Abstract
As part of a project aimed at obtaining selective inhibitors and drug-like compounds targeting tRNA synthetases from trypanosomatids, we have elucidated the crystal structure of human cytosolic histidyl-tRNA synthetase (Hs-cHisRS) in complex with histidine in order to be able to compare human and parasite enzymes. The resultant structure of Hs-cHisRS·His represents the substrate-bound state (H-state) of the enzyme. It provides an interesting opportunity to compare with ligand-free and imidazole-bound structures Hs-cHisRS published recently, both of which represent the ligand-free state (F-state) of the enzyme. The H-state Hs-cHisRS undergoes conformational changes in active site residues and several conserved motif of HisRS, compared to F-state structures. The histidine forms eight hydrogen bonds with HisRS of which six engage the amino and carboxylate groups of this amino acid. The availability of published imidazole-bound structure provides a unique opportunity to dissect the structural roles of individual chemical groups of histidine. Remarkably, the analysis revealed the importance of the amino and carboxylate groups, of the histidine in leading to these dramatic conformational changes of the H-state. Further, comparison with previously published trypanosomatid HisRS structures reveals a pocket in the F-state of the parasite enzyme that may provide opportunities for developing specific inhibitors of Trypanosoma brucei HisRS.
Keywords: histidyl-tRNA synthetase, drug design, trypanosome, sleeping sickness, Chagas disease
1. Introduction
Aminoacyl-tRNA synthetases (aaRS) form a group of ubiquitous and essential enzymes which are of critical importance involved in protein synthesis. Generally, each specific aaRS catalyzes the reaction of attaching the carboxylate group of a specific amino acid to the 2′-OH or 3′-OH of its cognate tRNA. Subsequently, the charged tRNA will recognize the anticodon of the mRNA at the ribosome for the incorporation of amino acids into the growing polypeptide chain [1]. Therefore, the coupling of amino acids to the correct tRNA by aaRS is one of the most important events in ensuring the efficiency and fidelity of protein synthesis. Usually, the charging of a tRNA by aaRS occurs in two steps: (1) recognition of the amino acid and ATP to form an active aminoacyl-adenylate intermediate, and (2) recognition of the cognate tRNA followed by transfer of the aminoacyl group to the terminal ribose 2′- or 3′-OH of acceptor stem. Interference with aaRS function leads to the disruption of protein chain elongation during translation. Consequently, aaRS enzymes are validated drug targets in the development of new anti-infectives [2–6].
During the aminoacylation reaction, the active site of aaRS enzymes need to specifically recognize multiple substrates (amino acid, ATP, tRNA), catalyze at least two reactions and release two products (pyrophosphate and charged tRNA). Structural requirements to ensure sufficient binding affinity, specificity and catalytic efficiency are often achieved through conformational changes as the aaRS enzymes proceed through different conformational states when catalyzing the steps mentioned above. Snapshots of these states are typically captured by crystallization of the enzyme in complex with different substrates or products. For example, multiple structures of methionyl-tRNA synthetase described a series of active site rearrangements and domain motions ranging from ligand-free, to substrate-bound, intermediate product-bound, and to inhibitor-bound states (see for examples [7–11]). Similarly, crystal structures of tryptophanyl-tRNA synthetases revealed transitions between multiple conformational states depending on the bound ligands. Crucially, bacterial and eukaryotic (cytoplasmic) tryptophanyl-tRNA synthetases appear to follow two different paths of conformational changes during the aminoacylation reactions (see for examples [12–18]). Therefore, structures of the aaRS in complex with different ligands are important in understanding how functional states of the enzymes are tied to conformational changes. In addition, information gained from studies of bacterial aaRS may not always be extended to their eukaryotic counterparts.
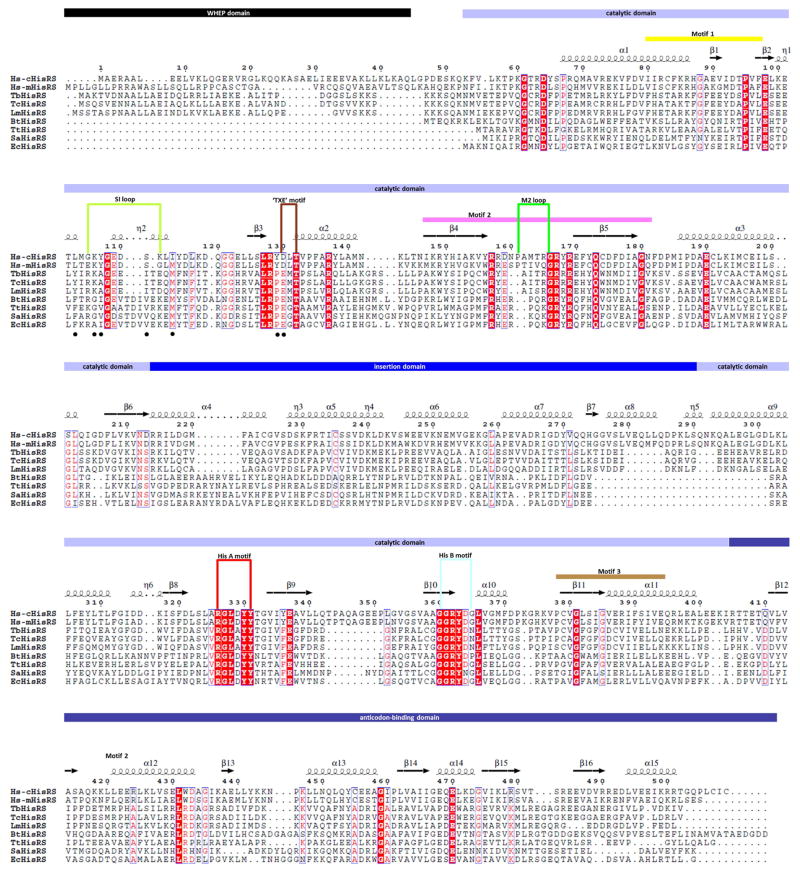
AaRS enzymes are grouped into two classes, class I and class II, based on the degree of sequence similarity and consensus structural motifs [19]. Class II catalytic domains share an antiparallel β-sheet core and three conserved motifs (motifs 1, 2, and 3) [19]. Here we focus on histidyl-tRNA synthetases (HisRSs), a class II enzyme. HisRSs are grouped into the IIa subclass of aaRS based on sequence characteristics in the anticodon-binding domain [20]. Unique to HisRS are two conserved motifs, HisA (RGLDYY) and HisB (GGRYDG) [21] (Fig. 1). The HisRS subunit contains at least three domains: (1) the catalytic domain with an antiparallel β-sheet core; (2) the C-terminal anticodon binding domain with a α/β-fold; and (3) the insertion domain. The folds of the catalytic and anticodon binding domains are conserved across species but the insertion domain is less so (see for examples [22, 23]). In addition to these three domains, a 50-residue WHEP domain is appended to some eukaryotic HisRS [24] (Fig. 1). In higher eukaryotes like humans, there are at least two HisRS genes, called HARS and HARS2, presumably encoding for the cytosolic and mitochondrial versions of the enzyme, respectively [25].
