Abstract
The KCNH2 gene encodes the Kv11.1 potassium channel that conducts the rapidly activating delayed rectifier current in the heart. KCNH2 pre-mRNA undergoes alternative processing; intron 9 splicing leads to the formation of a functional, full-length Kv11.1a isoform, while polyadenylation within intron 9 generates a non-functional, C-terminally truncated Kv11.1a-USO isoform. The relative expression of Kv11.1 isoforms plays an important role in the regulation of Kv11.1 channel function and the pathogenesis of long QT syndrome. In this study, we identified cis-acting elements that are required for KCNH2 intron 9 poly(A) signal activity. Mutation of these elements decreased Kv11.1a-USO expression and increased the expression of Kv11.1a mRNA, protein and channel current. More importantly, blocking these elements by antisense morpholino oligonucleotides shifted the alternative processing of KCNH2 intron 9 from the polyadenylation to the splicing pathway, leading to the predominant production of Kv11.1a and a significant increase in Kv11.1 current. Our findings indicate that the expression of the Kv11.1a isoform can be upregulated by an antisense approach. Antisense inhibition of KCNH2 intronic polyadenylation represents a novel approach to increase Kv11.1 channel function.
Keywords: potassium channels, hERG, alternative polyadenylation, morpholino, long QT syndrome
1. Introduction
KCNH2 or human ether-a-go-go-related gene 1 (hERG1) encodes the Kv11.1 channel that conducts the rapidly activating delayed rectifier K+ current (IKr) in the heart [1–4]. Kv11.1 channels are essential for cardiac action potential repolarization and mutations in KCNH2 cause long QT syndrome type 2 (LQT2) [5]. Alternative intronic polyadenylation has been shown to direct the expression of two Kv11.1 C-terminal isoforms, the functional Kv11.1a isoform and the non-functional Kv11.1a-USO isoform [6]. Kv11.1a is produced by splicing from exon 9 to exon 10 and use of a distal poly(A) site in exon 15, whereas Kv11.1a-USO is generated by the activation of a proximal poly(A) site within intron 9. The last 359 amino acids of Kv11.1a are absent in Kv11.1a-USO and the truncated isoform fails to form functional channels when expressed in mammalian cells [7–9]. We recently reported a novel LQT2 mutation that disrupted the alternative processing of KCNH2 intron 9 and resulted in switching the expression of Kv11.1 isoforms from Kv11.1a to Kv11.1a-USO [10]. Thus, the relative expression of Kv11.1a and Kv11.1a-USO isoforms plays an important role in the regulation of Kv11.1 channel function and the pathogenesis of LQT2.
The alternative processing of KCNH2 pre-mRNA is regulated by the relative efficiencies of RNA splicing and polyadenylation events. These events depend on interactions between transacting splicing and polyadenylation factors and cis-acting elements present in KCNH2. The poly(A) signal within KCNH2 intron 9 consists of a weak, noncanonical hexamer, AGUAAA [6]. When this poly(A) signal is changed to the strong, canonical poly(A) signal, AAUAAA, polyadenylation becomes the dominant reaction, resulting in the predominant expression of Kv11.1a-USO. The elimination of the intron 9 poly(A) signal by the AGUAAA to CGCAAA mutations results in predominant expression of Kv11.1a and an increase in channel current. Therefore, it is possible to modulate the relative expression of Kv11.1a and Kv11.1a-USO by manipulating the strength and the usage of the intron 9 poly(A) site.
In this study, we identified cis-acting elements that are required for the intron 9 poly(A) signal activity. We then tested the hypothesis that inhibition of these elements using antisense morpholino oligonucleotides (MO) would decrease Kv11.1a-USO expression, allowing a given transcript to be processed to the functional Kv11.1a isoform. Our findings indicate that the expression of the functional Kv11.1a isoform can be upregulated by antisense inhibition of KCNH2 intronic polyadenylation.
2. Materials and Methods
2.1 Generation of the minigene luciferase reporter construct
The minigene luciferase reporter construct was generated by subcloning the Renilla luciferase gene downstream of a splicing competent KCNH2 minigene composed of KCNH2 genomic DNA from exon 8 to exon 11. The construction of the KCNH2 minigene has been previously described [11]. The N-terminus of the minigene was tagged by the Myc epitope, which was inserted in-frame with the KCNH2 and luciferase translation sequence. Expression of the minigene luciferase reporter is driven by a CMV promoter. The vector also contains the firefly luciferase gene driven by the SV40 promoter, which was used as a control for transfection efficiency. The deletion and mutations of U/GU-rich elements downstream of the KCNH2 intron 9 poly(A) signal were performed using the pAlter in vitro mutagenesis system (Promega, Madison, WI). HEK293 cells were transiently transfected with the minigene luciferase reporter construct using the Effectene method (Qiagen, Valencia, CA). After 24 h, cells were harvested and assayed for both firefly and Renilla luciferase activity using the Dual-Luciferase assay kit (Promega). Data were analyzed by normalizing Renilla luciferase activity to firefly luciferase activity and presented as mean ± SEM.
