3 cells (purple line), Col2creER-P3 cells (red line) or OsxcreER-P3 cells (blue line) among CD45−Ter119−CD140a+Sca1+ (PαS) (g) or CD45−Ter119−CD140a+Sca1− (PαS−) (k) cells during the chase was plotted. X-axis represents the duration of the chase, and Y-axis represents the percentage of Tomato+GFP+ cells among total GFP+ cells. n=3–9 mice per group (see Methods for precise n values). All data are represented as mean ± SD.
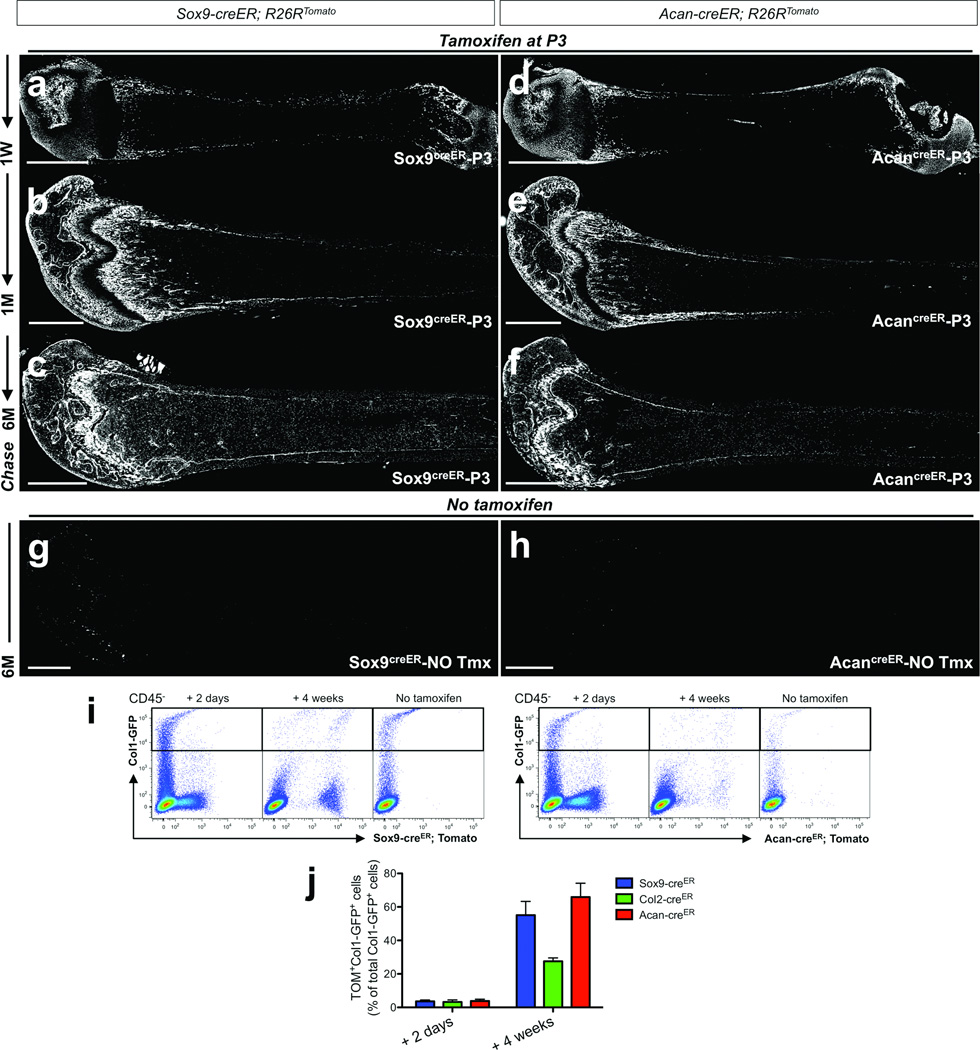
We asked further whether cells defined by promoter/enhancer activities of other genes involved in early chondrocyte development could, like Col2-creER, be used to mark early cells of the osteoblast lineage in postnatal bone development. For this purpose, we took advantage of Sox9-creER26 and aggrecan (Acan)-creER27 knock-in alleles and performed similar pulse-chase experiments. Shortly after the chase, Sox9-creER marked populations comparable to those marked by Col2-creER (Supplementary Figure 5a), while Acan-creER marked a larger number of perichondrial cells (Supplementary Figure 5c). After a week of chase, both Sox9creER-P3 and AcancreER-P3 cells increased in the metaphyseal spongiosa and endocortical surface (Figure 6a,b, see also Supplementary Figure 5b,d). We further conducted flow cytometry analysis using mice carrying Col1-GFP, Sox9-creER or Acan-creER and R26R-tomato reporter. A small fraction of cells expressed Col1-GFP shortly after the pulse (Sox9-creER; 3.6±0.8%, Acan-creER; 5.0±2.1%, n=3 per group, data represented as mean ± SD), and the fraction among osteoblasts increased to 55.1±8.2% (Sox9-creER, n=8) and 65.9±8.2% (Acan-creER, n=5, data represented as mean ± SD) after 4 weeks of chase (Figure 6i,j). Both Sox9creER-P3 and AcancreER-P3 cells continued to generate osteoblasts and stromal cells at least for 6 months in a pattern that resembled those of Col2creER-P3 cells (Figure 6c,f). Little to no tamoxifen-independent activities was noted with the use of these knock-in lines (Figure 6g,h).