Figure 1. Sequence alignment of HisRS.
Sequences of HisRS from various species discussed in the manuscript are aligned with Clustal Omega [52] and corresponding secondary structures are annotated based on the current Hs-cHisRS·His structure. Figure is prepared using the ESPript 3.0 server [53]. Domain boundary and structural features are indicated based on previous reports [23, 26]. The black dots under the aligned sequences indicate residues contributing side chains that delineate the pocket F as identified in TbHisRS structure (see Section 3.4). Sequence names are represented as following: human cytosolic HisRS: Hs-cHisRS; human mitochondrial HisRS: Hs-mHisRS; Trypanosoma brucei HisRS: TbHisRS; Trypanosoma cruzi HisRS: TcHisRS; Leishmania major HisRS: LmHisRS; Thermus thermophilus HisRS: TtHisRS; Staphylococcus aureus HisRS: SaHisRS; Escherichia coli HisRS: EcHisRS; Burkholderia thailandensis HisRS: BtHisRS.
Available crystal structures reveal a homodimer arrangement of HisRS. Structures of three different major HisRS states have been published earlier: (i) The ligand-free enzymes from Staphylococcus aureus, Thermus thermophilus and Trypanosoma brucei (hereafter called the F-state) [22, 26, 27]; (ii) the enzymes from T. thermophilus, Trypanosoma.cruzi, Escherichia coli and Burkholderia thailandensis in complex with the substrate histidine or histidinol (H-state) [22, 28–30], and (iii) HisRS from T. thermophilus, E. coli and T. cruzi in complex with the intermediate product histidyl-adenylate (HAMP, A-state) [21, 22, 28, 29]. These structures provide detailed information about the complexes and the conformational changes occurring during the first step of the HisRS aminoacylation reaction. F-state HisRS has an open active site. Binding of histidine results in the H-state, characterized by a compact histidine binding pocket, that is closed by the HisA motif and a nearby 11 residues-long loop, called the ‘small interface loop’ (SI loop) [26] or the ‘ordering loop’ [27]. These changes provide not only a well-defined binding site for histidine, but also place the catalytic Arg (Arg326 in human cytosolic HisRS) in a proper position to accelerate the reaction. Subsequently, changes in the active site are propagated throughout the HisRS dimer by co-operative and concerted movements, stabilizing and positioning the ATP-binding motif 2 loop (M2 loop), the insertion domain and the anticodon binding domain in the dimer for subsequent steps of the overall reaction [26].
The understanding of conformational changes in HisRS so far has been mainly based on crystal structures from prokaryotes and lower eukaryotes as described above. Recently, two crystal structures of human cytosolic HisRS (Hs-cHisRS) have been reported [23]. Crystals used to solve the structures were crystallized without addition of ligands for HisRS (PDB: 4G84 and 4G85). In one of the structures (PDB: 4G84), imidazole, present in the crystallization buffer, is bound in the open active site typical of F-state HisRS structures and hence denoted here as the “F-state Hs-cHisRS·imidazole” complex. The position of the imidazole appeared to correspond with the imidazole group histidine when compared to other HisRS·His structures. No ligand was present in the other reported Hs-cHisRS structure (PDB: 4G85) and will be denoted here as “F-state Hs-cHisRS”. The Hs-cHisRS·imidazole complex exhibits the structural features of a typical F-state of HisRS, namely the open HisA motif, disordered parts of the SI loop, and flexible segments of the M2 loop [26, 27]. No Hs-cHisRS structure in complex with any of its natural substrates has been reported. It is not clear if structural changes occurring in prokaryotic HisRS enzymes upon histidine binding are relevant in eukaryotic Hs-cHisRS.
In our efforts to develop specific inhibitors of trypanosomatid aaRS through structure-guided designs [7, 8, 18, 22, 31, 32], we determined the structure of Hs-cHisRS enzyme in complex with histidine (Hs-cHisRS·His) for comparison with trypanosomatid aaRS. Here we report the crystal structure of the H-state Hs-cHisRS·His complex and examine the structural plasticity of Hs-cHisRS during the first step of the aminoacylation reaction. We also arrive at suggestions for selective inhibitor design of trypanosomatid HisRS by comparing this histidine-containing complex of human cytosolic HisRS with the histidine-containing complex of T. cruzi HisRS (TcHisRS·His) and by comparing the ligand-free structures of human cytosolic HisRS and T. brucei HisRS (TbHisRS).
2. Materials and methods
2.1. Protein expression and purification
The cDNA clone of human cytosolic HisRS (NCBI Reference sequence NM_002109) was purchased from Origene (Rockville, MD, USA) and PCR amplified with primers designed to clone the open-reading frame into the vector AVA0421 by ligation-independent cloning (LIC). AVA0421 is a LIC-ready, pET14b modified expression vector, with N-terminal hexa-histidine tag and a modified human rhinovirus 3C protease recognition site [33, 34]. Protein was expressed in E. coli BL21 (DE3) host cells in autoinduction media [35] and purified using a Ni-NTA affinity column. The N-terminal hexa-histidine tag was cleaved off using human rhinovirus 3C protease at 4°C. Cleaved protein, collected in the flow through during a second Ni-NTA affinity purification step, was further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare Bio-Sciences, USA) in a buffer containing 25 mM HEPES, 500 mM NaCl, 2 mM DTT, 5% glycerol, and 0.025% NaN3 at pH 7.0. Purified protein retained four residues of the 3C protease cleavage site (GPGS) at the N-terminus.
2.2. Crystallization and data collection
Purified Hs-cHisRS, at a concentration of 8.2 mg/ml, was used for crystallization trials by vapor diffusion in the presence of 10 mM histidine. An initial hit was obtained when the protein was mixed at 1:1 ratio with a reservoir solution condition containing 0.1 M Tris pH 8.5, 0.2 M MgCl2 and 25% PEG 3350 at both room temperature and 4°C. Subsequent optimization of the conditions between pH 7 to 9, and 20 to 32.5 % of PEG 3350, yielded many chisel-shape crystals often with split ends. Crystals were cryo-protected in the mother liquor solution supplemented with 20 % glycerol and 10 mM histidine, and flash-frozen in liquid nitrogen. Diffraction data were collected under cryogenic conditions at SSRL beam line 12-2. Only a few crystals diffracted to better than 3 Å showing multiple lattices presumably related to the split ends of the crystals. Eventually the best dataset was collected using the 12-2 microfocus beam from a segment identified by raster-scanning the crystal. Data were integrated using XDS [36] and scaled with Scala to 2.84 Å resolution [37].
