Abstract
Yersinia pestis biovar Orientalis isolates have lost the capacity to ferment glycerol. Herein we provide experimental validation that a 93 bp in-frame deletion within the glpD gene encoding the glycerol-3-phosphate dehydrogenase present in all biovar Orientalis strains is sufficient to disrupt aerobic glycerol fermentation. Furthermore, the inability to ferment glycerol is often insured by a variety of additional mutations within the glpFKX operon which prevents glycerol internalization and conversion to glycerol-3-phosphate. The physiological impact of functional glpFKX in the presence of dysfunctional glpD was assessed. Results demonstrate no change in growth kinetics at 26°C and 37°C. Mutants deficient in glpD displayed decreased intracellular accumulation of glycerol-3-phosphate, a characterized inhibitor of cAMP receptor protein (CRP) activation. Since CRP is rigorously involved in global regulation Y. pestis virulence, we tested a possible influence of a single glpD mutation on virulence. Nonetheless, subcutaneous and intranasal murine challenge was not impacted by glycerol metabolism. As quantified by crystal violet assay, biofilm formation of the glpD-deficient KIM6+ mutant was mildly repressed; whereas, chromosomal restoration of glpD in CO92 resulted in a significant increase in biofilm formation.
Keywords: Yersinia pestis, glycerol, biofilm, glpD, glpFKX
1. INTRODUCTION
Yersinia pestis, the causative agent of plague, is historically accountable for greater than 200 million deaths throughout three pandemics [1]. Zoonotic maintenance of plague occurs through circulation amongst rodent reservoir hosts and flea vectors. Upon consumption of an infected mammalian blood meal by a naïve flea, Y. pestis proliferates in the flea midgut. Y. pestis forms a biofilm in the flea proventriculus which prevents the passage of blood during subsequent feeding attempts. Moreover, the biofilm enhances regurgitation of bacteria into the dermis of the mammalian host, thereby promoting the natural transmission of plague [2].
Y. pestis strains are subdivided into four major biovars (Microtus, Antiqua, Medievalis, and Orientalis) distinguished by phenotypic characteristics [3, 4]. Biovar Orientalis, the most recent evolutionary divergent responsible for the 3rd plague pandemic, has lost the capacity to ferment glycerol [5]. Y. pestis anaerobic metabolism of glycerol is facilitated by the glpABC operon; whereas, glpD and the glpFKX operon enable aerobic glycerol fermentation. Under aerobic conditions, glycerol is internalized by the GlpF (KIM10, y0046) facilitator. The GlpK (y0047) aerobic glycerol kinase converts glycerol into glycerol-3-phosphate (G3P). Bioinformatics analyses suggest the presence of an additional glycerol kinase in Y. pestis chromosome (y0876); however, it is uncertain whether this putative protein is functional [6]. Glycerol-3-phosphate can be converted into dihydroxyacetone phosphate (DHAP) by the sn-glycerol-3-phosphate dehydrogenase, GlpD (y3891). Both G3P and DHAP serve as precursors for phospholipid biosynthesis [7, 8]. Alternately, the TpiA (y0052) triose phosphate isomerase can convert DHAP into glyceraldehyde-3-phosphate, thereby enabling the glycolytic catabolism of glycerol [9].
The inability for biovar Orientalis isolates to ferment glycerol is presumed to result from a 93 bp in-frame deletion within the glpD gene encoding the aerobic glycerol-3-phosphate dehydrogenase [10]. However, genomic analyses of biovar Orientalis isolates indicate a variety of disruptions within the glpK gene, potentially preventing the formation and accumulation of the G3P in the absence of functional glpD. In E. coli, G3P prevents stimulation of adenylyl cyclase, thereby obstructing synthesis of the small-molecule inducer 3'–5'-cyclic adenosine monophosphate (cAMP) [11]. The cAMP receptor protein (CRP), a primary modulator of carbon catabolite repression which enables the catabolism of alternate carbon sources when available glucose is scarce, is allosterically activated by cAMP [12]. Furthermore, CRP is a global regulator which differentially modulates greater than 6% of Y. pestis total protein capacity, including prominent virulence factors [13, 14]. Y. pestis mutants deficient in crp have been shown to be attenuated for virulence [14, 15]. The interplay amongst glycerol metabolism and CRP activation has not been assessed in Y. pestis.