2.2 Generation of the short KCNH2 gene and stable transfection in Flp-In HEK293 cells
The generation of a short KCNH2 gene construct, in which the two longest introns, intron 2 (14.9 kb) and intron 5 (4.4 kb), were shortened to 600 bp has been previously described [10]. Mutations were introduced into the short KCNH2 gene by the pAlter in vitro mutagenesis system (Promega). Stably transfected Flp-In HEK293 cells were generated by the co-transfection of the KCNH2 gene constructs (0.1 μg) with the Flp recombinase expression vector pOG44 (0.9 μg) using the Effectene method and selected with 100 μg/ml hygromycin.
2.3 Generation of the tandem KCNH2 poly(A) signal construct
The generation of the tandem poly(A) signal construct was previously described [6]. The construct contained the SV40 promoter, the firefly luciferase gene, and 308 bp of KCNH2 intron 9 followed by a synthetic poly(A) signal. HEK293 cells were transiently transfected with the tandem poly(A) construct as previously described [6].
2.4 Antisense MO treatment
MOs were synthesized by Gene Tools (Philomath, OR). The KCNH2 antisense MO was designed to target a 25 nt sequence within KCNH2 intron 9 containing U/GU-rich elements (underlined) essential for the activation of the intron 9 poly(A) signal, 5′-CAGAACACAGTAGTGAATCAAAACC-3′. The invert MO with the same sequence but in a reverse orientation was used as a control. The Endo-Porter delivery system (Gene Tools) was used to deliver antisense and invert MOs into the cells.
2.5 RNase protection assay
RNA isolation and the RNase protection assay (RPA) were performed as previously described [6]. Briefly, antisense RNA riboprobes were transcribed in vitro in the presence of biotin-14-CTP. Yeast RNA was used as a control for the complete digestion of the probes by RNase. The relative intensity of each band was quantified using ImageJ software and adjusted for the number of biotin-labeled cytidines in each protected fragment. The expression level of the hygromycin B resistance gene from the KCNH2 gene constructs was used to normalize the relative expression of Kv11.1 isoforms.
2.6 Immunoblot analysis
Immunoblot analysis was performed as previously described [10]. The cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and then electrophoretically transferred onto nitrocellulose membranes. The membranes were incubated with an anti-Kv11.1 antibody against the N-terminus of Kv11.1a and Kv11.1a-USO proteins (H-175, Santa Cruz, Santa Cruz, CA) at a 1:600 dilution and visualized with the ECL detection kit (Amersham, Piscataway, NJ). The expression level of hygromycin B phosphotransferase (HPH) encoded by hygromycin B resistance gene was used as loading control [6].
2.7 Patch-clamp recordings
Membrane currents were recorded in whole cell configuration using suction pipettes as previously described [4]. The bath solution contained (in mM) 137 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES (pH 7.4 with NaOH). The pipette solution contained (in mM) 130 KCl, 1 MgCl2, 5 EGTA, 5 MgATP, and 10 HEPES (pH 7.2 with KOH). All patch-clamp experiments were performed at 22–23°C. Kv11.1 current was activated by depolarizing steps between −70 and +50 mV from a holding potential of −80 mV and Kv11.1 tail current was recorded following repolarization to −50 mV. The patch-clamp data are presented as mean ± SEM and analyzed by Student’s t-test. P<0.05 is considered statistically significant.
