Abstract
While it is thought that short-term memory arises from changes in protein dynamics that increase the strength of synaptic signaling, many of the underlying fundamental molecular mechanisms remain unknown. Our lab developed a C. elegans assay of positive olfactory short-term associative memory (STAM), in which worms learn to associate food with an odor and can remember this association for over one hour. Here we use this massed olfactory associative assay to identify regulators of C. elegans short-term and intermediate-term associative memory (ITAM) processes. We show that there are unique molecular characteristics for different temporal phases of STAM, which include: learning, which is tested immediately after training, short-term memory, tested 30 minutes after training, intermediate-term memory, tested 1 hour after training, and forgetting, tested 2 hours after training. We find that, as in higher organisms, C. elegans STAM requires calcium and cAMP signaling, and ITAM requires protein translation. Additionally, we found that STAM and ITAM are distinct from olfactory adaptation, an associative paradigm in which worms learn to disregard an inherently attractive odor after starvation in the presence of that odor. Adaptation mutants show variable responses to short-term associative memory training. Our data distinguish between shorter forms of a positive associative memory in C. elegans that require canonical memory pathways. Study of STAM and ITAM in C. elegans could lead to a more general understanding of the distinctions between these important processes and also to the discovery of novel conserved memory regulators.
Keywords: Short-term Memory, Intermediate-term Memory, C. elegans, cAMP, adaptation
1. Introduction
Learning and memory allow animals to navigate, find food, and survive in a changing environment. In humans, memory declines with age, and memory deficits are often a hallmark of neurodegenerative disorders, such as Alzheimer’s Disease (Hodges & Patterson, 1995; Salthouse, 1991). Therefore, an understanding of the molecular bases of different forms of memory is essential to develop treatments for memory loss.
Short-term memory lasts from minutes to hours, resulting from changes in synaptic strength mediated by modifications at the synapse in the appropriate neurons (Kandel, 2001). cAMP signaling and calcium signaling are required for short-term memory in many organisms (Hawkins et al., 2006; Kandel, 2001). These pathways are activated in response to neurotransmitter receptor activity, can modulate receptor activity, and are thought to activate synaptic proteins to facilitate memory formation and maintenance. Several kinases (Giese & Mizuno, 2013; Kandel, 2012) and synaptic cellular adhesion proteins (Cheng et al., 2001; Grotewiel et al., 1998) have also been identified as regulators of short and intermediate-term memory in higher organisms. However, downstream targets or parallel regulatory pathways remain largely unknown. Therefore, it is important to establish a model system in which regulation of short-term memory is conserved and new genetic regulators of short-term memory can be rapidly identified.
C. elegans has a simple nervous system comprised of just 302 neurons, and its stereotypic neural connections (the “connectome”) have been mapped (Varshney et al., 2011; White et al., 1986). This small nematode can learn and form both associative and non-associative memories, lasting as long as 24 hours (Ardiel & Rankin, 2010; Kauffman et al., 2010). We developed a protocol that pairs a relatively neutral odor (butanone at a specific concentration) with food to create a positive association, resulting in strong attraction to the odor (Kauffman, et al., 2010). Short-term associative memory (STAM) after one conditioning period decays by 2 hours after training, while long-term memory after spaced training (7 conditioning cycles) lasts more than 16 hours (Kauffman, et al., 2010). Importantly, because the worms display an increased response to odor after training, our positive associative assay allows clear distinction from non-associative forms of learning, such as habituation and adaptation, which repress behavioral response to a stimulant and therefore could be interpreted as negative associative learning.
We previously found that positive associative learning requires conserved receptors, and that long-term associative memory requires transcription as well as the activity of the Zn finger transcription factor CREB (Kauffman, et al., 2010). Here we show that C. elegans STAM requires cAMP and calcium signaling pathways. We also find that protein translation is required at two different steps: it is first required during massed associative memory training to extend memory past 30 minutes, and subsequently after memory training to ensure proper decay of the associative memory, or forgetting. Intermediate-term memory requires translation but not transcription (Ashraf et al., 2006; Ghirardi et al., 1995); our results show that massed training results in both short-term (30 minute) and intermediate-term (1 hour) associative memory (STAM and ITAM). Our data show that shorter-term memory mechanisms in C. elegans are conserved with higher organisms and further establish C. elegans as a good model to study the genetic regulation of short-term and intermediate-term associative memory.
C. elegans has been shown to associate starvation with a myriad of sensory cues including olfactory, gustatory, thermosensory, and pathogenic cues (Colbert & Bargmann, 1995; Mohri et al., 2005; Saeki et al., 2001; Zhang et al., 2005). Adaptation is a behavior in which worms display reduced responsiveness to an odor after a one-hour odor exposure in the absence of food (Colbert & Bargmann, 1995). Stetak et al. (2009) use the same odor/starvation conditioning paradigm to induce what they refer to as a negative associative short-term memory. Our massed associative memory assay also involves a one-hour exposure to an odor, but in the presence of food rather than the absence of food, resulting in a positive association between food and odor. Therefore, one hypothesis that we set out to test is whether STAM is simply a response occurring in the opposite direction of adaptation that requires the same molecular machinery. However, we find that mutants that are defective for adaptation have varying STAM/ITAM phenotypes, including prolonged, reduced, and normal memory, establishing positive olfactory STAM/ITAM as a distinct memory process in C. elegans.
2. Methods
2.1. Worm Cultivation
C. elegans were cultivated at 20°C on High Growth Media (HG) or Nematode Growth Media (NGM) seeded with OP50 E. coli using standard methods (Brenner, 1974). Animals were synchronized by hypochlorite treatment and tested for learning and memory at Day 1 of adulthood at room temperature.
2.2. Strains
Wild type: (N2 Bristol); mutant strains: KP1182 (acy-1(nu329)), KG518 (acy-1(ce2)), KG532 (kin-2(ce179)), KG744 (pde-4(ce268)), VC1052 (unc-43(gk452)), VC1408 (magi-1(gk657)), JH1270 (nos-1(gv5)), JK3022 (fbf-1(ok91)), CX20 (adp-1(ky20)), RB995 (hpl-2(ok916)), NL917 (mut-7(pk204)), and DR466 (him-5(e1490)) were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). egl-4(ky95);him-5(e1490) was a kind gift from N. L’Etoile (University of California, San Francisco, San Francisco, CA).