2.3. Structure solution and refinement
A search model was prepared using the structure of TcHisRS (28 % sequence identity) in complex with histidine (PDB: 3LC0). 3LC0 has one half of the HisRS dimer in the asymmetric unit and contains the catalytic domain, insertion domain and anticodon binding domain. A biological dimer of TcHisRS was downloaded from the PDBePISA server [38]. Non-identical residues were truncated beyond the Cβ atom using CHAINSAW [39] and solvent and ligand coordinates were removed to generate the search model. The structure was solved by molecular replacement using Phaser [40] in space group P212121 with four subunits in one asymmetric unit. Manual model building/rebuilding was carried out using Coot [41]. Refinement was carried out with REFMAC5 [42], utilizing global non-crystallographic symmetry restraints and translational/libration/screw (TLS) groups identified by the TLS motion determination server [43]. Throughout the model building process, the structure validation server MolProbity [44] was used to monitor the geometry of the model. The final crystallographic refinement statistics are given in the Table 1. Figures were created and rendered with Pymol [45]. Superposition of structures was carried out using the secondary structure matching algorithm [46]. Relative motion of protein domains are analyzed by DynDom [47]. Coordinates and structure factors are deposited in the Protein Data Bank under PDB entry ID 4PHC.
Table 1.
Crystallographic data collection and refinement statistics.
| Data collection & processing | |
|
| |
| Beamline | SSRL BL12-2 |
| Wavelength (Å) | 0.98 |
| Space group | P212121 |
| Unit cell parameters [a, b, c (Å); α=β= γ (°)] | 89.1, 93.0, 261.1; 90 |
| Resolution (Å) | 39.4 – 2.84 |
| Unique reflections | 49252 (4201) |
| Completeness (%) | 95.3 (89.4) |
| Rmerge | 0.15 (0.98) |
| Rpim | 0.075 (0.51) |
| CC1/2 | 0.99 (0.57) |
| Mean I/σ(I) | 9.0 (1.6) |
| Multiplicity | 4.3 (3.9) |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 39.4 – 2.84 |
| Rwork | 0.200 |
| Rfree | 0.238 |
| No. reflections; refinement/test set | 46705 / 2504 |
| RMSD bonds (Å) | 0.007 |
| RMSD angles (°) | 1.15 |
| No. atoms (protein/histidine/glycerol/water) | 13960/44/55/24 |
| Residues in favoured regions (%)# | 98.9 |
| Residues in allowed regions (%) | 1.1 |
| Residues in disallowed regions (%) | 0 |
| Average B factors for atoms (protein/histdine/glycerol/water) (Å2) | 62.9/48.2/34.3/54.1 |
| PDB entry | 4PHC |
Values in parenthesis are for the highest resolution shell
Ramachandran Plot statistics are as reported by the Molprobity server
Well-defined electron density is present for most parts of the protein in all four chains of Hs-cHisRS except the N-terminal WHEP domains (52 residues). In addition, in all four chains, five residues at the C-termini, and between eight to twelve residues in two internal loops (Pro344 to Gly355; Glu401 to Glu402), are disordered. These loops are not directly involved in histidine or ATP binding.
2.4 Thermal stability assay
The fluorescence-based thermal stability assay was performed using 2000x diluted thermofluor SYPRO® orange dye (Sigma), 0.5 mg/ml of Hs-cHisRS, TcHisRS or TbHisRS, in the presence or absence of 10 mM L-histidine. Assay solutions were heated in steps of 0.2 °C from 20 °C to 90 °C in a real-time PCR machine. Fluorescence readings (excitation window, 470–505 nm; absorption window, 540–700 nm) were taken to monitor the change in fluorescence intensity. The resulting curves were smoothen using the Opticon Monitor software (Bio-Rad) and melting temperatures (Tm) were determined as the temperature where the first derivative of the curves were the largest. The assays were performed twice in each condition and average Tm values are reported.
3. Results and discussion
3.1. Overall structure
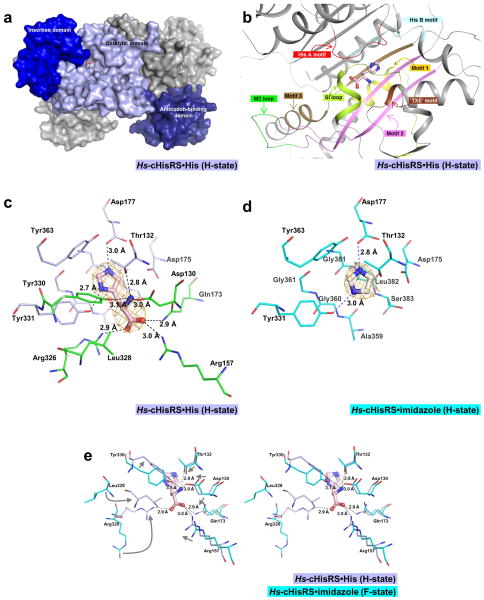
H-state Hs-cHisRS is a homodimer with 509 residues per subunit (Fig. 2a). The crystal structure at 2.8 Å resolution has good crystallographic and model quality (Table 1). The current crystal structure contains two dimers per asymmetric unit. Chains A and B form one dimer, which superimposes onto the second dimer in the asymmetric unit, formed by chains C and D, with an rmsd of 0.77 Å. Among the four chains in the asymmetric unit, pairwise values rmsd vary between 0.30 and 0.90 Å. Chain A differs most from the other three chains with rmsd values between 0.79 and 0.90 Å, while chains B, C and D are very similar to each other, with rmsd values between 0.30 and 0.54 Å. The main difference between chain A and others is in the insertion domain (Arg214 to Ser290), which in chain A rotate ~10° away from the active site compared to chain B. Chain B will be used as the representative structure to compare with other HisRS structures.
Figure 2. Histidine binding pocket of Hs-cHisRS·His.
a. Surface representation of an Hs-cHisRS·His dimer. The three domains of a monomer are colored in light blue (catalytic domain), blue (insertion domain) and deep blue (anticodon-binding domain). Bound histidine is shown in ball and stick model to indicate the location of the active site. The second monomer is colored gray.
b. Important structural features of HisRS involved in the binding of histidine, with their relative positions with respect to the bound histidine as described in section 3.2. Corresponding sequences of these structural features are annotated in Fig. 1 using the same color scheme.
c. Representative σA-weighted Fobs-Fcalc difference electron density map calculated by omitting histidine is shown at 3σ level. Residues within 4.5Å radius of the bound histidine are shown in sticks. The bound histidine forms eight H-bonds with the protein as indicated by the dash line. Residues that form the imidazole moiety binding site, and are not shifted in positions upon histidine binding, as compared to Fig. 2d, are colored light blue. Residues that are shifted in positions to fully form the histidine binding pocket are colored in green.
d. The σA-weighted Fobs-Fcalc difference electron density map for Hs-cHisRS·imidazole (PDB: 4G84) calculated by omitting imidazole is shown at 3σ level. Residues within 4.5Å radius of the bound imidazole are shown in sticks. Residues that form the imidazole binding site are colored cyan. Compared to Fig. 2c, residues that interact with imidazole moiety do not display significant changes in conformation.
e. Stereo pair showing the superpositions of Hs-cHisRS·imidazole (PDB: 4G84, cyan) and Hs-cHisRS·His (light blue) to indicate the movement of residues (indicated by arrows in the left view) in the active site when the protein changes from F-state to H-state. Significant movements occurred in residues that interact with the amino and carboxylate groups of the bound histidine, mainly through six H-bonds.