3. Results
3.1 Intron 9 poly(A) signal downstream elements are required for Kv11.1 alternative polyadenylation
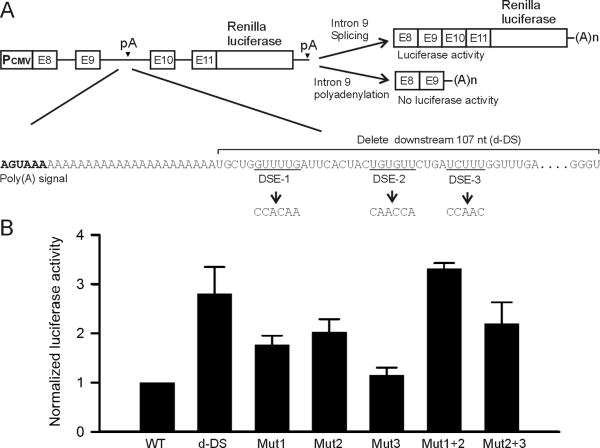
Polyadenylation sites are primarily defined by a hexameric poly(A) signal AAUAAA or other close variants [12]. In addition to the poly(A) signal, the cis-acting, U/GU-rich downstream element (DSE) is also required for the formation of the poly(A) tail. We have reported that alternative polyadenylation within KCNH2 intron 9 is directed by a noncanonical poly(A) signal AGUAAA [6]. To identify cis-acting elements required for intron 9 polyadenylation, we designed a reporter construct by subcloning the Renilla luciferase gene downstream of a splicing competent minigene composed of KCNH2 genomic DNA from exon 8 to exon 11. In this KCNH2 reporter construct, the splicing of intron 9 generated active luciferase and polyadenylation of intron 9 resulted in no luciferase activity (Fig. 1A). We first deleted 107 nt downstream of the poly(A) signal in the minigene luciferase reporter construct. The deletion significantly increased the luciferase activity, suggesting that the downstream region contains elements that are required for polyadenylation (Fig. 1B, d-DS). Analysis of the intron 9 sequence revealed several putative U/GU-rich elements that we refer to as DSE-1, DSE-2 and DSE-3 (Fig. 1A). We mutated DSE-1 from GUUUUG to CCACAA (Mut1), DSE-2 from UGUGUU to CAACCA (Mut2) and DSE-3 from UCUUU to CCAAC (Mut3) in the minigene luciferase reporter construct. Mut1 and Mut2, but not Mut3, significantly increased the luciferase activity (Fig. 1B). When both DSE-1 and DSE-2 were mutated (Mut1+2), the luciferase activity was further increased compared to Mut1 and Mut2 alone. When both DSE-2 and DSE-3 were mutated (Mut2+3), the luciferase activity was increased, but similar to Mut2 alone. Together, these results suggest that DSE-1 and DSE-2 play a key role in KCNH2 intron 9 polyadenylation.
Fig. 1.
Identification of cis-acting elements required for intron 9 polyadenylation using a luciferase reporter construct. (A) Diagram of the KCNH2 minigene luciferase reporter construct and sequence of downstream elements of the KCNH2 intron 9 poly(A) signal. Downstream element deletion and mutations are indicated. (B) Histogram showing the effect of downstream element deletion and mutations on luciferase activity (n=3–6).
3.2 Regulation of Kv11.1 isoform expression by intron 9 poly(A) signal downstream elements
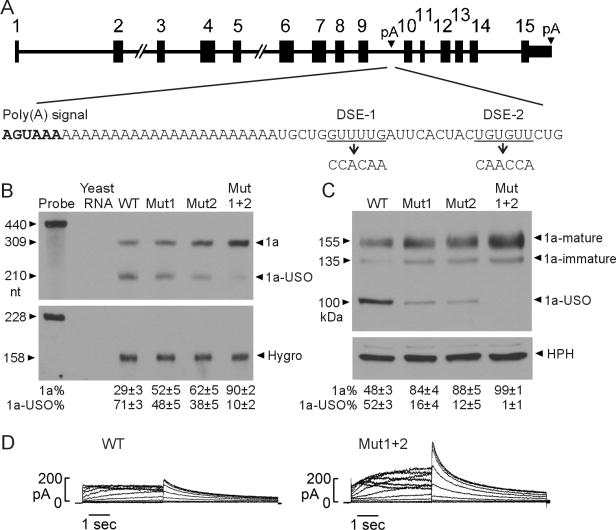
To test the effects of the downstream elements in the regulation of Kv11.1 isoform expression, we introduced the DSE-1 and DES-2 mutations into the short KCNH2 gene construct (Fig. 2A) [10]. Wild-type (WT) and mutant short gene constructs were stably transfected into Flp-In HEK293 cells. The Mut1 and Mut2 mutations increased Kv11.1a expression and decreased Kv11.1a-USO expression in RPA and immunoblot analyses (Fig. 2B, 2C). When both DSE-1 and DSE-2 were mutated (Mut1+2) Kv11.1a was predominantly expressed. Patch-clamp studies showed that Kv11.1 channel current was significantly increased in Mut1+2 (Fig. 2D). The maximum tail current densities of WT and Mut1+2 were 7.0 ± 0.6 pA/pF (n=9) and 17.8 ± 1.7 pA/pF (n=10, P<0.001), respectively. These results suggest that DSE-1 and DSE-2 are important downstream elements of the intron 9 poly(A) signal, and that disruption of these elements leads to a shift in KCNH2 pre-mRNA processing toward the production of the Kv11.1a isoform and an increase in Kv11.1 channel function.
Fig. 2.