2.3. Behavioral Assays
2.3.1. Short-term Associative Memory Training
STAM assays were performed as previously described (Kauffman, et al., 2010; Kauffman et al., 2011). Briefly, synchronized day 1 hermaphrodites were starved for 1 hr in M9 buffer. Worms were then transferred to 6 cm NGM plates with 500 μL of OP50 and 2 μL of 10% butanone in ethanol on the lid and conditioned for 1 hr. After conditioning, 100–500 worms from the trained population of worms were tested once for chemotaxis to butanone either immediately (0 hr) or after being transferred to 10 cm NGM plates with 900 μL fresh OP50 for specified intervals before testing (30 mins-4 hrs). Graphs display the average chemotaxis index at each time-point for 3 or more separate biological replicate experiments, with each experiment displayed as a pale colored curve behind this average line. Each experiment included at least 3 technical replicates per time-point, with the exception of one of four biological replicate experiments for acy-1(nu329) and kin-2(ce179) mutants that had only one technical replicate per time-point. Significance was calculated by comparing the experimental and control groups using two-way ANOVAs (p ≤ 0.05 for significant experiments) followed by Bonferroni-corrected post-hoc unpaired t-tests comparing the experimental and control at each time-point (Bonferroni-correction per t-test: pbonf ≤ 0.05).
2.3.2. Chemotaxis Assay
Chemotaxis assays were performed as previously described (Bargmann et al., 1993). Briefly, 100–500 day 1 adult worms were placed at the origin on a 10 cm NGM plate with butanone (1 μL 10% butanone in ethanol + 1 μL NaN3) and ethanol control (+ 1 μL NaN3) equidistant from the origin. After 1 hr., black and white images of each plate were taken using a Basler A fire wire camera (Basler AG, Ahrensburg, Germany) using “Measurement and Automation” software (National Instruments) to capture images. Images were analyzed using count_worms_v7.3 (Kauffman, et al., 2011). The chemotaxis index =[(nattractant)-(ncontrol)]/[(Total-norigin)].
2.3.3. Drug Treatments
200 μg/mL Actinomycin D ≥ 95% (Sigma Aldrich, Saint Louis, Missouri) was added to M9 buffer for the final 30 minutes of starvation and added to S basal during conditioning along with 1:1000 Butanone and OP50 E. coli bacteria that had been grown overnight. Cycloheximide ≥ 94% (Sigma Aldrich, Saint Louis, Missouri) was added to NGM at 0.8 mg/mL. Plates were poured and solidified overnight at 4°C, seeded with OP50 E. coli, and then used for Short-term associative memory training either during conditioning or after training. To ensure 1 hour of drug treatment for 30 minute conditioning, worms were added to cycloheximide plates with no food for the last 30 minutes of starvation followed by 30 minutes of conditioning on cycloheximide with OP50 and 10% butanone.
3. Results
3. 1 C. elegans STAM and ITAM Require Conserved Memory Mechanisms
3.1.1 Blocking translation, but not transcription, inhibits both memory maintenance and forgetting
We and others have shown that long-term memory in C. elegans requires both transcription and translation (Kauffman, et al., 2010; Timbers & Rankin, 2011; Vukojevic et al., 2012), as is required for long-term memory in other organisms. While transcription is not required for temporal phases other than long-term memory, protein translation is required for intermediate-term (1–3 hr.) phases of memory (H. P. Davis & Squire, 1984; Ghirardi, et al., 1995; Hawkins, et al., 2006; Jin et al., 2011; Michel et al., 2012; Parvez et al., 2005; Sutton et al., 2001) and for what is termed negative associative short-term memory in C. elegans that may actually be intermediate-term memory (Hadziselimovic et al., 2014). As in our previously published short-term associative memory assay, several thousand day 1 adult worms are starved for 1 hour, conditioned with food and 10% butanone for 1 hour, and then held on food before subsets of the trained population are tested for memory using a chemotaxis assay at specific time-points after training (Fig. 1A)(Kauffman, et al., 2010).
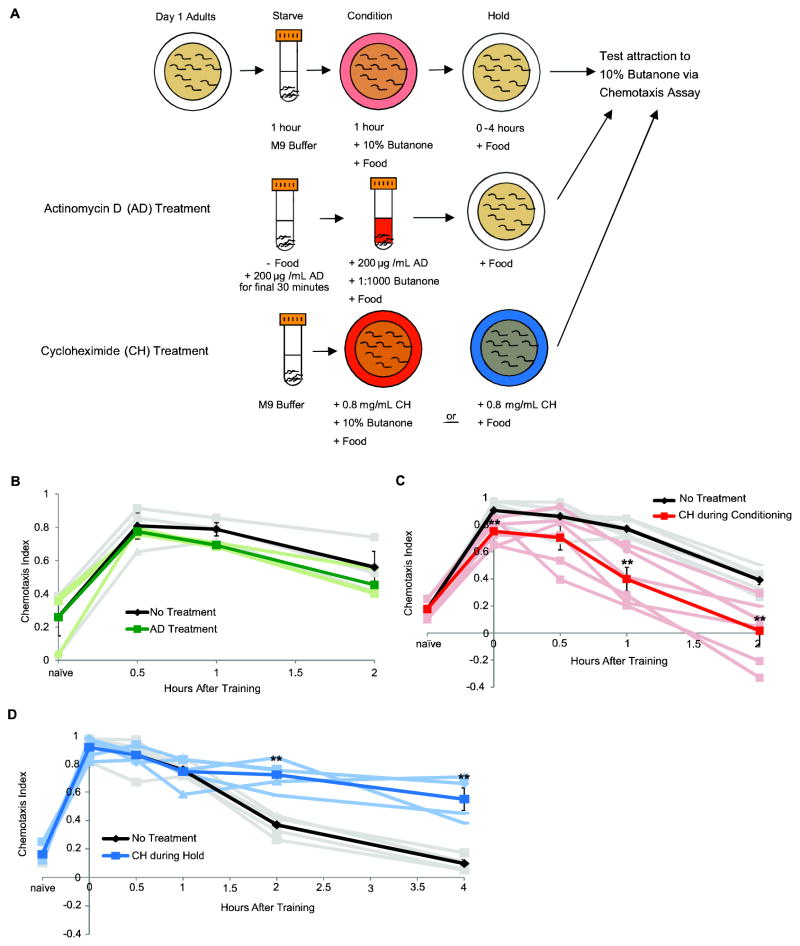
Figure 1.