Globally, the dimer organization and the overall fold of the Hs-cHisRS·His structure is similar to other reported HisRS structures, especially in the highly conserved catalytic domain and anticodon binding domain (Fig. 2a). The largest difference between H-state Hs-cHisRS·His with other HisRS structures occurs in the insertion domain. Structural conservation in this domain is low among different species, reflecting the low sequence similarity [22, 23]. The orientation of the domain in different structures is also highly variable, without correlations to the ligands bound in the active site of the protein, similarly noted in previous reports [22, 23].
3.2. Histidine-binding pocket
Important structural features of HisRS involved in the binding of histidine, specifically motifs 1, 2, 3, HisA and HisB, the loops SI, M2 and ‘TXE’, are well defined in the electron density and depicted in Fig. 2b. The bound histidine is visible with clear density in all four chains (Fig. 2c). In the Hs-cHisRS·His complex, the histidine is bound in a deep pocket formed by residues of (i) motifs 2 and 3 at the ‘bottom’; (ii) the Class II aaRS signature ‘TXE’ loop (DLT in Hs-cHisRS) and the HisRS specific HisB motif on the ‘sides’; and (iii) another HisRS specific motif, HisA, which partially covers the bound histidine on the “top” (Fig. 2b). Histidine makes eight H-bonds with the protein. These H-bonds are between: (i) the histidine imidazole nitrogens and the side chains of Asp177 and Tyr331; (ii) the amino group of the histidine and the side chains of Asp130, Thr132 and Tyr330; and, (iii) the carboxylate group of the histidine and the side chains of Arg157, Gln173 and Arg326. In addition, the imidazole ring of histidine stacks against a flat surface formed by residues Ala359, Gly360 and Gly361 in β-strand 10, and residues Gly381, Leu382 and Ser383 in β-strand 11 (Fig. 2c). Clearly, each of the three moieties of the bound histidine, imidazole, amino and carboxylate groups, make specific interactions with the enzyme.
In the current Hs-cHisRS·His structure, the interactions of the amino and carboxylate groups of histidine with the enzyme are very extensive compared to other structures of histidine bound to HisRS. For example, the histidine amino group makes one more H-bond to Hs-cHisRS than observed in the T. thermophilus HisRS·His (TtHisRS·His) structure [28]. Similarly, the histidine carboxylate group makes more H-bonds with Hs-cHisRS (three) than any other known structure of H-state HisRS; i.e. TtHisRS·His (one water mediated H-bond) [28], TcHisRS·His (one H-bond) [22] and B. thailandensis HisRS (BtHisRS·His, two H-bonds) [30] complexes. Unique to Hs-cHisRS·His is the interaction between the guanidinium group of Arg157 and the carboxylate group of histidine. This H-bond stabilizes Arg157 in its position in the current Hs-cHisRS·His. Histidine increases the melting temperature (Tm) of Hs-cHisRS by 5.6 °C compared to increments of 3.3 °C and 4 °C in TcHisRS and TbHisRS, respectively. Therefore, at least in comparison to TcHisRS·His, the extra H-bonds between the carboxylate group of histidine and guanidinium groups of Arg157 and Arg326 appear to strengthen the binding between histidine and Hs-cHisRS and stabilizing the enzyme. The stabilization of Arg157 in its position is an important structural feature that has implications for deriving selective inhibitors of trypanosomatid HisRS, which will be discussed later in this manuscript.
3.3. Comparison between F-state and H-state Hs-cHisRS conformations
The four crystallographically independent subunits of the current H-state Hs-cHisRS·His structure superpose onto the ligand-free Hs-cHisRS (PDB: 4G85) and Hs-cHisRS·imidazole (PDB: 4G84) structures with quite substantial rmsd values ranging from 1.4 to 2.3 Å. In contrast, the ligand-free Hs-cHisRS (PDB: 4G85) and Hs-cHisRS·imidazole (PDB: 4G84) structures are similar to each other, with rmsd values between 0.86 to 1.1 Å. Interactions between the protein and the imidazole moiety of histidine are essentially the same as between protein and free imidazole in the respective complexes (Fig. 2d). However, the presence of imidazole alone is not enough to fully form the histidine binding pocket observed in the H-state of Hs-cHisRS. Comparing Hs-cHisRS·His, ligand-free Hs-cHisRS (PDB: 4G85) and Hs-cHisRS·imidazole (PDB: 4G84) in the active site, conformational changes occurred in Hs-cHisRS·His, bringing six residues, i.e. Asp130, Arg157, Gln173, Arg326, Leu328 and Tyr330, within a 4.5 Å radius of the histidine. The shifts of the six residues towards the bound histidine result in new H-bonds between these residues (except Leu328) and the amino and carboxylate groups of histidine (Fig. 2e). While Hs-cHisRS·imidazole is similar to ligand-free Hs-cHisRS, it differs from Hs-cHisRS·His in active site conformation. Therefore, both Hs-cHisRS·imidazole and ligand-free Hs-cHisRS can be denoted as F-state Hs-cHisRS.
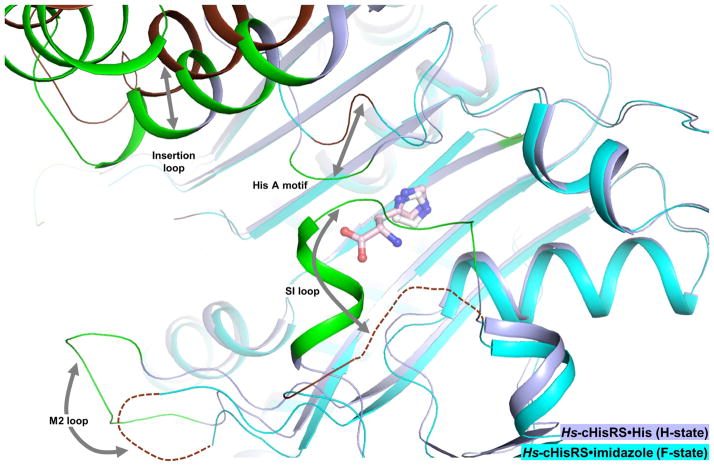
Globally, in addition to the flexible insertion domain, there are major differences between these F-state and H-state structures, mainly occurring in three motifs and loops surrounding the active site (Fig. 3):
Figure 3. Global changes in Hs-cHisRS conformations of F- and H-states.