Prior transcriptomic analyses indicate that the components of the aerobic glycerol metabolic pathways are induced during multiple aspects of the Y. pestis infectious cycle. Aerobic glycerol metabolism is simulated during temperature shifts representing the transition from the flea vector (26°C) to the human host (37°C) and amid survival in the macrophage intraphagosomal environment [16, 17]. Alternately, glycerol metabolic pathways are upregulated during infection of the flea vector as well as in flowcell biofilms relative to planktonic culture [18]. However, the physiological implications of Y. pestis glycerol metabolism remain unclear.
The objective of this study was to discern the molecular mechanism defining the glycerol fermentation deficiency in biovar Orientalis isolates. Moreover, we seek to establish the physiological relevance of glycerol metabolism during the Y. pestis infectious cycle. We provide formal proof that the 93 bp in-frame deletion in the glpD gene of biovar Orientalis isolates is sufficient to impair glycerol fermentation; however, the inability to ferment glycerol is often ensured by additional impairments of the glpFKX operon. The natural prevalence of glpFKX inactivation in the presence of glpD dysfunction observed in biovar Orientalis isolates may be suggestive of negative selection. Therefore, we assessed the impact of functional glpFKX in the presence of dysfunctional glpD. Mutants expressing functional glpK in the presence of dysfunctional glpD did not alter growth kinetics at either 26°C or 37°C. Inactivation of glpD in a biovar Medievalis background (KIM6+) resulted in decreased concentrations of G3P. Conversely, restoration of functional glpD in biovar Orientalis background (CO92L) had no impact upon intracellular G3P concentration. Respective inactivation and restoration of glpD in KIM6+ and CO92 backgrounds did not alter persistence in RAW 264.7 murine macrophage-like cells. Moreover, subcutaneous and intranasal murine challenge did not reveal alterations in the survival of mice inoculated with the Y. pestis KIM6+ glpD deficient mutant in respect to the isogenic control. Biovar Medievalis strain KIM6+ expressing non-functional glpD resulted in a slight, yet significant, defect in biofilm formation. Conversely, expression of functional glpD promotes enhanced biofilm formation of biovar Orientalis strain CO92L.
2. MATERIALS AND METHODS
2.01 Bacterial strains, plasmids, and primers
Bacterial strains and plasmids used in this study are described in Table 1. Oligonucleotides are detailed in Supplemental Data Table S1. Strains utilized for biofilm quantification lacked the pCD1 virulence plasmid. Complementation of functional KIM6+ derived glpK and glpD were afforded by independent plasmid expression. The genetic composition of mutants and plasmids prepared in this study were verified via sequencing. For virulent studies, Y. pestis KIM6+ and derivatives were transformed with the pCD1Ap plasmid representing pCD1 containing the ampicillin resistance marker inserted into the yadA pseudogene (kindly provided by Dr. Robert Perry).
TABLE 1.
Bacterial strains and plasmids
| Y. pestis strains | Characteristics* | Reference |
|---|---|---|
| CO92 | Pgm+ pCD1+ pPCP+ Gly− | [49] |
| CO92L | Pgm+ pCD1− pPCP+ Gly− (cured of virulence plasmid pCD1) |
This study |
| CO92L glpD+ | Pgm+ pCD1− pPCP+ Gly− (defective glpD’ allele replaced with functional glpD gene in the chromosome) |
This study |
| KIM 5 (D27) | Pgm− pCD1+ pPCP+ Gly+ | Brubaker collection |
| KIM 6+ | Pgm+ pCD1− pPCP+ Gly+ | Brubaker collection |
| KIM 6+ glpD’ | Pgm+ pCD1− pPCP+ Gly− (functional glpD gene replaced with defective glpD’ allele in the chromosome) |
This study |
| KIM 6+ ΔglpABC |
Pgm+ pCD1− pPCP+ Gly+ (glpABC operon deletion) |
This study |
| KIM 6+ glpD’ΔglpABC |
Pgm+ pCD1− pPCP+ Gly− (functional glpD gene replaced with defective glpD’ allele in the chromosome; glpABC operon deletion) |
This study |
| 195-P2 | Pgm− pCD1+ pPCP+ Gly− | Hinnebusch collection |