Figure 6. Sox9-creER+ and Acan-creER+ cells encompass early growth skeletal progenitor cells in postnatal growing bones.
a–f. Lineage-tracing of postnatal Sox9-creER+ and Acan-creER+ cells was performed by injecting 0.1mg tamoxifen into P3 mice. Sox9-creER; R26RTomato (a–c) or Acan-creER; R26RTomato (d–f) mice were chased for a week (a,d), a month (b,e) and 6 months (c,f). Shown are femur sections with the distal side on the left. Sox9creER-P3 represents descendants of Sox9-creER+ cells at P3, and AcancreER-P3 represents descendants of Acan-creER+ cells at P3. White indicates tdTomato. Scale bars: 1mm.
g,h. No tamoxifen controls of Sox9-creER; R26RTomato (g) or Acan-creER; R26RTomato (h) mice at 6 months of age. Shown are femur sections with the distal side on the left. White indicates tdTomato. Scale bars: 1mm.
i,j. Flow cytometry analysis was performed using dissociated bone cells harvested from mice that received 0.1mg tamoxifen at P3 (except no tamoxifen controls) and were chased for indicated periods. CD45− fraction was gated for GFP. (i) Representative dot plots of cells from Col1-GFP; Sox9-creER; R26RTomato (left panels) and Col1-GFP; Acan-creER; R26RTomato (right panels) mice. Left subpanels: 2 days chase, middle subpanels: 4 weeks chase, right subpanels: no tamoxifen controls. X-axis represents tdTomato, and Y-axis represents GFP. (j) Percentage of Sox9creER-P3 cells (blue bars), Col2creER-P3 cells (green bars) or AcancreER-P3 cells (red bars) among Col1-GFP+ osteoblasts after 2 days of chase (left group) or 4 weeks of chase (right group) was enumerated. n=3 mice for 2 days chase of Sox9creER-P3 and AcancreER-P3 cells, n=4 mice for 2 days chase of Col2creER-P3 cells, n=8 mice for 4 weeks chase of Sox9creER-P3 cells, n=5 mice for 4 weeks chase of Col2creER-P3 and AcancreER-P3 cells. All data are represented as mean ± SD.