Hs-cHisRS·imidazole (PDB: 4G84, cyan) and Hs-cHisRS·His (light blue) are superimposed to indicate global changes in Hs-cHisRS conformations of F- and H-states. Residues that are significantly shifted in positions are colored brown in Hs-cHisRS·imidazole (F-state) and green in Hs-cHisRS·His (H-state) with relative movements indicated by arrows.
The HisA motif (Arg326 to Tyr331), open in the F-state, but closed in the H-state.
The SI loop (Gly105 to Lys112), disordered in the F-state, but ordered in the H-state.
The M2 loop (Pro161 to Gly166), disordered in the F-state, but ordered in the H-state.
These conformational changes are all inter-related, involving a large network of interactions, with the bound histidine at the center in the H-state.
Changes within the HisA motif resulted in a closed active site, covering the ‘top’ of the active site in the H-state (Fig. 3). Arg326 and Tyr330 form H-bonds with the carboxylate and amino groups of histidine as described above (Fig. 2e). Arg326 also forms an H-bond with Tyr331 to stabilize the HisA motif. The guanidinium group of Arg326 moved by an average of 7.5 Å in H-state. The movement of Arg326 into the proximity of carboxylate group of histidine is particularly important, since this residue is proposed to act as the electrophile during catalysis [29].
Another key characteristic of the current structure is that, on the opposite side of the HisA motif, the SI loop is ordered in the H-state (Fig. 3). Residues in the SI loop are not directly in contact with bound histidine. Instead, the stabilization of the SI loop is mainly due to: (i) contacts between the tip of the loop with the HisA motif and the TXE loop; (ii) a H-bond between the backbone oxygen of Tyr107 (SI loop) and the backbone nitrogen of Asp329 (HisA motif); and (iii) a H-bond between the side chain carboxylate of Asp110 (SI loop) and the backbone oxygen of Gly327 (HisA motif). The insertion of SI loop residue Tyr107 into a hydrophobic pocket formed by Leu328 and Tyr330 (both from the HisA motif) and Tyr129, Asp130, Val133 (all part of the ‘TXE’ loop) also appears to be important. In contrast, in F-state Hs-cHisRS, the HisA motif is open, and Tyr129 in the TXE loop adopts a different rotamer that would directly clash with Tyr107 of an ordered SI loop in H-state. Therefore, the stabilization of the SI appears to be a second shell effect of histidine binding, resulting from interactions with histidine-contacting HisA motif and the ‘TXE’ loop.
Not all known H-states of HisRS exhibit the ordered M2 loop. While the M2 loops of Hs-cHisRS·His and TtHisRS·His are ordered, the M2 loops of TcHisRS·His and BtHisRS·His are not. In the Hs-cHisRS·His structure, the M2 loop, connecting strands β4 and β5 of the catalytic domain, is located within the Class II aaRS motif 2 and is next to the SI loop. Many residues flanking the M2 loop, such as Arg157, Asp167 and Phe171 are important for interactions with ATP or HAMP, as observed in A-state HisRS structures [21, 22, 28, 29]. Therefore, the ordering of the M2 loop appears to be a structural hallmark of the A-state when ATP or HAMP is bound [21, 22, 28, 29], but not necessarily in the H-state when histidine is bound [22, 28, 30].In the current Hs-cHisRS·His structure, crystal contacts are involved in the stabilization of the M2 loops in two of the four subunits in the asymmetric unit, but not the other two subunits (chains B and C). No direct contacts can be observed between residues in the M2 loop and other parts of the protein, nor with the bound histidine. Therefore, residues flanking the M2 loop might be important in anchoring the loop in its ordered conformation in H-state (Fig. 3). For example, in the current H-state structure, residues Arg157 and Gln173 each make one H-bonds with the carboxylate group of the bound histidine. In contrast, in F-state Hs-cHisRS, the H-bond between the guanidinium group of residues equivalent to Arg157 and the carboxylate group of histidine is absent and the M2 loop is disordered. Therefore, while the ordering of the M2 loop in the H-state of Hs-cHisRS remains intriguing, its functional implication is not clear.
The closed conformation of the HisA motif and the ordered SI loop in H-state Hs-cHisRS is similar to characteristics of prokaryotic HisRS in complex with histidine. In bacterial HisRS, the ordering of the SI loop was proposed to affect the HisRS dimer interface, thereby repositioning the C-terminal anticodon-binding domain for tRNA binding through long-range cooperativity [26]. The formation of the His-binding pocket through the stabilization of the HisA motif and the SI loop as well as the correct positioning of catalytic residue Arg326 in the H-state was therefore suggested to be the fidelity mechanism of bacterial HisRS [26]. Analysis of the F- and H-states of Hs-cHisRS structures shows agreement with this model. Therefore, it is likely that eukaryotic Hs-cHisRS exhibits the same fidelity mechanism as bacterial HisRS.
Our structure shows that essentially every atom of the bound histidine is interacting with Hs-cHisRS. Interestingly, several interactions, especially the multiple H-bonds that stabilize the HisA motif and the TXE loop that in turn stabilize the SI loop, are mediated through the amino and carboxylate groups of the bound histidine but not by the imidazole moiety (Fig. 2e). In other words, although the imidazole moiety is unique in histidine among natural amino acids, the H-state of Hs-cHisRS is not stabilized by imidazole alone (Fig. 2c & d). Interestingly, HisRS is not known to be particularly error prone compared to other aaRS, and is able to discriminate histidine from other smaller amino acid such as Ala or Ser without utilization of an editing domain to correct for mis-acylation [48]. This implies that although binding of imidazole by itself does not result in conformational changes postulated to be an important fidelity mechanism, the imidazole ring is nonetheless needed, in combination with the amino and carboxylate group, for obtaining the correct enzyme conformation for the next steps in the aminoacylation reaction in HisRS.
3.4. Opportunities for structure-based drug design
The availability of H- and F-state structures of Hs-cHisRS now enables comparison with H- and F-state structures of trypanosomatid HisRS to identify differences that can possibly be exploited for structure-based design of selective inhibitors, forming a starting point for the development of compounds useful in the battle against sleeping sickness and Chagas disease, two neglected tropical diseases [49].