Effect of DSE mutations on Kv11.1 isoform expression using the short KCNH2 gene construct. (A) The structure of the short KCNH2 gene construct and sequence of downstream elements of the KCNH2 intron 9 poly(A) signal. DSE-1 and DSE-2 mutations are indicated. (B) RPA analysis of mRNA in Flp-In HEK293 cells stably expressing WT and DSE mutant short KCNH2 genes. Signals of Kv11.1 isoforms were quantified and shown as a percentage of the total signal (1a+1a-USO) (mean±SD, n=3). (C) Immunoblot analysis of Kv11.1 protein expressed in cells expressing WT and DSE mutant short KCNH2 genes. The expression level of hygromycin B phosphotransferase (HPH) encoded by hygromycin B resistant gene served as a loading control. Signals were quantified and shown as isoform percentage of total (1a+1a-USO) Kv11.1 protein (mean±SD, n=3). (D) Representative currents recorded from cells stably expressing WT and Mut1+2 short KCNH2 genes.
3.3 Antisense MO inhibition of the KCNH2 intron 9 polyadenylation
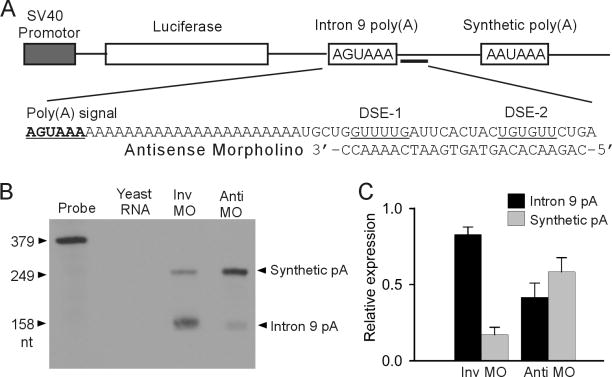
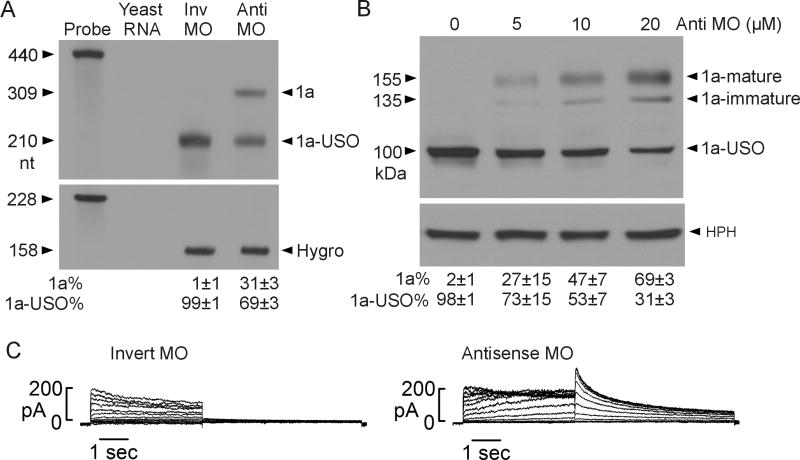
Because DSE-1 and DSE-2 are important for KCNH2 intron 9 polyadenylation, we reasoned that blocking these elements using an antisense approach would inhibit KCNH2 intron 9 polyadenylation and lead to the upregulation of the functional Kv11.1a isoform. We designed a 25-mer antisense MO complementary to the KCNH2 intron 9 downstream elements including DSE-1 and DSE-2 (Fig. 3A). To determine whether the antisense MO can inhibit the poly(A) signal in intron 9, we performed competition assay using a tandem poly(A) signal construct as previously described [6]. The KCNH2 intron 9 poly(A) signal AGUAAA and flanking sequences (−130/+172 nt) were positioned upstream of a relatively strong synthetic poly(A) signal (Fig. 3A). We treated the cells expressing the tandem poly(A) construct with 5 μM invert or antisense MO and performed RPA analysis using a probe specific to 249 nt of KCNH2 intron 9 [6]. This probe will generate a 158 nt fragment if the intron 9 poly(A) signal is used and a 249 nt fragment if the synthetic poly(A) signal is utilized. In the presence of the invert MO, the KCNH2 intron 9 poly(A) signal was predominantly used (Fig. 3B). In the presence of the antisense MO against DSE-1 and DSE-2, the transcription was predominantly terminated at the synthetic poly(A) signal. Treatment with antisense MO resulted in a decreased usage of the intron 9 poly(A) site from 83% to 41% and an increased usage of the synthetic poly(A) site from 17% to 59% (n=3, Fig. 3C). These results indicate that antisense MO treatment inhibits the poly(A) signal in KCNH2 intron 9.