Discussion
Our data collectively indicated that cells defined by promoter/enhancer activities of genes associated with chondrocytes and their precursors, such as Sox9, Col2 and Acan encompass early mesenchymal progenitors that continue to become chondrocytes, osteoblasts, stromal cells and adipocytes during endochondral bone development. Cre/creER-recombinases driven by the Osx promoter mark a downstream population with limited lineage potential to generate osteoblasts and stromal cells (Supplementary Figure 6, see the diagram). Cells marked by Osx-creER at P3 are not continually generated in the metaphysis, in contrast to cells marked by Sox9-creER, Col2-creER and Acan-creER, but do persist as stromal cells in metaphyseal bone marrow. Any inconsistency of our data with Mizoguchi et al28 is likely due to the length of the chase, because we chased these cells for a longer period of time, for up to 18 months. In addition, when postnatal Col2-creER+ cells generated osteoblasts and stromal cells progressively in the metaphysis, there was no comparable increase of Col2-creER+ descendants among a PαS fraction. We speculate that this latter observation may be relevant to findings recently published by Zhou et al7. These investigators found that, in adult mice, PαS cells overlapped considerably with those marked by leptin receptor (LepR)-cre and that cells marked with the LepR-cre; R26R-tomato progressively marked osteoblasts, but only starting at two months of age. We suggest that the growth-associated progenitors that we have identified here may be the source of osteoblasts during the rapid phase of bone growth before LepR-cre-marked cells provide precursors for the osteoblast lineage. The demonstration of Col2-cre; R26R-tomato mice mark a large fraction of adult osteoblasts and stromal cells, including PαS cells, suggests that the adult mesenchymal precursor cells might derive from the progenitors that we have identified. Transition of these growth-associated progenitors into adult BMSCs appears to be infrequent and requires continuous inputs to make an easily discernable contribution. Further experiments will be required to establish the relationship between the growth-related mesenchymal progenitors identified here and adult mesenchymal precursors. The existence of these two populations provides opportunities for distinct regulation of the explosive growth of bone in early life and the much slower remodeling that occurs in adulthood.
Methods
Mice
Col1(2.3kb)-GFP15, Oc-GFP15, Nestin-GFP29, Cxcl12-GFP/null20, Col2a1-cre13, Osx-cre∷GFP16, Col2a1-creERT225, Osx-creERT22, Nestin-creERT230, Sox9-creERT226, Acan-creERT227, Runx2-LacZ/null19 mice have been described elsewhere. Rosa26-loxP-stop-loxP-tdTomato (R26R-tomato, JAX7914) mice were acquired from Jackson laboratory. All procedures were conducted in compliance with the Guideline for the Care and Use of Laboratory Animals approved by Massachusetts General Hospital’s Institutional Animal Care and Use Committee (IACUC). All mice were housed in a specific pathogen-free condition, and analyzed in a mixed background. Mice were identified by micro-tattooing or ear tags. Tail biopsies of mice were lysed by a HotShot protocol (incubating the tail sample at 95C° for 30 minutes in an alkaline lysis reagent followed by neutralization) and used for PCR-based genotyping (GoTaq Green Master Mix, Promega, and C1000 Touch Cycler, Bio-rad). Perinatal mice were also genotyped fluorescently (BLS miner’s lamp) whenever possible. Mice were sacrificed by over-dosage of carbon dioxide or decapitation under inhalation anesthesia in a drop jar (Aerrane isoflurane, Henry Schein).
For experiments in Figure 1a–f, male mice (Col1-GFP; Col2-cre; R26RtdTomato/tdTomato or Osx-cre∷GFP; R26RtdTomato/tdTomato mice) were mated to female CD1 mice (>8 weeks old, Charles River Laboratories) and the vaginal plug was checked in the morning. For cell proliferation assay, 1mg of 5-ethynyl-2’-deoxyuridine (EdU) (Invitrogen A10044) dissolved in PBS was administered to pregnant mice three hours before sacrificed at indicated embryonic days. Embryos were used for analysis regardless of the sex. At least three embryos of the indicated genotype were examined at each time point shown in the figure.