| TS | Pgm+ pCD1− pPCP+ Gly− | Brubaker collection |
| Salazar | Pgm− pCD1− pPCP+ Gly− | Brubaker collection |
| EV76H | Pgm+ pCD1− pPCP+ Gly− (derivative of the live plague vaccine strain EV76) |
Brubaker collection |
| M23 | Pgm− pCD1+ pPCP+ Gly− | Brubaker collection |
| Kimberley | Pgm+ pCD1− pPCP+ Gly− | Perry collection |
| PEXU-2 | Pgm+ pCD1+ pPCP+ Gly− | CDC collection |
| E. coli strains | Characteristics | Reference |
| DH5α | F-φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) gal− phoA supE44 λ− thi-1 gyrA96 relA1 |
Invitrogen (Carlsbad, CA) |
| BW25141 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB- phoR)580, λ−, galU95, ΔuidA3::pir+, recA1, endA9(del- ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 |
[19] |
| Plasmids | Characteristics | Reference |
| pBluescript SK+ | Cloning vector, ApR | Stratagene (La Jolla, CA) |
| pBBR1Tp | Cloning vector, TpR | ATCC (Manassas, VA) |
| pKD46 | Mutagenesis helper plasmid, source of Lambda Red Recombinase, ApR |
[19] |
| pKD4 | Mutagenesis helper plasmid, source of kan cassette, KmRApR |
[19] |
| pCVD442 | Source of sacB gene, ApR | [50] |
| pKD4_Km-sacB | Cloned sacB from pCVD442 in NgoMIV site of pKD4, source of kan-sacB cassette, KmRApR,SucS |
This study |
| pBGLPD5 | Cloned glpD from KIM 6+ in Hind III and BamHI sites of pBluescript SK+ |
This study |
| pTpGLPFKX6 | Cloned glpFKX from KIM 6+ in XbaI and XhoI sites of pBBR1Tp |
This study |
| pCD1Ap | Virulence plasmid pCD1 of Y. pestis KIM labeled with ApR marker |
Provided by R. Perry |
All strains contained a large plasmid pMT, encoding capsular antigen F1 and murine toxin Ymt. Pgm: pigmentation locus, pCD1: virulence plasmid encoding T3SS, pPCP: small plasmid encoding pesticin and plasminogen activator Pla, Gly: aerobic glycerol fermentation. glpD: functional glpD gene of KIM. glpD’: defective glpD gene of CO92. KmR, ApR, TpR, and SucS are markers of resistance to kanamycin, ampicillin, trimethoprim, and sensitivity to sucrose, respectively. kan and sacB are genes for kanamycin resistance and sucrose sensitivity, respectively.
2.02 Chromosomal allelic exchange of glpD and anaerobic glycerol metabolism (glpABC) operon deletion
Chromosomal allelic exchange to replace the functional glpD gene of Y. pestis strain KIM6+ (biovar Medievalis) with the disrupted glpD’ allele of Y. pestis strain CO92 (biovar Orientalis), yielding strain KIM6+ glpD’, was performed through modification of the Lambda Red Recombinase technique (Supplemental Fig. S1) [19]. Furthermore, a reciprocal exchange repaired the defective glpD’ of CO92L with the functional glpD of KIM6+, yielding strain CO92L glpD+. To do so, the counter-selective sacB gene was incorporated alongside the kan gene for the kanamycin resistance into pKD4 to obtain pKD4_Km-sacB plasmid (Table 1). Crossover homologous recombination was utilized to replace the native glpD gene in both CO92L and KIM6+ with a kanamycin-resistant/sucrose sensitive cassette via Lambda Red Recombinase afforded by the pKD46 helper plasmid. A secondary recombination event scarlessly replaced the deletion cassette with the desired glpD sequence determined by 10% sucrose counter-selection.
The glpABC operon which facilitates the anaerobic metabolism of glycerol was deleted from KIM6+ and KIM6+ glpD’. Briefly, a single crossover recombination event replaced the glpABC operon with a kanamycin-resistant deletion construct in electrocompetent Y. pestis expressing Lambda Red Recombinase encoded by the pKD46 helper plasmid. FLP-mediated recombination was utilized for excision of the glpABC deletion cassette.
2.03 Phenotypic assessment of glycerol fermentation
To determine the capacity for the various mutants to ferment glycerol, strains were cultured on MacConkey Agar Base indicator plates (Difco 281810) supplemented with 0.2% glycerol and incubated for 18–24 hours at both 26°C and 37°C. Results were verified via growth on acid-fuchsin plates utilizing 0.2% glycerol as the sole carbon source [20].