C. elegans positive olfactory short-term associative memory (STAM) requires translation but not transcription. (A) Illustration of short-term associative memory assay and changes for drug treatment protocols. For Actinomycin D (AD) treatment, worms were starved in M9 with 200 μg/mL AD added during the final 30 minutes of starvation then conditioned in OP50 in S-Basal with 200 μg/mL AD and 0.1% butanone. Untreated worms were tested using the same protocol without AD. For Cycloheximide (CH) treatment, 0.8 mg/mL CH was added to nematode growth media (NGM) when plates were poured. OP50 was grown overnight on NGM plates with or without drug and worms were treated either during the STAM conditioning period or during the holding period after training. Otherwise the protocol followed the normal STAM assay. (B) There is no change in memory retention with Actinomycin D treatment. Worms were either not treated (black), or treated during starvation and conditioning (green). n = 3 experiments (pale lines). (C) Cycloheximide treatment during training causes a slight decrease in learning (red; pbonf = 0.046), at at 1 hr (pbonf = 0.007), but 30-minute memory does not require translation. Chemotaxis index of treated worms at two hours was also significantly below that of controls (black; pbonf = 0.023). n = 6 experiments. (D) Cycloheximide treatment after training (blue) shows that translation is required for normal forgetting (pbonf = 0.001 at 2 hrs. pbonf = 0.010 at 4 hrs.). n = 5 experiments. Error bars represent the mean +/− SEM. Asterisks indicate significance as determined by an unpaired student T-test with a Bonferroni correction for multiple comparisons.
In order to assess the requirement for transcription and translation in C. elegans short-term associative memory, we used actinomycin D and cycloheximide, respectively, during conditioning or after training to inhibit these processes in wild-type day 1 adult worms (Fig. 1A). To test whether STAM requires transcription, we treated worms with 200 μg/mL actinomycin D during the final 30 minutes of starvation and in S Basal + OP50 and 1:1000 butanone during conditioning. We tested worms 30 minutes after training, allowing them to regain motility after submersion (Fig. 1A). While treatment with actinomycin D during spaced training was sufficient to inhibit long-term associative memory (Kauffman, et al., 2010), we found that inhibiting transcription during training does not inhibit short-term memory formation or retention (Fig. 1B). This is consistent with our previous observation that short-term associative memory is not dependent on the cAMP Response Element-Binding Protein (CREB) transcription factor (Kauffman, et al., 2010).
We then tested whether protein translation is required for short-term associative memory. We treated worms with 0.8 mg/ml cylcoheximide in growth medium during or after STAM training (Fig. 1A). Worms treated with cycloheximide during conditioning had a small reduction in learning (p = 0.046) that may be due to the stress of the treatment (Fig. 1C). However, 30 minute memory was normal. 1 hour memory was significantly reduced compared to untreated controls. These data suggest that memory 30 minutes after training is a translation-independent short-term memory, and that by 1 hour after training C. elegans have formed an intermediate-term memory, since intermediate-term memory requires translation but not transcription (Ghirardi, et al., 1995). (For continuity, we will continue to refer to our training as “STAM training”, although the same training also induces ITAM.) We also tested whether conditioning worms for only 30 minutes would result in a fully translation-independent memory, since we had previously shown that 30 minutes of conditioning is enough to maximize learning (Kauffman, et al., 2010). We treated worms for 1 hour with cycloheximide, as in our initial experiment, by treating for the last 30 minutes of starvation followed by the full 30 minutes of conditioning. Interestingly, we found that duration of memory after 30 minutes of conditioning was equivalent to that of 1 hour of conditioning, and 1 hour memory was dependent on translation (Sup. Fig. 1).
Unexpectedly, worms treated with cycloheximide after training showed increased chemotaxis at both at 2 and 4 hours, after untreated worms have lost their attraction to butanone; in fact, memory is still close to 0 hr post-training levels after 4 hours, suggesting a decrease in forgetting (blue trace, Fig. 1D). (The full duration of memory for C. elegans treated after cycloheximide training could not be tested due to the impairment of worm motility after 4+ hours of cycloheximide exposure.) Forgetting a negative associative memory also requires translation (Hadziselimovic, et al., 2014). Taken together, these data indicate that forgetting is an actively regulated process that requires protein translation.
3.1.2 Conserved Genetic Pathways Regulate STAM and ITAM
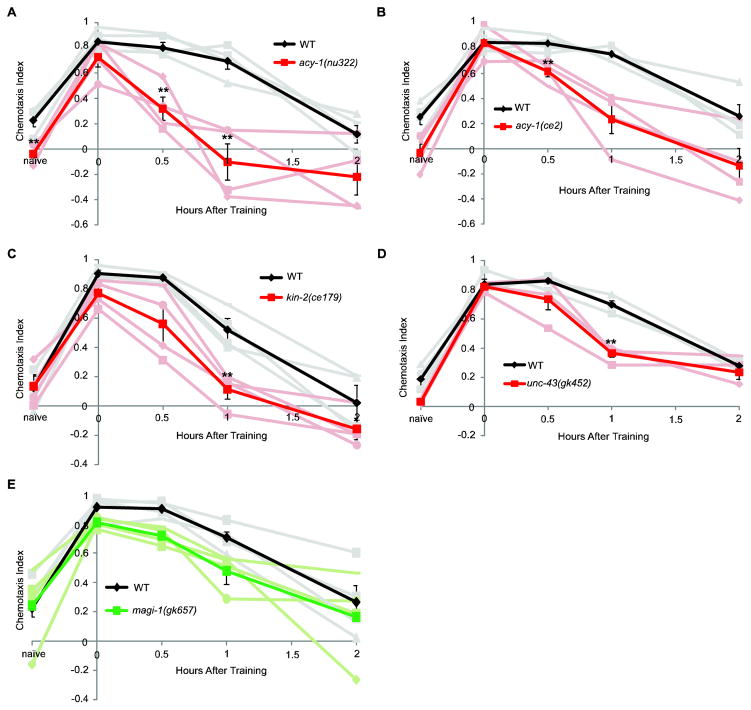