Fig. 3.
Inhibition of the KCNH2 intron 9 poly(A) site by antisense MO. (A) Diagram of the tandem poly(A) signal construct and the sequence of the antisense MO. (B) RPA analysis of relative usage of intron 9 poly(A) signal and synthetic poly(A) signal following the treatment with invert or antisense MO. (C) Quantitative analysis of RPA results (mean±SD, n=3).
3.4 Modulation of Kv11.1 isoform expression by antisense MO
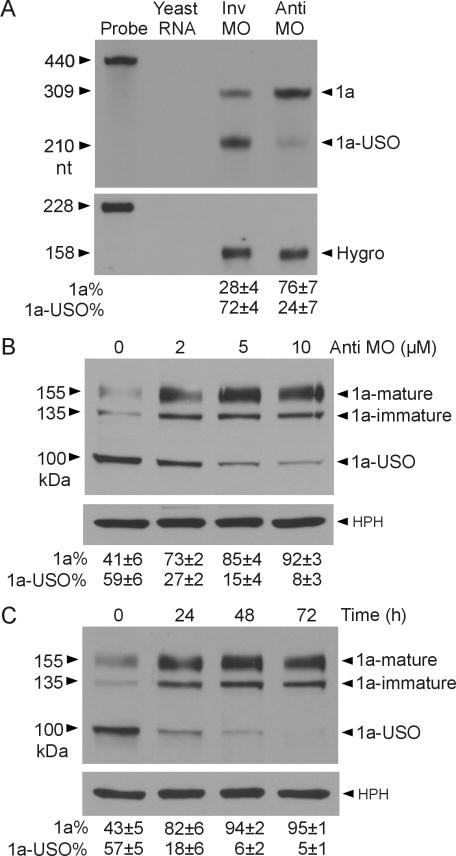
To test whether the inhibition of intron 9 polyadenylation by MO is able to modulate the relative expression of Kv11.1 isoforms, we treated Flp-In HEK293 stably expressing short KCNH2 gene with antisense MO against DSE-1 and DSE-2. RPA analysis showed that treatment with 5 μM antisense MO for 48 h resulted in an increase in the Kv11.1a transcript and a decrease in the Kv11.1a-USO transcript compared to the control invert MO (Fig. 4A). This result suggests that the relative expression of Kv11.1 C-terminal isoforms can be modulated by an antisense approach that targets the downstream elements of intron 9 poly(A) signal. To determine whether antisense MO-induced modulation of Kv11.1 C-terminal isoform expression leads to the isoform switch at the protein level, we analyzed Kv11.1 protein expression by immunoblot. Flp-In HEK293 cells stably expressing short KCNH2 gene were treated with different concentrations of antisense MO for 48 h or with 5 μM for different times (Fig. 4B, 4C). Treatment with antisense MO significantly increased the level of Kv11.1a protein and decreased Kv11.1a-USO protein levels. Antisense MO treatment exhibited concentration and time dependence. The level of Kv11.1a protein increased with 2 μM antisense MO and reached a maximum at 10 μM antisense MO (Fig. 4B). The increase of the Kv11.1a protein level was observed by 24 h and reached a maximum by 72 h following antisense MO treatment (Fig. 4C).
Fig. 4.
Modulation of Kv11.1 isoform expression by antisense MO. (A) RPA analysis of the effect of antisense MO on Kv11.1 isoform expression. Signals were quantified and shown as an isoform percentage of the total signal (1a+1a-USO, mean±SD, n=3). (B) Immunoblot analysis showing concentration dependence of antisense MO effect. Signals were quantified and shown as an isoform percentage of total (1a+1a-USO) Kv11.1 protein (mean±SD, n=3). (C) Time course of antisense MO effect. Signals were quantified and shown as an isoform percentage of total (1a+1a-USO) Kv11.1 protein (mean±SD, n=3).