2.04 Phylogenetic analyses
Full-length genomic sequences of glpD, glpK, and glpX of 41 biovar Orientalis isolates obtained from GenBank were evaluated with SeaView version 4.5.0 [21]. Sequence read archive (SRA) reads were pre-aligned using NCBI BLAST. A maximum parsimony tree was generated with 200 bootstrap replicates. The tree was rooted to biovar Orientalis predicted ancestral strain, E1979001 [22]. The glpFKX operon of stain Salazar was sequenced and submitted to GenBank as accession number KJ719254.
2.05 Growth kinetics
Bacteria derived from glycerol stocks were pre-grown on Heart Infusion Agar (HIA) plates for 18–24 hours at 26°C. Bacteria were sub-cultured in 25 ml Heart Infusion Broth (HIB) at 26°C for 12–18 hours shaking at 250 rpm. 70 ml of fresh HIB was inoculated with OD600 = 0.2 and grown for 3 hours at 26°C whilst shaking at 250 rpm to obtain actively growing bacteria. Cultures were centrifuged at 4.5K rpm for 15 min at 26°C. Residual media was aspirated and the bacteria were washed in 0.033 M K-phosphate buffer (pH = 7.2). Bacteria were re-suspended to a OD600 = 1in K-phosphate buffer. For each strain, 25 ml of BCS chemically defined media [23] containing 4 mM CaCl2 and supplemented with either 0.2% glycerol, 0.2% glucose, or both 0.2% glycerol and 0.2% glucose were inoculated to OD600 = 0.1 and incubated at either 26°C or 37°C while shaking at 250 rpm. One milliliter aliquots were extracted at 0, 2, 6, 12, and 24 hours post-inoculation for determination of optical density.
2.06 Semi-quantitative reverse transcriptase PCR
The Access RT-PCR System (Promega, Madison, WI) was utilized to characterize the expression of the aerobic glycerol metabolic pathway. Y. pestis KIM5 (D27) derived from glycerol stock was streaked on HIA plates and incubated at 26°C for 36 hours. Resulting cultures were pre-grown in 50 ml of chemically defined BCS medium supplemented with 0.2% K-gluconate in a 500 ml flask at an initial OD600 = 0.1 for 12 hours at 26°C whilst shaking at 250 rpm. 170 ml of chemically defined BCS medium supplemented with 0.2% K-gluconate was again inoculated with OD600 = 0.1 of pre-grown bacteria in 1 L flask and incubated for 4 hours at 26°C while shaking at 250 rpm. Twenty-five milliliters of chemically defined BCS media with 4 mM CaCl2 containing either 0.2% K-gluconate; 0.2% K-gluconate and 0.2% glycerol; or 0.2% K-gluconate, 0.2% glycerol, and 0.2% glucose were inoculated with OD600 = 0.3 in 250 ml flasks and incubated at 26°C or 37°C. Approximately 8×108 total cells as determined by optical density were extracted at 1 and 4 hours post-inoculation and immediately added to an equal volume of cold (4°C) RNAlater Stabilization Solution (Ambion, Austin, TX). Total RNA was isolated using the RNAeasy mini column kit (Qiagen, Hilden, Germany) and processed by the TURBO DNA-free kit (Ambion) for DNase treatment. RNA concentration was determined through application of the Synergy HT Take3 Multi-Volume Plate (BioTek, Winooski, VT). RT-PCR reactions were performed in accordance with manufacturer guidelines using 25 ng of total RNA for 25 PCR cycles. Three microliter aliquots of the RT-PCR reactions were visualized on 2% agarose gels infused with ethidium bromide (Figs. S1 & S2). Semi-quantitative analyses were performed by ImageJ v1.47 (http://rsb.info.nih.gov/ij/) with normalization to the gyrB gene [24–26].
2.07 Intracellular glycerol-3-phosphate quantification
Y. pestis strains KIM6+/pCD1Ap and KIM6+ glpD’/pCD1Ap were cultured on HIA plates containing 50 µg/mL carbenicillin (Cb50) and incubated at 26°C for 18–24 hours. Bacteria were re-streaked on fresh HIA/Cb50 plates and incubated for an additional 12–18 hours at 26°C. 50 ml of HIB/Cb50 were inoculated with OD600 = 0.1 and incubated for 12 hours at 26°C whilst shaking at 250 rpm. Bacteria were sub-cultured to an OD600 = 0.1 in seventy-five milliliters of fresh HIB/Cb50 supplemented with 0.2% glycerol and incubated at either 26°C or 37°C whilst shaking at 250 rpm. At 3 hours post-inoculation, approximately 2×109 cells of either strain as determined by optical density readings were harvested. Following centrifugation at 4.5K rpm for 10 min at 4°C, growth media was aspirated.