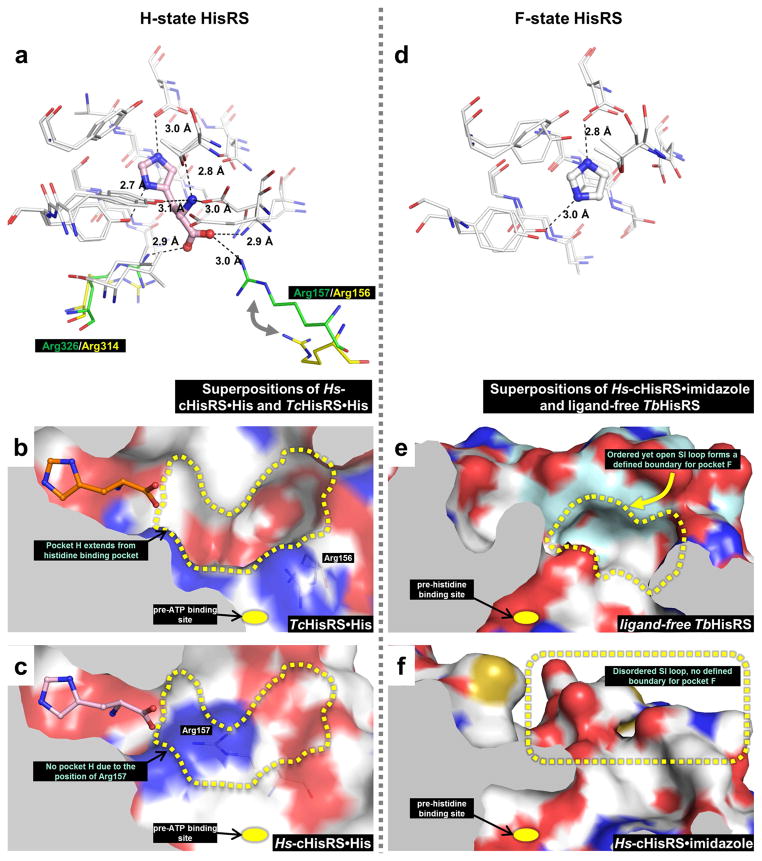
When comparing the H-states of Hs-cHisRS and TcHisRS (PDB: 3LC0) [22], most residues in contact with histidine appear to adopt similar positions (Fig. 4a). However, Arg326 (Arg314 in TcHisRS) and Arg157 (Arg156 in TcHisRS) are exceptions. Both residues form H-bonds with the carboxylate group of histidine in Hs-cHisRS, but not in TcHisRS, as discussed in Section 3.2. In TcHisRS, a guanidinium nitrogen of Arg314 in is 4.1 Å away from the closest histidine carboxylate oxygen and a guanidinium nitrogen of Arg156 is 8.5 Å from the closest carboxylate oxygen. Most interestingly, Arg156 in TcHisRS adopts a different rotamer conformation compared to the equivalent Arg157 in Hs-cHisRS, resulting in a shift of ~ 6.5 Å in the guanidinium group, creating space near the end of the histidine carboxylate group (Fig. 4a). Indeed, superposition of H-states of Hs-cHisRS and TcHisRS reveals a deep pocket extending from the bound histidine to the SI loop in TcHisRS·His but this pocket is not present in Hs-cHisRS·His (Fig. 4b & c). The rotation of Arg156 away from the active site in TcHisRS·His is key to the formation of this pocket. This pocket in TcHisRS, named hereafter ‘pocket H’, points away from where ATP would bind (pre-ATP pocket), and is not a conserved pocket between TcHisRS and Hs-cHisRS. Pocket H in TcHisRS could be a good site for inhibitor binding. Unfortunately, further examination of pocket H reveals a limited druggability as it is largely delineated by charged atoms (Fig. 4b).
Figure 4. Opportunities for structure-based drug design targeting trypanosomatid HisRS.
a. Superposition of H-state Hs-cHisRS·His and TcHisRS·His (PDB: 3LC0) indicates that most of the active site residues adopts similar positions (white) except for two residues. Arg326 (Hs-cHisRS·His, green) and Arg157 (Hs-cHisRS·His, green) both forms H-bonds with carboxylate group of bound histidine (only histidine bound to Hs-cHisRS is shown). The equivalent residues in TcHisRS·His are Arg314 and Arg156 (both colored yellow), respectively, and do not form H-bond with bound histidine.
b. In TcHisRS·His, the difference in the position of Arg156 contributed to the formation of a continuous, deep pocket extending from the carboxylate group of the bound histidine (orange). The pocket, named pocket H (yellow dash line), is away from the site where ATP is going to bind to (yellow dot). However, pocket H is largely delineated by charged atoms (carbon: white; nitrogen: blue; oxygen: red).
c. In Hs-cHisRS·His, Arg157 occupies a large part of the equivalent position pocket H (yellow dash line). Therefore, an inhibitor that binds to the pocket H in TcHisRS may bind to Hs-cHisRS with lower affinity, resulting in selectivity for TcHisRS.
d. Superposition of F-state Hs-cHisRS·imidazole (PDB: 4G84) and ligand-free TbHisRS (PDB: 3HRI) indicates that most of the active site residues adopts similar positions (white). The bound imidazole of F-state Hs-cHisRS·imidazole is shown.
e. In ligand-free TbHisRS, the ordered yet open SI loop (light blue) forms a defined boundary for a pocket next to where histidine is going to bind (yellow dot). The pocket, named pocket F (yellow dash line), is largely delineated by non-polar atoms (carbon: white or light blue; nitrogen: blue; oxygen: red).
f. In Hs-cHisRS·imidazole, the SI loop is disordered (note the lack of surface colored light blue which is used to indicate SI loop). This results in a lack of defined boundary for the equivalent of pocket F. A pocket F-binding inhibitor for TbHisRS will not only perturb movement of SI loop required during aminoacylation, but is also not likely to bind well to Hs-cHisRS due to the lack of pocket F, possibly resulting in selectivity for TbHisRS.
Comparison of F-state Hs-cHisRS (PDB: 4G84) [23] and F-state T. brucei HisRS (TbHisRS, PDB: 3HRI) [22] shows that the most striking difference (Fig. 4d–f) is the conformation of the SI loop (comprising Gly105 to Lys112 in Hs-cHisRS and Arg99 to Glu107 in TbHisRS). This loop is disordered in F-state Hs-cHisRS but is ordered, yet in an open conformation, in F-state TbHisRS. The ordered conformation of F-state TbHisRS is unusual, since a disordered SI loop is a hallmark of ligand-free HisRS in other structures [22, 26, 27]. This unique feature of F-state TbHisRS may thus be exploited for development of selective inhibitors. Considering the motions that occur within, and are propagated through the SI loop during aminoacylation (such as transitions between F- and H-states), the conformational differences between the F-states of Hs-cHisRS and TbHisRS in the SI loop present an opportunity for development of selective inhibitors that interfere with the motion of this loop in TbHisRS, thereby possibly decreasing substrate binding.