We and others have shown that C. elegans associative learning is dependent on AMPA type receptors (Kauffman, et al., 2010; Lau et al., 2013; Vukojevic, et al., 2012), as in other organisms (Cui et al., 2011; Q. Li et al., 2005; Sanderson et al., 2008; Zamanillo et al., 1999). In order to identify genes that are required for memory, we examined calcium signaling genes downstream of receptor activity, specifically calcium-activated CaMKII and components of the cAMP pathway that are required for learning and memory in many organisms (Hawkins, et al., 2006; Squire L; Kandel, 2008). cAMP activates Protein Kinase A (PKA) and modulates memory in a concentration-dependent manner in Drosophila (Feany, 1990). Adenylate cyclase catalyzes the conversion of ATP to cAMP and is required for short-term memory in mice, Drosophila, and Aplysia (R. L. Davis et al., 1995; Hawkins, et al., 2006; Wang et al., 2011). Protein Kinase A is activated by cAMP and is required for memory in many organisms (W. Li et al., 1996; Michel, et al., 2012; Nguyen & Woo, 2003; Squire L; Kandel, 2008). We found that loss-of-function mutants of adenylate cyclase, acy-1(nu322), have reduced chemotaxis compared to WT, but can learn to associate butanone and food. Memory after training is significantly reduced by 30 minutes compared to wild-type worms (Fig. 2A). To take the naïve chemotaxis index (CI) of acy-1(nu322) mutants into account, we calculated the learning index after training (Learning Index = CIafter training – CInaïve) (Sup. Fig. 2). We find that memory at one hour after training is still significantly reduced in acy-1(nu322) mutants compared to WT. acy-1(ce2) gain-of-function mutants have normal naïve chemotaxis, but reduced 30 minute and 1-hour memory (Fig. 2B). Taken together, these data indicate that cAMP signaling is required for STAM and ITAM. kin-2(ce179) mutants are mutated in the PKA inhibitory domain and have increased PKA activity; kin-2(ce179) mutants have reduced one-hour memory compared to WT (Fig. 2C). cAMP phosphodiesterase (PDE), is required for memory in Drosophila (Dudai et al., 1976). However, pde-4(ce268) mutants have normal short-term associative memory (Sup. Fig. 3) though this may be due to the partial loss-of-function of available mutants. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is required for memory in many organisms (Ashraf, et al., 2006; Giese et al., 1998; Hawkins, et al., 2006) and required for forgetting adaptation in C. elegans (Inoue et al., 2013). Mutants of the C. elegans CaMKII homolog unc-43(gk452) have reduced memory by 1 hr. (Fig. 2D).
Figure 2.
C. elegans STAM requires canonical memory genes. (A) acy-1(nu329) mutants (red) have impaired naïve chemotaxis as compared to wild type (black) (pbonf = 0.025). acy-1(nu329) can learn but impaired memory by 30 min. WT (pbonf = 0.012 at 30 min. and pbonf = 0.011 at 1 hr.). n = 4 experiments. (B) acy-1(ce2) gain-of-function mutants (red) have reduced 30 min. and 1 hr. memory as compared to WT (black) (pbonf = 0.031 at 30 min. and pbonf = 0.021 at 1 hour). n = 4 experiments. (C) kin-2(ce179) mutants (red) have reduced 1 hr. memory as compared to wild-type (black) (pbonf = 0.018). n = 4 experiments. (D) unc-43(gk452) mutants (red) have reduced 1 hr. memory as compared to WT (black) (pbonf = 0.0007). n = 4 experiments. (E) magi-1(gk657) mutants (red) are not significantly different from WT (black) n = 4 experiments. Error bars represent the mean +/− SEM. Asterisks indicate significance as determined by an unpaired student T-test with a Bonferroni correction for multiple comparisons.
In addition to cAMP and calcium signaling pathways, short-term memory requires changes at the synapse (Grotewiel, et al., 1998; Hawkins, et al., 2006; Stetak et al., 2009). We have previously shown that the AMPA receptor subunit GLUR1 homolog, glr-1, is required for learning (Kauffman, et al., 2010). Membrane-associated guanylate cyclase (MAGUK) proteins regulate receptor distribution at the synapse in many organisms, including C. elegans (Deng et al., 2006; Dobrosotskaya et al., 1997; Hirao et al., 1998; Stetak, et al., 2009), and MAGI-1 regulates GLR-1 and is required for C. elegans negative associative memory (Stetak, et al., 2009). We tested the role of MAGI-1 in our STAM assay, and interestingly, found that magi-1(gk657) mutants have positive short-term associative memory that is similar to wild-type’s (Fig. 2E), indicating that negative and positive associative memories in C. elegans may be regulated differently at the synapse.
Together, these data indicate that C. elegans STAM requires cAMP signaling, and ITAM requires both cAMP and CaMKII signaling, as is required for short and intermediate-term memory in other organisms.
3.2 C. elegans STAM and Adaptation are molecularly distinct behavioral paradigms
Adaptation is a plastic response in which C. elegans learn to ignore the presence of odors persisting in their environment (Colbert & Bargmann, 1995). Many of the molecular mechanisms underlying this behavioral response have been previously elucidated using a negative associative assay that pairs odor with starvation to induce adaptation (Colbert et al., 1997; Juang et al., 2013; Kaye et al., 2009; L’Etoile et al., 2002). Adaptation to butanone and benzaldehyde requires AWCon, the sensory neuron required for sensing butanone (Kaye, et al., 2009). Because our short-term associative memory assay tests C. elegans’ response to butanone after one hour of butanone exposure with food as opposed to without food, as in adaptation studies, and butanone is also sensed by AWCon, one hypothesis is that adaptation and our short-term associative memory use the same molecular machinery. If this were true, we would expect that mutants that cannot adapt to butanone would extend memory after STAM training.