3.5 Antisense MO treatment increased Kv11.1 channel current
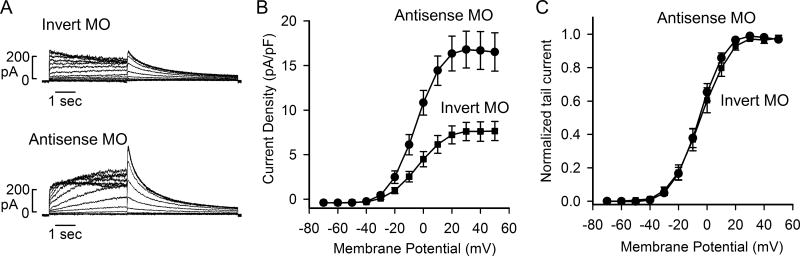
To study the functional consequence of antisense MO-induced isoform switch, we performed patch-clamp recordings of Kv11.1 channel current. Cells stably expressing the short KCNH2 gene were treated with 10 μM antisense or invert MO for 48 h. Treatment with antisense MO significantly increased Kv11.1 current compared to the invert MO control (Fig. 5). The maximum tail current densities in invert and antisense MO-treated cells were 7.6 ± 1.0 pA/pF (n=8) and 16.8 ± 2.1 pA/pF (n=9, p<0.001, Fig. 5B), respectively. The voltage dependence of Kv11.1 channel activation was determined by fitting the normalized tail currents with a Boltzmann function (Fig. 5C). The half maximal activation voltages (V1/2) for control and treatment with antisense MO were −4.2 ± 2.9 mV and −5.6 ± 2.0 mV, respectively, and the slope factor (k) for control and treatment with antisense MO were 8.7 ± 0.2 and 7.9 ± 0.3, respectively. These patch-clamp experiments demonstrated that inhibition of polyadenylation of intron 9 using antisense MO can increase the functional expression of Kv11.1 channel current.
Fig. 5.
Effect of antisense MO on Kv11.1 channel current. (A) Representative currents recorded from Flp-In HEK293 cells stably expressing short KCNH2 gene following treatment with 10 μM invert or antisense MO for 48 h. (B) I–V plot of tail current density measured at −50 mV following test voltages from −70 to +50 mV for invert (■) and antisense (●) MO. (C) Activation curves for invert (■) and antisense (●) MO treatment.
3.6 Effect of antisense MO on Kv11.1 isoform expression in canonical poly(A) signal construct
We have previously shown that the poly(A) signal in KCNH2 intron 9 is intrinsically weak due to the presence of the noncanonical hexamer AGUAAA. When the intron 9 poly(A) signal AGUAAA was changed to the canonical poly(A) signal AAUAAA, the Kv11.1a-USO isoform was predominantly expressed [6]. To test whether the antisense MO is able to mediate the switch of the expression from Kv11.1a-USO to the Kv11.1a isoform in the presence of a strong poly(A) signal, we treated HEK293 cells stably expressing the short KCNH2 gene construct containing the canonical poly(A) signal with inverted or antisense MO. As shown in RPA analysis (Fig. 6A), in the presence of the inverted MO, Kv11.1a-USO was predominantly expressed with no obvious expression of Kv11.1a mRNA. When treated with 10 μM antisense MO, the expression of the Kv11.1a isoform was significantly increased. Immunoblot analysis revealed that the expression of the Kv11.1a isoform increased with the increased antisense MO concentrations (Fig. 6B). Patch clamp studies showed that treatment with 10 μM antisense MO significantly increased Kv11.1 current compared to the invert MO control (Fig. 6C). The maximum tail current densities in invert and antisense MO-treated cells were 1.3 ± 0.3 pA/pF (n=8) and 11.5 ± 1.4 pA/pF (n=9, p<0.001), respectively. These results indicate that antisense MO can mediate Kv11.1 isoform switch in the presence of a strong intron 9 poly(A) signal. However, the relative efficiency of Kv11.1 isoform switch is markedly reduced compared with the weak, non-canonical poly(A) signal.
Fig. 6.
Effect of antisense MO on Kv11.1 isoform expression in in canonical poly(A) signal construct. (A) RPA analysis of mRNA from cells stably expressing the short KCNH2 gene containing the canonical poly(A) signal following the treatment with 10 μM invert or antisense MO. Signals were quantified and shown as an isoform percentage of the total signal (1a+1a-USO, n=3). (B) Immunoblot analysis showing concentration dependence of antisense MO effect. Signals were quantified and shown as an isoform percentage of total (1a+1a-USO) Kv11.1 protein (mean±SD, n=3). (C) Representative currents recorded from Flp-In HEK293 cells stably expressing short KCNH2 gene containing the canonical poly(A) signal following treatment with 10 μM invert or antisense MO for 48 h.
4. Discussion
In the present study, we identified two downstream U/GU-rich elements important for the intron 9 poly(A) signal activity. Mutations of these elements resulted in the predominant production of Kv11.1a and a marked increase in channel current. Furthermore, we found that blocking these elements by antisense MO decreased the relative expression of the non-functional Kv11.1a-USO isoform and increased the expression of the functional Kv11.1a isoform. These results suggest that the relative expression of Kv11.1 C-terminal isoforms can be modulated by inhibition of the intron 9 poly(A) signal with an antisense approach. Our findings highlight antisense MO-mediated Kv11.1 isoform switch as a potential therapeutic approach for LQT2.