For experiments in Figure 1g, male mice (Col1-GFP; Col2-cre; R26RtdTomato/tdTomato; Runx2+/−) were mated to female Runx2+/− mice. Embryos were used for analysis regardless of the sex. At least three embryos of the indicated genotype were examined at the time point shown in the figure.
For experiments in Figure 1h–m, male mice (Col1-GFP; Col2-cre; R26RtdTomato/tdTomato or Oc-GFP; Col2-cre; R26RtdTomato/tdTomato or Cxcl12-GFP; Col2-cre; R26RtdTomato/tdTomato or Col1-GFP; Osx-cre∷GFP; R26RtdTomato/tdTomato or Oc-GFP; Osx-cre∷GFP; R26RtdTomato/tdTomato or Cxcl12-GFP; Osx-cre∷GFP; R26RtdTomato/tdTomato) were mated to female CD1 mice. Pups at postnatal day 3 were used for analysis regardless of the sex. Three pups of the indicated genotype were examined at the time point shown in the figure. n=3 mice per group.
For experiments in Figure 2, male mice (Col1-GFP; Col2-creER; R26RtdTomato/tdTomato or Col2-creER; R26RtdTomato/tdTomato or Osx-creER; R26RtdTomato/tdTomato) were mated to female CD1 mice and the vaginal plug was checked in the morning. Pregnant mice received 1mg tamoxifen (Sigma T5648) and progesterone (Sigma P3972) intraperitoneally at an indicated embryonic day. Embryos or pups were used for analysis regardless of the sex. At least three embryos or pups of the indicated genotype were examined at the time point shown in the figure.
For experiments in Figure 3 and Figure 4, male mice (Col2-creER; R26RtdTomato/tdTomato or Col1-GFP; Col2-creER; R26RtdTomato/tdTomato or Cxcl12-GFP; Col2-creER; R26RtdTomato/tdTomato or Osx-creER; R26RtdTomato/tdTomato or Col1-GFP; Osx-creER; R26RtdTomato/tdTomato or Cxcl12-GFP; Osx-creER; R26RtdTomato/tdTomato) were mated to female FVB/N mice (Charles River Laboratories) to generate pups. Three day-old mice received 0.1mg of tamoxifen intraperitoneally using a method reported previously31. Mice were used for analysis regardless of the sex. At least three mice of the indicated genotype were examined at the time point shown in the figure. For no tamoxifen controls, the same amount of sunflower oil (without tamoxifen) was injected into three day-old mice. For experiments shown in Figure 4i and 4j, the number of mice used to plot the data was as following: Col1-GFP; Col2-creER; R26RTomato; n=4 (2 days chase), n=3 (1 week chase), n=5 (2 weeks chase), n=3 (3 weeks chase) and n=6 (4 weeks chase) mice; Cxcl12-GFP; Col2-creER; R26RTomato; n=3 (2 days chase), n=4 (1 week chase), n=4 (2 weeks chase), n=4 (3 weeks chase) and n=4 (4 weeks chase) mice; Col1-GFP; Osx-creER; R26RTomato; n=4 (2 days chase), n=7 (1 week chase) and n=3 (4 weeks chase) mice; Cxcl12-GFP; Osx-creER; R26RTomato; n=3 (2 days chase), n=3 (2 weeks chase) and n=4 (4 weeks chase) mice.
For experiments in Figure 5a,b, male mice (Col2-cre; R26RtdTomato/tdTomato or Osx-cre∷GFP; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Mice were used for analysis regardless of the sex. Four mice of the indicated genotype were examined.
For experiments in Figure 5c, male mice (Col2-creER; R26RtdTomato/tdTomato or Osx-creER; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Three day-old mice received 0.1mg of tamoxifen intraperitoneally. Mice were used for analysis regardless of the sex. Four mice of the indicated genotype were examined.