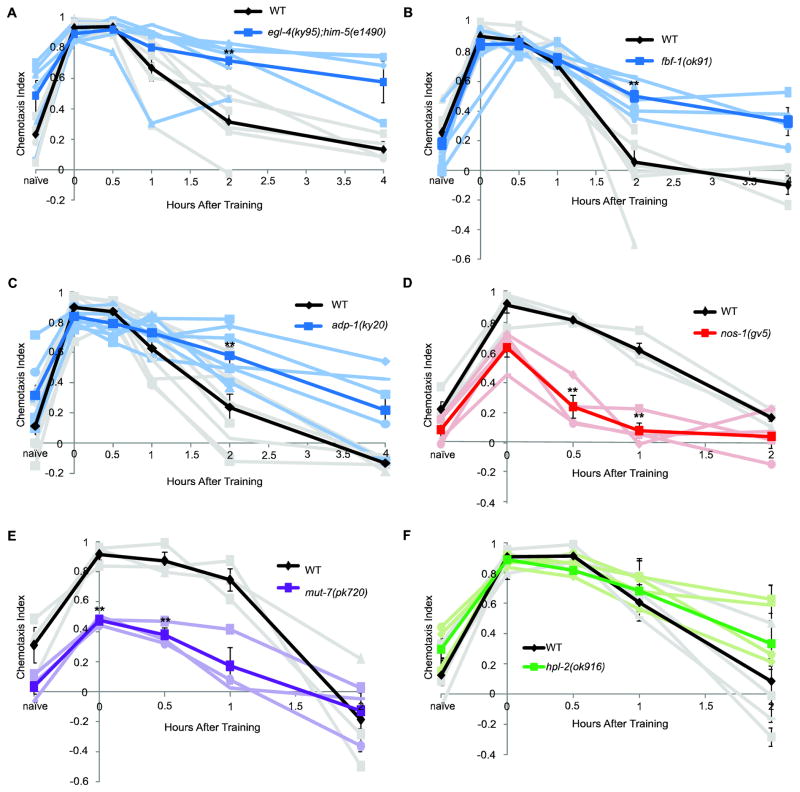
To test this hypothesis, we examined adaptation mutants. Loss-of-function mutations in nos-1, fbf-1, egl-4, mut-7, hpl-2, and adp-1 result in the inability to adapt to an attractive odor after exposure (Colbert & Bargmann, 1995; Juang, et al., 2013; Kaye, et al., 2009; L’Etoile, et al., 2002). FBF-1, a Pumilio homolog, facilitates the translation of EGL-4, a cyclic GMP-dependent protein kinase (Kaye, et al., 2009). Both fbf-1(ok91) and egl-4(ky95), which is mutated only at the site of FBF-1 binding, have prolonged memory after STAM training, decaying by 4 hrs instead of 2 hrs (Fig. 3A, B). Similarly, adp-1(ky20), which was identified in a screen for adaptation mutants but remains uncloned (Colbert & Bargmann, 1995), displayed extended memory after STAM training that decays by 4 hrs (Fig. 3C). Thus, fbf-1, egl-4, and adp-1 mutants all extend STAM, as would be expected of a mutant that cannot adapt.
Figure 3.
C. elegansSTAM and Adaptation are molecularly distinct behavioral paradigms. (A) egl-4(ky95);him-5(e1490) mutants (blue) have extended memory compared to WT (black) (pbonf = 0.013 at 2 hrs.). n = 3 experiments for naive-2 hrs. and 3 experiments for naive-4 hrs. (egl-4(ky95) mutants were also mutant for him-5, but him-5(e1490) has normal STAM (Sup. Fig. 4).) (B) fbf-1(ok91) mutants (blue) have extended memory compared to WT (black) (pbonf = 0.036 at 2 hrs.). n = 4 experiments for naïve-2 hrs. and 3 experiments for naïve-4 hrs. (C) adp-1(ky20) mutants (blue) have extended memory compared to WT (black) (pbonf = 0.037 at 2 hrs.). n = 3 experiments for naïve-2 hrs. and 5 experiments for naïve-4 hrs. (D) nos-1(gv5) mutants (red) have no memory by 30 minutes after training (pbonf = 0.001 at 30 min. and pbonf = 0.001 at 1 hr.). n = 4 experiments. (E) mut-7(pk720) mutants (purple) have reduced learning and memory as compared to WT (black) (pbonf = 0.002 at 0 hr. and pbonf = 0.013 at 30 min.). n = 3 experiments. (F) hpl-2(ok916) mutants (green) have normal memory compared to WT (black). n = 5 experiments. Error bars represent the mean +/− SEM. Stars indicate significance as determined by an unpaired student T-test with a Bonferroni correction for multiple comparisons.
By contrast, nos-1(gv5), a mutant of one of three C. elegans Nanos homologs, has reduced memory by 30 minutes as compared to wild-type (Fig. 3D). Adaptation requires EGL-4 translocation into the nucleus, where it phosphorylates MUT-7 and HPL-2 and regulates the expression of odr-1, a transmembrane guanylyl cyclase required for normal olfaction (Bargmann, et al., 1993; Juang, et al., 2013). After STAM training, mut-7(pk20) mutants of the RNAse III homolog have reduced learning and memory ability (Fig. 3E). Mutants of heterochromatin protein 1 homolog hpl-2(ok196) have normal learning and memory (Fig. 3F). Together, these data indicate that disrupting genes required for adaptation causes varying STAM phenotypes. While FBF-1, EGL-4, and ADP-1 may have overlapping functions between adaptation and STAM, NOS-1, HPL-2, and MUT-7 play different roles in adaptation and STAM, suggesting that the two paradigms are at least partially distinct.
4. Discussion
While many learning and memory paradigms have been described and several genes that regulate these plastic processes have been identified, many aspects of the process remain unknown. We previously developed a novel C. elegans positive olfactory short-term associative memory assay. Here we show that STAM does not require gene transcription, but that protein translation is required for longer memory after training and for forgetting. When we inhibited translation during training, memory was established, but was impaired by 1 hour post-training. We have therefore identified a form of C. elegans intermediate-term associative memory. Though intermediate-term memory is a less well-defined phase of memory, translation is required for later memory time-points after massed training in Drosophila, as well as for intermediate-term memory in Aplysia (Ashraf, et al., 2006; Hawkins, et al., 2006). Recently, a non-translation dependent intermediate-term memory had been identified for habituation in C. elegans and a translation-dependent negative-associative memory has been identified but was not characterized as intermediate-term memory (Hadziselimovic, et al., 2014; C. Li et al., 2013). Furthermore, we found that protein translation is necessary for normal forgetting. Little is known about the process of forgetting in any organism. Previously, forgetting has been considered a passive process, possibly related to basal phosphatase deactivation of kinases that establish short-term memory (Jonides et al., 2008; Wixted, 2004). However, recent experiments in both flies and C. elegans suggest that forgetting involves active processes, including remodeling of the actin cytoskeleton at the synapse (Hadziselimovic, et al., 2014; Shuai et al., 2010). Forgetting of a negative benzaldehyde odor association in C. elegans is regulated by an UNC-43-activated MAP kinase pathway in AWC (Inoue, et al., 2013). However, in our positive associative assay, unc-43 is required for memory maintenance, which may mask any potential role in forgetting.
We also show that as in higher organisms, C. elegans uses the conserved cAMP and calcium signaling pathways for the formation and maintenance of short-term and intermediate-term memory. Interestingly, our data show that both loss-of-function and gain-of-function adenylate cyclase (acy-1) mutants have reduced memory, suggesting that, as in Drosophila, a balanced (or adjustable) level of cAMP is required for normal memory. In addition to adenylate cyclase, ITAM requires PKA and CaMKII activity. Though the PKA regulatory subunit mutant, kin-2, has an ITAM defect, STAM may be affected as it is in other organisms (Dudai, 1988; Kandel, 2012) by knocking down the catalytic domain of PKA, kin-1, an experiment we have not been able to do due to the essential requirement of kin-1.