The KCNH2 intron 9 poly(A) site consists of a relatively weak, noncanonical hexamer, AGUAAA. In addition to the noncanonical hexamer, the KCNH2 intron 9 poly(A) site also contains a 22 nt “A” stretch immediately following the AGUAAA hexamer. It has been reported that many human noncanonical poly(A) sites contain A-rich sequences and have a higher frequency of U and GU nucleotides in the downstream region compared with canonical poly(A) sites [13]. These U/GU-rich downstream elements are critical for the efficient polyadenylation of noncanonical poly(A) signals. In support of this notion, we identified two U/GU rich downstream elements that play a critical role in KCNH2 intron 9 polyadenylation and showed that antisense MO targeting these elements can effectively inhibit KCNH2 intron 9 polyadenylation, leading to the isoform switch from Kv11.1a-USO to Kv11.1a. The fact that antisense MO is much less effective in the presence of a strong, canonical poly(A) signal indicates that the downstream elements of the weak, noncanonical poly(A) signal serve as ideal targets for antisense MO-mediated inhibition of KCNH2 intron 9 polyadenylation.
Despite the presence of the weak poly(A) signal in KCNH2 intron 9, polyadenylation is the dominant event in the heart, as about two-thirds of KCNH2 pre-mRNA are processed to the nonfunctional Kv11.1a-USO isoform in the heart [6,7]. We have shown previously that the balance between splicing and polyadenylation, mediated by intrinsically weak splicing and weak poly(A) signals, is important for the alternative processing of KCNH2 pre-mRNA and the relative expression of Kv11.1a and Kv11.1a-USO isoforms. Our present results demonstrate that inhibition of intron 9 polyadenylation by the antisense approach can shift the balance toward the splicing pathway, thereby leading to the predominant expression of the full-length Kv11.1a isoform. A limitation of this study is the use of HEK293 cells rather than cardiomyocytes. Although the short KCNH2 gene construct expressed in HEK293 cells exhibited a similar expression pattern of Kv11.1 isoforms as in the heart, future studies using cardiomyocytes will be necessary to confirm the antisense MO-induced regulation of KCNH2 alternative polyadenylation.
The significance of this work is highlighted by our recent finding that a disease-causing KCNH2 mutation, IVS9-2delA, resulted in the switch from the functional Kv11.1a isoform to the non-functional Kv11.1a-USO isoform [10]. The antisense approach shown in the present work would be an ideal strategy for correcting the isoform switching defect caused by IVS9-2delA, as it can switch the isoform expression from Kv11.1a-USO to Kv11.1a in the WT allele. The antisense MO strategy is also suitable for the majority of nonsense and frameshift mutations that are sensitive to the nonsense-mediated mRNA decay [14] as well as many trafficking defective mutations that do not have a dominant negative effect or have an incomplete dominant negative effect on the WT channels [15,16]. For those mutations that have severe dominant negative effects, the antisense approach is effective if mutant mRNA can be downregulated by allele-specific RNA interference [17,18]. Thus, the antisense inhibition of intron 9 polyadenylation represents a novel approach for the upregulation of Kv11.1 channel current which may have therapeutic applications across a wide spectrum of LQT2 mutations.
Alternative polyadenylation is increasingly recognized as an important mechanism of gene regulation [12,19]. Alternative polyadenylation can lead to the generation of alternate transcripts with different coding sequences and/or variable 3′-untranslated regions. Furthermore, aberrant alternative polyadenylation has been associated with many human diseases [19]. Thus, modulation of alternative polyadenylation is of particular interest for the regulation of isoform expression with potential therapeutic applications. Although antisense approaches have been widely used in the downregulation of gene expression and modulation of alternative splicing [20], antisense modulation of alternative polyadenylation is only reported in a few cases including E-selectin and receptor tyrosine kinases [21,22]. In the case of E-selectin, inhibition of the 3′-most polyadenylation site by antisense oligonucleotides results in the redirection of polyadenylation to one of two upstream poly(A) sites and shortens the 3′-untranslated region. The shorter transcripts generated following antisense treatment have fewer destabilization sequences, thereby increasing mRNA stability [21]. Many receptor tyrosine kinase genes undergo alternative intronic polyadenylation that generates dominant negative, secreted isoforms. Inhibition of U1 snRNA binding by targeting the upstream 5′ splice sites with antisense oligonucleotides can activate the intronic poly(A) sites and induce the switch from the full-length, membrane bound receptor to the truncated, soluble isoforms of receptor tyrosine kinases [22]. In contrast, antisense inhibition of intronic poly(A) sites results in a decrease in the expression of the soluble isoforms [22]. Our results demonstrate that antisense MO-induced inhibition of intronic poly(A) site not only reduces the truncated Kv11.1a-USO isoform but also increases the full-length Kv11.1a isoform. The present work is the first demonstration of using the antisense approach to modulate alternative polyadenylation that results in a switch from the truncated isoform to the full-length isoform. Our findings suggest that antisense MO may represent a useful strategy to upregulate the expression of full-length isoforms of other genes that are regulated by alternative intronic polyadenylation.