For experiments in Figure 5d,e,h,i male mice (Cxcl12-GFP; Col2-cre; R26RtdTomato/tdTomato or Cxcl12-GFP; Osx-cre∷GFP; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate mice. Mice were used for analysis regardless of the sex. Four mice of the indicated genotype were examined.
For experiments in Figure 5f,g,j,k, male mice (Nes-GFP; Nes-creER; R26RtdTomato/tdTomato or Cxcl12-GFP; Col2-creER; R26RtdTomato/tdTomato or Cxcl12-GFP; Osx-creER; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Three day-old mice received 0.1mg of tamoxifen intraperitoneally. Mice were used for analysis regardless of the sex. For experiments shown in Figure 5g,k, the number of mice used to plot the data was as following: Nes-creER; R26RTomato; n=5 (2 days chase), n=3 (2 weeks chase) and n=4 (4 weeks chase) mice; Col2-creER; R26RTomato; n=4 (2 days chase), n=5 (1 week chase), n=7 (2 weeks chase), n=5 (3 weeks chase) and n=9 (4 weeks chase) mice; Osx-creER; R26RTomato; n=6 (2 days chase), n=5 (2 weeks chase) and n=8 (4 weeks chase) mice.
For experiments in Figure 6, male mice (Sox9-creER; R26RtdTomato/tdTomato or Col1-GFP; Sox9-creER; R26RtdTomato/tdTomato or Acan-creER; R26RtdTomato/tdTomato or Col1-GFP; Acan-creER; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Three day-old mice received 0.1mg of tamoxifen intraperitoneally. Mice were used for analysis regardless of the sex. At least three mice of the indicated genotype were examined at the time point shown in the figure. For no tamoxifen controls, the same amount of sunflower oil (without tamoxifen) was injected into three day-old mice. For experiments shown in Figure 6j, the number of mice used to plot the data was as following: Sox9-creER; R26RTomato; n=3 (2 days chase) and n=8 (4 weeks chase) mice; Col2-creER; R26RTomato; n=4 (2 days chase) and n=5 (4 weeks chase) mice; Acan-creER; R26RTomato; n=3 (2 days chase) and n=5 (4 weeks chase) mice.
For experiments in Supplementary Figure 4a, male mice (Nes-GFP; Col2-cre; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Mice were used for analysis regardless of the sex. Five mice of the indicated genotype were examined.
For experiments in Supplementary Figure 4b, male mice (Cxcl12-GFP; Col2-creER; R26RtdTomato/tdTomato) were mated to female CD1 mice and the vaginal plug was checked in the morning. Pregnant mice received 1mg tamoxifen and progesterone intraperitoneally at E13.5. Pups were used for analysis regardless of the sex. One pup of the indicated genotype was examined at P7.
For experiments in Supplementary Figure 4c, male mice (Cxcl12-GFP; Nes-creER; R26RtdTomato/tdTomato) were mated to female FVB/N mice to generate pups. Three day-old mice received 0.1mg of tamoxifen intraperitoneally. Mice were used for analysis regardless of the sex. At least three mice of the indicated genotype were examined at the time point shown in the figure.
Tamoxifen
Tamoxifen (Sigma T5648) was mixed with 100% ethanol until completely dissolved. Subsequently, a proper volume of sunflower seed oil (Sigma S5007) was added to the tamoxifen-ethanol mixture and rigorously mixed. The tamoxifen-ethanol-oil mixture was incubated at 60C° in a chemical hood until ethanol evaporated completely. The tamoxifen-oil mixture was stored at room temperature until use.