In C. elegans, CaMKII regulates neuronal cell fate and, through a separate pathway, regulates forgetting after adaptation (Chuang & Bargmann, 2005; Inoue, et al., 2013). In higher organisms, different CaMKII alpha and beta mutations specifically affect behaviors from learning to long-term memory (Giese & Mizuno, 2013), and CaMKII autophosphorylation is essential for learning after massed training in mice (Irvine et al., 2005). Further research could determine whether our CaMKII mutant intermediate-term memory defect is specific to our positive olfactory associative assay, the particular CaMKII isoforms involved, and/or C. elegans behavior.
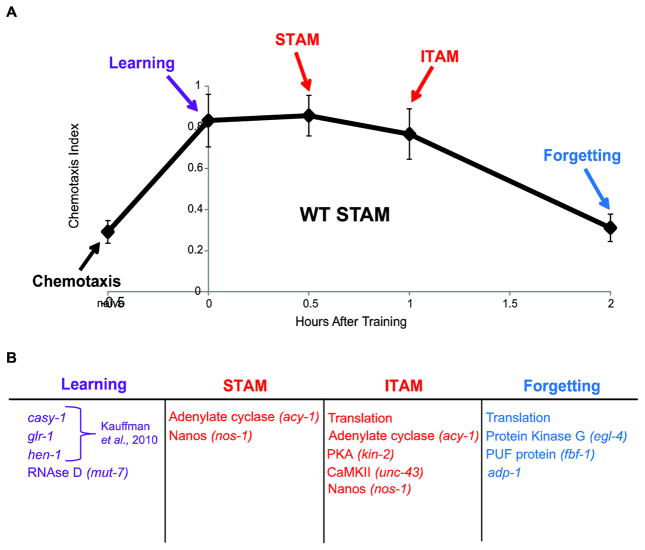
Our behavioral assay has allowed us to determine the temporal points at which particular gene products affect different phases of positive associative memory after massed training (Figure 4A, B). Specifically, we find that mut-7, casy-1, glr-1, and hen-1 (Kauffman, et al., 2010), are required for learning, while acy-1 and nos-1 are required for STAM. kin-2, acy-1 and unc-43 are required for ITAM, and finally, egl-4 and fbf-1 are required for forgetting.
Figure 4.
Molecular regulation of phases of memory after massed training. (A) Schematic of normal shorter-term associative memory phases. Before training, a test is run to determine naïve attraction to butanone (chemotaxis ability). Following training, learning is immediately tested. Short-term memory (STAM) is tested 30 minutes following training, and Intermediate-term memory (ITAM) at 1 hour. Forgetting is tested 2 hours after training when wild-type memory has decayed. (B) Genes and molecular processes that regulate each phase of memory based on our current data and our previously published learning data (Kauffman, et al., 2010).
The fact that forgetting requires protein translation is also interesting to consider in the context of the relationship between STAM and adaptation. We found that a mutation in the 3′UTR of the egl-4 Protein Kinase G that inhibits its binding to translational activator FBF-1 (Kaye, et al., 2009) inhibits forgetting, as does deletion of fbf-1. These genes are uniquely co-expressed in the AWB and AWC neurons (Kaye, et al., 2009), so forgetting may occur in the sensory neuron through translation of EGL-4. The memory inhibitor MSI-1 has been found to reduce synapse size after negative STAM training through its translational repression of the ARP2/3 complex in command motor neurons downstream of the sensory neuron, and translation is required for forgetting a negative short-term associative memory (Hadziselimovic, et al., 2014). Taken together, these data suggest that forgetting is an active process involving translation, possibly of multiple proteins that alter cellular machinery in both the sensory and command motor neurons.
Adaptation involves starvation-induced desensitization to a previously attractive odor after long-term exposure. Since our assay involves a 60-minute exposure to butanone in the presence rather than the absence of food, we tested whether STAM and adaptation are regulated by the same molecular mechanisms. Adaptation results in part from the translation of EGL-4, mediated by FBF-1, and subsequent EGL-4 translocation into the nucleus, where it phosphorylates HPL-2 and MUT-7, which then reduce the levels of odr-1 guanylyl cyclase (Juang, et al., 2013; Kaye, et al., 2009). While all of the adaptation mutants have defective adaptation to AWC-sensed odors (Juang, et al., 2013; Kaye, et al., 2009; L’Etoile, et al., 2002; Lee et al., 2010), we found here that they have varying STAM phenotypes (Table 1). The Nanos homolog mutant nos-1 could learn, but could not remember, while adp-1 had impaired forgetting. The cytoplasmic components of the adaptation pathway, egl-4 and fbf-1, have forgetting defects, as one might have expected if STAM and adaptation are the same process. However, HPL-2, a nuclear component of the adaptation pathway, had normal memory when mutated, suggesting that it is only required for adaptation, not STAM. MUT-7, a nuclear component of the adaptation pathway that is expressed in both the cytoplasm and nucleus, is required for learning after STAM training, so its role in memory is not clear. Our data suggest that memory after massed training may only share the cytoplasmic components of the adaptation pathway, and that EGL-4 may have unidentified targets outside of the nucleus that regulate forgetting. Therefore, STAM and adaptation are separate but related learning and memory paradigms. Since adaptation is either non-associative or a result of a negative association (Pereira & van der Kooy, 2012), STAM that is formed after a positive association may be regulated differently by similar pathways. Indeed, Insulin/IGF-1-like signaling differentially regulates negative vs. positive associative learning and memory (Kauffman, et al., 2010; Stein & Murphy, 2012). By testing adaptation mutants, we were able to identify conserved genes that have not previously been known to regulate associative memory (nos-1) and forgetting (egl-4 and fbf-1) in higher organisms, though their mechanisms of action remain to be studied.
Table 1.