This hydrophobic pocket is delineated mainly by side chains of Tyr97, Lys100, Ala101, Ile105 and Met109 in and around the ordered TbHisRS SI loop, and side chains of Pro124 and Glu125 in and around the “TXE” motif (Fig. 4e). This pocket is called ‘pocket F’ hereafter. In contrast, this pocket is not present in F-state Hs-cHisRS due to lack of a well-defined boundary since the SI loop is disordered (Fig. 4f). In addition, only one out of these seven residues is identical between TbHisRS and Hs-cHisRS. For example, Ile105 in TbHisRS is replaced by a small, polar side chain of Ser111 in Hs-cHisRS; and Pro124 in TbHisRS is replaced by a large side chain of Tyr129. The largely hydrophobic nature of pocket F in the parasite enzyme (Fig. 4e) suggests that drug-like molecules could bind here. Both the difference in the stability of SI loop and the difference in the identity of side chains demarking pocket F suggest the possibility of deriving selective inhibitors by targeting this pocket. The importance of this pocket for drug design is under investigation. We are currently using a fragment cocktail crystallographic screening approach to hopefully obtain fragments that bind to this pocket that can subsequently be developed into useful inhibitors.
Interestingly, superpositions of TcHisRS·His, TcHisRS·HAMP and ligand-free TbHisRS structures reveal pockets where histidine and ATP would bind but are not yet completely formed (which could be called “pre-histidine” and “pre-ATP” pockets) (Fig. 4b & e). The presence of these nearly “nascent” pockets presents opportunities to develop inhibitors which mainly utilize pocket F as selectivity pocket, and possibly “expending” into pre-histidine and/or pre-ATP pockets of TbHisRS and thereby increase affinity. It is worth noting that inhibitors targeting multiple substrate binding pockets in aaRS are common. For example, naturally occurring LeuRS inhibitor mupiricin [50] or synthetic inhibitors of ThrRS [51] bind to both amino acid and ATP pockets to derive high binding affinity, and could serve as inspiration to develop HisRS inhibitors.
4. Conclusions
The current structure of Hs-cHisRS in complex with histidine revealed structural differences with respect to recently reported ligand-free or imidazole-bound Hs-cHisRS structures. Comparing these structures with other available HisRS structures ascertained that the current structure of Hs-cHisRS·His represents the H-state of the enzyme, while both the ligand-free Hs-cHisRS and Hs-cHisRS·imidazole structures represent the F-state of the enzyme. The structural changes that define H- and F-states Hs-cHisRS are shown to be largely due to the interactions between amino and carboxylate groups of histidine and Hs-cHisRS. Comparisons between the F- and H-states of Hs-cHisRS structures with that of trypanosomatid HisRS, i.e. F-state TbHisRS and H-state TcHisRS, reveal at least two binding pockets that may be exploited to selectively inhibit trypanosomatid HisRS. Crystallographic screenings for fragments targeting these pockets are currently underway.
Highlights.
Crystal structure of human cytoplasmic HisRS in complex with histidine
First structure representing the substrate-bound state of the enzyme (H-state)
H-state conformation is distinct from ligand-free/imidazole-bound state (F-state)
The NH3+ and COO− groups of histidine are important in stabilizing the H-state
Two specificity pockets on trypanosomatid HisRS are identified through comparisons
Acknowledgments
We thank Stewart Turley, Frank Zucker and Jonathan Kay for providing support for the X-ray data collection, database management and computing environment at the Biomolecular Structure Center of the University of Washington and Ms. Jessica E. Kim for technical assistance. We also thank the staff of Stanford Synchrotron Radiation Lightsource for assistance during data collection. Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R56AI084004 and RO1AI084004.
Abbreviations
- Hs-cHisRS
human cytosolic histidyl-tRNA synthetase
- H-state
substrate (histidine)-bound state
- F-state
ligand-free state
- aaRS
aminoacyl-tRNA synthetases
- A-state
intermediate product (histidyl-adenylate)-bound state
- HAMP
histidyl-adenylate
- rmsd
root-mean-square deviations
- TcHisRS
Trypanosoma cruzi histidyl-tRNA synthetase
- TbHisRS
Trypanosoma brucei histidyl-tRNA synthetase
- LIC
ligation-independent cloning
- TtHisRS
Thermus thermophilus histidyl-tRNA synthetase
- BtHisRS
Burkholderia thailandensis histidyl-tRNA synthetase
- LeuRS
leucyl-tRNA synthetase
- ThrRS
threonyl-tRNA synthetase
- H-bond
hydrogen bond
- TLS
translational/libration/screw
- PDB
Protein Data Bank
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Shibata S, Gillespie JR, Kelley AM, Napuli AJ, Zhang Z, Kovzun KV, Pefley RM, Lam J, Zucker FH, Van Voorhis WC, Merritt EA, Hol WG, Verlinde CL, Fan E, Buckner FS. Selective inhibitors of methionyl-tRNA synthetase have potent activity against Trypanosoma brucei Infection in Mice. Antimicrob Agents Chemother. 2011;55:1982–1989. doi: 10.1128/AAC.01796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vondenhoff GH, Van Aerschot A. Aminoacyl-tRNA synthetase inhibitors as potential antibiotics. Eur J Med Chem. 2011;46:5227–5236. doi: 10.1016/j.ejmech.2011.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Kalidas S, Cestari I, Monnerat S, Li Q, Regmi S, Hasle N, Labaied M, Parsons M, Stuart K, Phillips MA. Genetic Validation of Aminoacyl-tRNA Synthetases as Drug Targets in Trypanosoma brucei. Eukaryot Cell. 2014;13:504–516. doi: 10.1128/EC.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham JS, Dawson KL, Jackson KE, Lim EE, Pasaje CF, Turner KE, Ralph SA. Aminoacyl-tRNA synthetases as drug targets in eukaryotic parasites. Int J Parasitol Drugs Drug Resist. 2014;4:1–13. doi: 10.1016/j.ijpddr.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibata S, Gillespie JR, Ranade RM, Koh CY, Kim JE, Laydbak JU, Zucker FH, Hol WG, Verlinde CL, Buckner FS, Fan E. Urea-based inhibitors of Trypanosoma brucei methionyl-tRNA synthetase: selectivity and in vivo characterization. J Med Chem. 2012;55:6342–6351. doi: 10.1021/jm300303e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh CY, Kim JE, Shibata S, Ranade RM, Yu M, Liu J, Gillespie JR, Buckner FS, Verlinde CL, Fan E, Hol WG. Distinct States of Methionyl-tRNA Synthetase Indicate Inhibitor Binding by Conformational Selection. Structure. 2012;20:1681–1691. doi: 10.1016/j.str.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson ET, Kim JE, Zucker FH, Kelley A, Mueller N, Napuli AJ, Verlinde CL, Fan E, Buckner FS, Van Voorhis WC, Merritt EA, Hol WG. Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate. Biochimie. 2011;93:570–582. doi: 10.1016/j.biochi.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingvarsson H, Unge T. Flexibility and communication within the structure of the Mycobacterium smegmatis methionyl-tRNA synthetase. FEBS J. 2010;277:3947–3962. doi: 10.1111/j.1742-4658.2010.07784.x. [DOI] [PubMed] [Google Scholar]