Histology
Samples were dissected under a stereomicroscope (Nikon SMZ-10A) to remove soft tissues, and fixed in 4% paraformaldehyde, overnight at 4C°, then decalcified in 15% EDTA for a proper period, typically ranging from 1 to 14 days. Decalcified samples were cryoprotected in 30% sucrose/PBS solutions then in 30% sucrose/PBS:OCT (1:1) solutions, each overnight at 4C°. Samples were embedded in an OCT compound (TissuTek, Sakura) under a stereomicroscope and transferred on a sheet of dry ice to solidify the compound. Embedded samples were cryosectioned at 15µm using a cryostat (Leica CM1850). Images were captured with a widefield fluorescence microscope (Nikon Eclipse E800) with prefigured triple-band filter settings for DAPI/FITC/TRITC, and merged with Spot Advanced Software (Spot Imaging) or an automated fluorescent microscope with a whole-slide scanning platform (TissueFAXS, TissueGnostics). Confocal images were acquired using LSM510 and Zen2009 software (Zeiss) with lasers and corresponding band-pass filters for DAPI (Ex.405nm, BP420-480), GFP (Ex.488nm, BP505-530), tdTomato (Ex.543nm, BP565-595) and Alexa633 (Ex.633nm LP650). LSM Image Viewer and Adobe Photoshop software were used to capture and align images. Representative images of at least three independent biological samples were shown in the figures.
Flow cytometry
Femurs, tibias, iliac crests and humeri were carefully dissected and gently crushed in 5ml Ca2+, Mg2+-free HBSS (Sigma) with a pestle and a mortar (Coors Tek), then supernatants were filtered through a 70µm cell strainer (Fisher) into a 50ml tube on ice. Bone fragments were crushed for two additional times in 5ml HBSS and supernatants were filtered into the same tube. Tissue remnants were incubated with 2 Wunsch units of Liberase TM (Roche) at 37°C for 45 minutes on a shaking incubator (Thermomixer, Eppendorf). 0.25% trypsin-EDTA (Gibco) was added for experiments to harvest osteoblasts. Cells were mechanically triturated using an 18-gauge needle and a 1ml syringe (BD) and filtered into the same tube. Cells were pelleted, resuspended and layered over Ficoll-Paque PLUS (GE Healthcare) to collect low-density cell fractions. Cells were stained with anti-mouse CD45-APC, CD45-eFlour 450, Ter119-eFlour 450, Sca1-Alexa Flour 700 (1:500, eBioscicence), PDGFRα (CD140a)-APC (1:250, eBioscience) or their isotype controls (1:500, eBioscience) in DPBS/2%FBS on ice for 30 min. Flow cytometry was performed using a four-laser BD LSRII (Ex.355/407/488/633nm) and FACSDiva, and analyzed on FlowJo (TreeStar). Representative plots of at least three independent biological samples were shown in the figures.
Immunohistochemisty
Cryosections were stored at −20C° freezers until use. Sections were postfixed in 4% paraformaldehyde for 15 minutes, blocked with 2% BSA/TBST for 30 minutes and incubated with rat anti-CD31 monoclonal antibody (1:100, AbD Serotec MCA2388), overnight at 4°C, and subsequently with Alexa Fluor 633-conjugated goat anti-rat IgG (1:400, Invitrogen A21087) for 3 hours at 4°C. For lipid staining, cryosections were gently rinsed with TBS and incubated with LipidTOX Deep Red (1:200, Invitrogen H34477) for 30 minutes at room temperature. Sections were further incubated with DAPI (4',6-diamidino-2-phenylindole, Invitrogen D1306) to stain nuclei. Stained samples were cover-slipped and mounted in a mounting medium for fluorescence imaging (Vectashield H-1000, Vector Labs). The edge of the cover slip was coated with commercially available transparent nail polish.
Cell proliferation assay
To evaluate cell proliferation, 1mg of 5-ethynyl-2’-deoxyuridine (EdU) (Invitrogen A10044) dissolved in PBS was administered to pregnant mice three hours before sacrificed at indicated embryonic days. Click-iT Imaging Kit (Invitrogen, C10337) with Alexa Flour 647-azide (Invitrogen A10277) was used to detect EdU in cryosections.