Adaptation and STAM phenotypes for mutants tested in both paradigms. (−) reduced compared to WT. (+) increased compared to WT. (Colbert & Bargmann, 1995; Juang, et al., 2013; Kaye, et al., 2009; L’Etoile, et al., 2002)
| Mutant | Homolog | Protein Cellular Localization | Adaptation Phenotype | STAM Phenotype |
|---|---|---|---|---|
| egl-4(ky95) | PKG | Cytoplasm | − | + |
| fbf-1(ok91) | PUF Protein | Cytoplasm | − | + |
| adp-1(ky20) | Uncloned | Unknown | − | + |
| nos-1(gv5) | Nanos | Unknown | − | − |
| mut-7(pk720) | RNAse III | Nucleus and Cytoplasm | − | - learning |
| hpl-2(ok916) | HPI | Nucleus | − | WT |
Here we have identified many genetic regulators of STAM and ITAM, including several conserved regulators, and we have determined when these regulators are temporally required. Importantly, we have established a positive olfactory associative paradigm that tests a conserved form of shorter-term memory. It is complicated to rule out possible habituation or adaptation defects in associative memory mutants using negative olfactory associative assays; a positive associative assay avoids these complications, allowing more precise determination of the temporal requirements of associative memory. Together, our data provide a molecular framework for future studies of short-term memory formation, maintenance, and forgetting.
Highlights.
Massed positive associative memory training induces both short-term and intermediate-term memory.
Forgetting is actively regulated and requires protein translation.
C. elegans memory requires canonical memory regulators.
C. elegans positive olfactory associative memory is distinct from olfactory adaptation.
Acknowledgments
We thank N. L’Etoile and the C. elegans Genetics Center and the International C. elegans Gene Knockout Consortium for strains. We thank A. Williams, V. Lakhina, G. Kleemann and A. Sylvain for comments and the entire Murphy Lab for helpful discussion. This work was funded by an NIH Cognitive Aging R01, as well as Keck Young Scholars and McKnight Scholars awards to CTM.
Abbreviations
- STAM
Short-Term Associative Memory
- ITAM
Intermediate-Term Associative Memory
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardiel EL, Rankin CH. An elegant mind: learning and memory in Caenorhabditis elegans. Learn Mem. 2010;17(4):191–201. doi: 10.1101/lm.960510. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74(3):515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Endo K, Wu K, Rodan AR, Heberlein U, Davis RL. Drosophila fasciclinII is required for the formation of odor memories and for normal sensitivity to alcohol. Cell. 2001;105(6):757–768. doi: 10.1016/s0092-8674(01)00386-5. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19(2):270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans. Neuron. 1995;14(4):803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17(21):8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Darby-King A, Grimes MT, Howland JG, Wang YT, McLean JH, Harley CW. Odor preference learning and memory modify GluA1 phosphorylation and GluA1 distribution in the neonate rat olfactory bulb: testing the AMPA receptor hypothesis in an appetitive learning model. Learn Mem. 2011;18(5):283–291. doi: 10.1101/lm.1987711. [DOI] [PubMed] [Google Scholar]
- Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96(3):518–559. [PubMed] [Google Scholar]
- Davis RL, Cherry J, Dauwalder B, Han PL, Skoulakis E. The cyclic AMP system and Drosophila learning. Mol Cell Biochem. 1995;149–150:271–278. doi: 10.1007/978-1-4615-2015-3_31. [DOI] [PubMed] [Google Scholar]
- Deng F, Price MG, Davis CF, Mori M, Burgess DL. Stargazin and other transmembrane AMPA receptor regulating proteins interact with synaptic scaffolding protein MAGI-2 in brain. J Neurosci. 2006;26(30):7875–7884. doi: 10.1523/JNEUROSCI.1851-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrosotskaya I, Guy RK, James GL. MAGI-1, a membrane-associated guanylate kinase with a unique arrangement of protein-protein interaction domains. J Biol Chem. 1997;272(50):31589–31597. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- Dudai Y. Neurogenetic dissection of learning and short-term memory in Drosophila. Annu Rev Neurosci. 1988;11:537–563. doi: 10.1146/annurev.ne.11.030188.002541. [DOI] [PubMed] [Google Scholar]
- Dudai Y, Jan YN, Byers D, Quinn WG, Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc Natl Acad Sci U S A. 1976;73(5):1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB. Rescue of the learning defect in dunce, a Drosophila learning mutant, by an allele of rutabaga, a second learning mutant. Proc Natl Acad Sci U S A. 1990;87(7):2795–2799. doi: 10.1073/pnas.87.7.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M, Montarolo PG, Kandel ER. A novel intermediate stage in the transition between short- and long-term facilitation in the sensory to motor neuron synapse of aplysia. Neuron. 1995;14(2):413–420. doi: 10.1016/0896-6273(95)90297-x. [DOI] [PubMed] [Google Scholar]
- Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279(5352):870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- Giese KP, Mizuno K. The roles of protein kinases in learning and memory. Learn Mem. 2013;20(10):540–552. doi: 10.1101/lm.028449.112. [DOI] [PubMed] [Google Scholar]
- Grotewiel MS, Beck CD, Wu KH, Zhu XR, Davis RL. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391(6666):455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- Hadziselimovic N, Vukojevic V, Peter F, Milnik A, Fastenrath M, Fenyves BG, Stetak A. Forgetting Is Regulated via Musashi-Mediated Translational Control of the Arp2/3 Complex. Cell. 2014;156(6):1153–1166. doi: 10.1016/j.cell.2014.01.054. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Kandel ER, Bailey CH. Molecular mechanisms of memory storage in Aplysia. Biol Bull. 2006;210(3):174–191. doi: 10.2307/4134556. [DOI] [PubMed] [Google Scholar]
- Hirao K, Hata Y, Ide N, Takeuchi M, Irie M, Yao I, Takai Y. A novel multiple PDZ domain-containing molecule interacting with N-methyl-D-aspartate receptors and neuronal cell adhesion proteins. J Biol Chem. 1998;273(33):21105–21110. doi: 10.1074/jbc.273.33.21105. [DOI] [PubMed] [Google Scholar]
- Hodges JR, Patterson K. Is semantic memory consistently impaired early in the course of Alzheimer’s disease? Neuroanatomical and diagnostic implications. Neuropsychologia. 1995;33(4):441–459. doi: 10.1016/0028-3932(94)00127-b. [DOI] [PubMed] [Google Scholar]