- 10.Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, Zelwer C. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J Mol Biol. 2001;306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 11.Mechulam Y, Schmitt E, Maveyraud L, Zelwer C, Nureki O, Yokoyama S, Konno M, Blanquet S. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. Journal of Molecular Biology. 1999;294:1287–1297. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 12.Ilyin VA, Temple B, Hu M, Li G, Yin Y, Vachette P, Carter CW., Jr 2.9 A crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci. 2000;9:218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Retailleau P, Huang X, Yin Y, Hu M, Weinreb V, Vachette P, Vonrhein C, Bricogne G, Roversi P, Ilyin V, Carter CW., Jr Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations. J Mol Biol. 2003;325:39–63. doi: 10.1016/s0022-2836(02)01156-7. [DOI] [PubMed] [Google Scholar]
- 14.Laowanapiban P, Kapustina M, Vonrhein C, Delarue M, Koehl P, Carter CW., Jr Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations. Proc Natl Acad Sci U S A. 2009;106:1790–1795. doi: 10.1073/pnas.0812752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen N, Zhou M, Yang B, Yu Y, Dong X, Ding J. Catalytic mechanism of the tryptophan activation reaction revealed by crystal structures of human tryptophanyl-tRNA synthetase in different enzymatic states. Nucleic Acids Res. 2008;36:1288–1299. doi: 10.1093/nar/gkm1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh CY, Kim JE, Napoli AJ, Verlinde CL, Fan E, Buckner FS, Van Voorhis WC, Hol WG. Crystal structures of Plasmodium falciparum cytosolic tryptophanyl-tRNA synthetase and its potential as a target for structure-guided drug design. Mol Biochem Parasitol. 2013;189:26–32. doi: 10.1016/j.molbiopara.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 20.Cusack S. Eleven down and nine to go. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 21.Arnez JG, Harris DC, Mitschler A, Rees B, Francklyn CS, Moras D. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J. 1995;14:4143–4155. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merritt EA, Arakaki TL, Gillespie JR, Larson ET, Kelley A, Mueller N, Napuli AJ, Kim J, Zhang L, Verlinde CL, Fan E, Zucker F, Buckner FS, van Voorhis WC, Hol WG. Crystal structures of trypanosomal histidyl-tRNA synthetase illuminate differences between eukaryotic and prokaryotic homologs. J Mol Biol. 2010;397:481–494. doi: 10.1016/j.jmb.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Z, Wei Z, Zhou JJ, Ye F, Lo WS, Wang F, Lau CF, Wu J, Nangle LA, Chiang KP, Yang XL, Zhang M, Schimmel P. Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure. 2012;20:1470–1477. doi: 10.1016/j.str.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 25.O’Hanlon TP, Miller FW. Genomic organization, transcriptional mapping, and evolutionary implications of the human bi-directional histidyl-tRNA synthetase locus (HARS/HARSL) Biochem Biophys Res Commun. 2002;294:609–614. doi: 10.1016/S0006-291X(02)00525-9. [DOI] [PubMed] [Google Scholar]
- 26.Qiu X, Janson CA, Blackburn MN, Chhohan IK, Hibbs M, Abdel-Meguid SS. Cooperative structural dynamics and a novel fidelity mechanism in histidyl-tRNA synthetases. Biochemistry. 1999;38:12296–12304. doi: 10.1021/bi990482v. [DOI] [PubMed] [Google Scholar]
- 27.Yaremchuk A, Tukalo M, Grotli M, Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J Mol Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- 28.Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase. Biochemistry. 1997;36:3084–3094. doi: 10.1021/bi9618373. [DOI] [PubMed] [Google Scholar]
- 29.Arnez JG, Augustine JG, Moras D, Francklyn CS. The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1997;94:7144–7149. doi: 10.1073/pnas.94.14.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, Armour B, Begley DW, Dieterich SH, Dranow DM, Abendroth J, Fairman JW, Fox D, 3rd, Staker BL, Phan I, Gillespie A, Choi R, Nakazawa-Hewitt S, Nguyen MT, Napuli A, Barrett L, Buchko GW, Stacy R, Myler PJ, Stewart LJ, Manoil C, Van Voorhis WC. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One. 2013;8:e53851. doi: 10.1371/journal.pone.0053851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh CY, Kim JE, Wetzel AB, de van der Schueren WJ, Shibata S, Ranade RM, Liu J, Zhang Z, Gillespie JR, Buckner FS, Verlinde CL, Fan E, Hol WG. Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness. PLoS Negl Trop Dis. 2014;8:e2775. doi: 10.1371/journal.pntd.0002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larson ET, Kim JE, Castaneda LJ, Napuli AJ, Zhang Z, Fan E, Zucker FH, Verlinde CL, Buckner FS, Van Voorhis WC, Hol WG, Merritt EA. The double-length tyrosyl-tRNA synthetase from the eukaryote Leishmania major forms an intrinsically asymmetric pseudo-dimer. J Mol Biol. 2011;409:159–176. doi: 10.1016/j.jmb.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehlin C, Boni E, Buckner FS, Engel L, Feist T, Gelb MH, Haji L, Kim D, Liu C, Mueller N, Myler PJ, Reddy JT, Sampson JN, Subramanian E, Van Voorhis WC, Worthey E, Zucker F, Hol WG. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 34.Choi R, Kelley A, Leibly D, Hewitt SN, Napuli A, Van Voorhis W. Immobilized metal-affinity chromatography protein-recovery screening is predictive of crystallographic structure success. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:998–1005. doi: 10.1107/S1744309111017374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. Journal of Applied Crystallography. 2008;41:641–643. [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–111. [Google Scholar]
- 44.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLano W. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
- 46.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 47.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 48.Perona JJ, Gruic-Sovulj I. Synthetic and Editing Mechanisms of Aminoacyl-tRNA Synthetases. Top Curr Chem. 2013 doi: 10.1007/128_2013_456. [DOI] [PubMed] [Google Scholar]
- 49.Gilbert IH. Drug discovery for neglected diseases: molecular target-based and phenotypic approaches. J Med Chem. 2013;56:7719–7726. doi: 10.1021/jm400362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakama T, Nureki O, Yokoyama S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J Biol Chem. 2001;276:47387–47393. doi: 10.1074/jbc.M109089200. [DOI] [PubMed] [Google Scholar]
- 51.Teng M, Hilgers MT, Cunningham ML, Borchardt A, Locke JB, Abraham S, Haley G, Kwan BP, Hall C, Hough GW, Shaw KJ, Finn J. Identification of bacteria-selective threonyl-tRNA synthetase substrate inhibitors by structure-based design. J Med Chem. 2013;56:1748–1760. doi: 10.1021/jm301756m. [DOI] [PubMed] [Google Scholar]
- 52.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]