In summary, we have demonstrated that the relative expression of Kv11.1 C-terminal isoform can be modulated using antisense MO, resulting in the upregulation of the full-length functional Kv11.1a isoform. These results suggest that antisense strategies targeting the alternative polyadenylation of the KCNH2 gene may represent a novel approach to increase Kv11.1 channel function.
Research Highlights.
Alternative polyadenylation of KCNH2 mRNA generates two Kv11.1 isoforms.
Intron 9 poly(A) signal activity depends on downstream U/GU-rich elements.
Antisense inhibition of these elements upregulates Kv11.1a isoform expression.
Increasing Kv11.1a expression may be a therapeutic strategy for long QT syndrome.
Acknowledgments
This study was supported by the NIH grant HL68854 (ZZ). Dr. Stump is supported by the NIH training grant T32HL094294.
Footnotes
Disclosures
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci US A. 1994;91:3438–42. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 3.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Z, Gong Q, Ye B, Fan Z, Makielski JC, Robertson GA, et al. Properties of HERG channels stably expressed in HEK 293 cells studied at physiological temperature. Biophys J. 1998;74:230–41. doi: 10.1016/S0006-3495(98)77782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 6.Gong Q, Stump MR, Dunn AR, Deng V, Zhou Z. Alternative splicing and polyadenylation contribute to the generation of hERG1 C-terminal isoforms. J Biol Chem. 2010;285:32233–41. doi: 10.1074/jbc.M109.095695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupershmidt S, Snyders DJ, Raes A, Roden DM. A K+ channel splice variant common in human heart lacks a C-terminal domain required for expression of rapidly activating delayed rectifier current. J Biol Chem. 1998;273:27231–35. doi: 10.1074/jbc.273.42.27231. [DOI] [PubMed] [Google Scholar]
- 8.Guasti L, Crociani O, Redaelli E, Pillozzi S, Polvani S, Masselli M, et al. Identification of a posttranslational mechanism for the regulation of hERG1 K+ channel expression and hERG1 current density in tumor cells. Mol Cell Biol. 2008;28:5043–60. doi: 10.1128/MCB.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stump MR, Gong Q, Zhou Z. Isoform-specific dominant-negative effects associated with hERG1 G628S mutation in long QT syndrome. PLoS ONE. 2012;7:e42552. doi: 10.1371/journal.pone.0042552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong Q, Stump MR, Deng V, Zhang L, Zhou Z. Identification of Kv11.1 isoform switch as a novel pathogenic mechanism of long QT syndrome. Circ Cardiovasc Genet. 2014;7:482–90. doi: 10.1161/CIRCGENETICS.114.000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Q, Zhang L, Moss AJ, Vincent GM, Ackerman MJ, Robinson JC, et al. A splice site mutation in hERG leads to cryptic splicing in human long QT syndrome. J Mol Cell Cardiol. 2008;44:502–9. doi: 10.1016/j.yjmcc.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–20. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunes NM, Li W, Tian B, Furger A. A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–36. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Q, Stump MR, Zhou Z. Position of premature termination codons determines susceptibility of hERG mutations to nonsense mediated mRNA decay in long QT syndrome. Gene. 2014;529:190–97. doi: 10.1016/j.gene.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ficker E, Thomas D, Viswanathan PC, Dennis AT, Priori SG, Napolitano C, et al. Novel characteristics of a misprocessed mutant HERG channel linked to hereditary long QT syndrome. Am J Physiol Heart Circ Physiol. 2000;279:H1748–56. doi: 10.1152/ajpheart.2000.279.4.H1748. [DOI] [PubMed] [Google Scholar]
- 16.Anderson CL, Delisle BP, Anson BD, Kilby JA, Will ML, Tester DJ, et al. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006;113:365–73. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, Yang X, Huang X, Huang C, Sun HH, Jin L, et al. RNA interference targeting E637K mutation rescues hERG channel currents and restores its kinetic properties. Heart Rhythm. 2013;10:128–36. doi: 10.1016/j.hrthm.2012.09.124. [DOI] [PubMed] [Google Scholar]
- 18.Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, et al. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J. 2014;35:1078–87. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 20.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–93. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 21.Vickers TA, Wyatt JR, Burckin T, Bennett CF, Freier SM. Fully modified 2′ MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 2001;29:1293–99. doi: 10.1093/nar/29.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vorlová S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 2011;43:927–39. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]