- Inoue A, Sawatari E, Hisamoto N, Kitazono T, Teramoto T, Fujiwara M, Ishihara T. Forgetting in C. elegans is accelerated by neuronal communication via the TIR-1/JNK-1 pathway. Cell Rep. 2013;3(3):808–819. doi: 10.1016/j.celrep.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Irvine EE, Vernon J, Giese KP. AlphaCaMKII autophosphorylation contributes to rapid learning but is not necessary for memory. Nat Neurosci. 2005;8(4):411–412. doi: 10.1038/nn1431. [DOI] [PubMed] [Google Scholar]
- Jin I, Kandel ER, Hawkins RD. Whereas short-term facilitation is presynaptic, intermediate-term facilitation involves both presynaptic and postsynaptic protein kinases and protein synthesis. Learn Mem. 2011;18(2):96–102. doi: 10.1101/lm.1949711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonides J, Lewis RL, Nee DE, Lustig CA, Berman MG, Moore KS. The mind and brain of short-term memory. Annu Rev Psychol. 2008;59:193–224. doi: 10.1146/annurev.psych.59.103006.093615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang BT, Gu C, Starnes L, Palladino F, Goga A, Kennedy S, L’Etoile ND. Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell. 2013;154(5):1010–1022. doi: 10.1016/j.cell.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294(5544):1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8(5):e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2011;(49) doi: 10.3791/2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61(1):57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, Bargmann CI. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36(6):1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- Lau HL, Timbers TA, Mahmoud R, Rankin CH. Genetic dissection of memory for associative and non-associative learning in Caenorhabditis elegans. Genes Brain Behav. 2013;12(2):210–223. doi: 10.1111/j.1601-183X.2012.00863.x. [DOI] [PubMed] [Google Scholar]
- Lee JI, O’Halloran DM, Eastham-Anderson J, Juang BT, Kaye JA, Scott Hamilton O, L’Etoile ND. Nuclear entry of a cGMP-dependent kinase converts transient into long-lasting olfactory adaptation. Proc Natl Acad Sci U S A. 2010;107(13):6016–6021. doi: 10.1073/pnas.1000866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Timbers TA, Rose JK, Bozorgmehr T, McEwan A, Rankin CH. The FMRFamide-related neuropeptide FLP-20 is required in the mechanosensory neurons during memory for massed training in C. elegans. Learn Mem. 2013;20(2):103–108. doi: 10.1101/lm.028993.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Roberts AC, Glanzman DL. Synaptic facilitation and behavioral dishabituation in Aplysia: dependence on release of Ca2+ from postsynaptic intracellular stores, postsynaptic exocytosis, and modulation of postsynaptic AMPA receptor efficacy. J Neurosci. 2005;25(23):5623–5637. doi: 10.1523/JNEUROSCI.5305-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tully T, Kalderon D. Effects of a conditional Drosophila PKA mutant on olfactory learning and memory. Learn Mem. 1996;2(6):320–333. doi: 10.1101/lm.2.6.320. [DOI] [PubMed] [Google Scholar]
- Michel M, Green CL, Gardner JS, Organ CL, Lyons LC. Massed training-induced intermediate-term operant memory in aplysia requires protein synthesis and multiple persistent kinase cascades. J Neurosci. 2012;32(13):4581–4591. doi: 10.1523/JNEUROSCI.6264-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri A, Kodama E, Kimura KD, Koike M, Mizuno T, Mori I. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics. 2005;169(3):1437–1450. doi: 10.1534/genetics.104.036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PV, Woo NH. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog Neurobiol. 2003;71(6):401–437. doi: 10.1016/j.pneurobio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Parvez K, Stewart O, Sangha S, Lukowiak K. Boosting intermediate-term into long-term memory. J Exp Biol. 2005;208(Pt 8):1525–1536. doi: 10.1242/jeb.01545. [DOI] [PubMed] [Google Scholar]
- Pereira S, van der Kooy D. Two forms of learning following training to a single odorant in Caenorhabditis elegans AWC neurons. J Neurosci. 2012;32(26):9035–9044. doi: 10.1523/JNEUROSCI.4221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J Exp Biol. 2001;204(Pt 10):1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- Salthouse TA. Theoretical perspectives on cognitive aging. Hillsdale, N.J: L. Erlbaum Associates; 1991. [Google Scholar]
- Sanderson DJ, Good MA, Seeburg PH, Sprengel R, Rawlins JN, Bannerman DM. The role of the GluR-A (GluR1) AMPA receptor subunit in learning and memory. Prog Brain Res. 2008;169:159–178. doi: 10.1016/S0079-6123(07)00009-X. [DOI] [PubMed] [Google Scholar]
- Shuai Y, Lu B, Hu Y, Wang L, Sun K, Zhong Y. Forgetting is regulated through Rac activity in Drosophila. Cell. 2010;140(4):579–589. doi: 10.1016/j.cell.2009.12.044. [DOI] [PubMed] [Google Scholar]
- Squire L, Kandel E. Memory: From Mind to Molecules. 1. Roberts and Company Publishers; 2008. [Google Scholar]
- Stein GM, Murphy CT. The Intersection of Aging, Longevity Pathways, and Learning and Memory in C. elegans. Front Genet. 2012;3:259. doi: 10.3389/fgene.2012.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A, Horndli F, Maricq AV, van den Heuvel S, Hajnal A. Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS One. 2009;4(6):e6019. doi: 10.1371/journal.pone.0006019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Masters SE, Bagnall MW, Carew TJ. Molecular mechanisms underlying a unique intermediate phase of memory in aplysia. Neuron. 2001;31(1):143–154. doi: 10.1016/s0896-6273(01)00342-7. [DOI] [PubMed] [Google Scholar]
- Timbers TA, Rankin CH. Tap withdrawal circuit interneurons require CREB for long-term habituation in Caenorhabditis elegans. Behav Neurosci. 2011;125(4):560–566. doi: 10.1037/a0024370. [DOI] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol. 2011;7(2):e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Gschwind L, Vogler C, Demougin P, de Quervain DJ, Papassotiropoulos A, Stetak A. A role for alpha-adducin (ADD-1) in nematode and human memory. EMBO J. 2012;31(6):1453–1466. doi: 10.1038/emboj.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Phan T, Storm DR. The type 3 adenylyl cyclase is required for novel object learning and extinction of contextual memory: role of cAMP signaling in primary cilia. J Neurosci. 2011;31(15):5557–5561. doi: 10.1523/JNEUROSCI.6561-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wixted JT. The psychology and neuroscience of forgetting. Annu Rev Psychol. 2004;55:235–269. doi: 10.1146/annurev.psych.55.090902.141555. [DOI] [PubMed] [Google Scholar]
- Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Sakmann B. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science. 1999;284(5421):1805–1811. doi: 10.1126/science.284.5421.1805. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438(7065):179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]