Colony-forming unit fibroblast (CFU-F) assay
Bone marrow cells were harvested as described in the Flow cytometry section. 106 bone marrow nucleated cells were plated into a 9.6cm2 glass bottom chamber slide (Lab-Tek II, Nunc) and cultured in MesenCult Proliferation medium for mouse cells (StemCell technologies) for 10 days. Cells were fixed in 4% paraformaldehyde, for 30 minutes at 4C° and counterstained with Wheat Germ Agglutinin Alexa Flour 488 conjugate (Invitrogen W11261) for 15 minutes. Representative images from at least three independent biological samples were shown in the figures.
Statistical Analysis
Results were represented as mean values ± S.D. Statistical evaluation was conducted using the Mann-Whitney’s U-test. A P-value of <0.05 was considered significant. No statistical method was used to predetermine sample size. Sample size was determined based on previous literature and our prior experience that give sufficient standard deviations of the mean so as not to miss a biologically important difference between groups. The experiments were not randomized. All the available mice of the desired genotypes were used for experiments. The investigators were not blinded to during experiments and outcome assessment. One femur from each mouse was arbitrarily chosen for histological analysis. Genotypes were not particularly highlighted during quantification.
Supplementary Material
Acknowledgments
The authors thank David Rowe for Col2.3-GFP and Oc-GFP mice and Susan Mackem for Col2a1-creERT2 mice, Haruhiko Akiyama and Benoit de Crombrugghe for Sox9-creERT2 and Acan-creERT2 mice, Laura Prickett, Kat Folz-Donahue and Meredith Weglarz of Harvard Stem Cell Institute/Massachusetts General Hospital flow cytometry core and Thomas Diefenbach of Ragon Institute imaging core for their expert assistance. This work was supported by National Institutes of Health Grants DE022564 to N.O. and DK056246 to H.M.K.
Footnotes
Author Contributions
NO and HMK conceived the project and designed the experiments. TN provided mice. NO and WO performed the experiments. NO and WO analyzed the data. NO and HMK wrote the manuscript; TN critiqued the manuscript.
Disclosures
The authors have declared that no conflict of interest exists.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Maes C, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama H, et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenette PS, Pinho S, Lucas D, Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu. Rev. Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- 5.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morikawa S, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-Receptor-Expressing Mesenchymal Stromal Cells Represent the Main Source of Bone Formed by Adult Bone Marrow. Cell. Stem Cell. 2014 doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacchetti B, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, et al. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Hum. Mol. Genet. 2011;20:3910–3924. doi: 10.1093/hmg/ddr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev. Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford-Hutchinson AF, et al. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J. Bone Miner. Res. 2007;22:1245–1259. doi: 10.1359/jbmr.070420. [DOI] [PubMed] [Google Scholar]
- 12.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev. Biol. 2007;308:93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- 14.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalajzic Z, et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–660. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 16.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 18.Ono N, et al. Vasculature-associated cells expressing nestin in developing bones encompass early cells in the osteoblast and endothelial lineage. Dev. Cell. 2014;29:330–339. doi: 10.1016/j.devcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 20.Ara T, et al. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- 21.Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, et al. Osterix-cre labeled progenitor cells contribute to the formation and maintenance of the bone marrow stroma. PLoS One. 2013;8:e71318. doi: 10.1371/journal.pone.0071318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, et al. Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One. 2014;9:e85161. doi: 10.1371/journal.pone.0085161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- 26.Soeda T, et al. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry SP, et al. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47:805–814. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizoguchi T, et al. Osterix Marks Distinct Waves of Primitive and Definitive Stromal Progenitors during Bone Marrow Development. Dev. Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignone JL, Kukekov V, Chiang AS, Steindler D, Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol. 2004;469:311–324. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- 30.Balordi F, Fishell G. Mosaic removal of hedgehog signaling in the adult SVZ reveals that the residual wild-type stem cells have a limited capacity for self-renewal. J. Neurosci. 2007;27:14248–14259. doi: 10.1523/JNEUROSCI.4531-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.