One set of samples was processed by methanol/chloroform extraction [27]. Briefly, the bacterial pellets were washed with 1 ml of 150 mM ammonium bicarbonate and re-suspended in 0.75 ml cold methanol (−20°C). The mixture was homogenized with approximately 300 µl of cold zirconia beads (−20°C) utilizing a microtube homogenizer (Benchmark, South Plainfield, NJ) for 3 series at 4.5K rpm for 30 sec followed by a resting stage on ice for 60 sec. 0.4 ml of molecular grade water and 0.25 ml of chloroform were added each sample. Approximately 0.75 ml of the aqueous phase was extracted following phase separation by centrifugation (5 min, 13K rpm).
Alternately, direct cell lysates of duplicate samples were prepared by suspending the bacterial pellets in 400 µl of G3P detection assay buffer (Biovision, Milpitas, CA). Samples were homogenization with 300 µl of cold zirconia beads (−20°C) utilizing a microtube homogenizer (Benchmark) for 3 series at 4.5K rpm for 30 sec followed by a resting stage on ice for 60 sec. Samples were centrifuged at 12K rpm for 10 min at 4°C and approximately 350 µl of the supernatant was extracted.
Two-fold dilution series of 50 µl, 25 µl, and 12.5 µl of sample extracts were assessed by G3P detection via colorimetric assay (Biovision) performed in accordance with manufacturer guidelines. In addition, direct lysates of CO92L and CO92L glpD+ grown at 37°C were analyzed as previously described. Statistical significance amongst relative sample G3P concentrations was determined by Student’s T test.
2.08 Macrophage infection
RAW 264.7 murine macrophage-like cells (ATCC TIB-71, Manassas, VA) seeded at 2×105 cells per well were grown in Dulbecco’s modified Eagle medium (DMEM, Gibco BRL, Grand Island, NY) supplemented with glutamine, 10% (v/v) FBS and 1 mM sodium pyruvate (tissue culture medium) at 37° C in the presence of 5% CO2. Following overnight incubation, the cells were washed twice with Dulbecco’s Phosphate-Buffered Saline lacking calcium and magnesium (DPBS, Corning, Manassas, VA) and reconstituted with 250 µl/well fresh tissue culture medium. Y. pestis CO92L, CO92L glpD+, KIM6+/pCD1Ap, KIM6+ glpD’/pCD1Ap, KIM6+ΔglpABC/pCD1Ap, and KIM6+ glpD’ΔglpABC/pCD1Ap were grown stationary overnight at 26°C in DMEM containing glutamine, 1% (v/v) FBS, 1 mM sodium pyruvate, and 30 mM HEPES (bacterial growth medium). Overnight cultures were then diluted and the bacteria were grown to the exponential phase. Macrophages were inoculated with an Multiplicity of Infection (MOI) = 50 of the bacterial suspension, and uptake was allowed for 1 hour at 37°C. The monolayer was washed twice with DPBS and then was incubated for an additional hour at 37°C with 1 ml of fresh tissue culture medium supplemented with 15 µg/ml gentamicin (Corning). Thereafter, coinciding with the 0 hour timepoint, the monolayer was washed twice with DPBS. The 18 hour timepoint samples were incubated at 37°C with tissue culture medium supplemented with 4 µg/ml gentamicin. Macrophages were washed twice with DPBS and lysed with 0.2 ml of 0.1% sodium deoxycholate in DPBS. The lysate was suspended in 1.8 ml of bacterial growth medium, serial dilutions were prepared, and 100 µl of each dilution was plated onto HIB agar plates supplemented with 0.5% (v/v) FBS in duplicate. Results reflect the average of two biological replicates.
2.09 Murine challenge
Y. pestis strains derived from glycerol stocks were cultured on HIA plates and grown at 26°C for 18–24 hours. Bacteria were streaked on fresh HIA plates and incubated again at 26°C for 12–18 hours. Strains were suspended and diluted in 0.033M K-phosphate buffer at 4°C. Groups of 5 Female 8 week Swiss Webster mice (Taconic Farms, Germantown, NY) were challenged by the subcutaneous (s.c.) route with 500 CFU (KIM6+/pCD1Ap s.c. LD50 = ~10 CFU) or by the intranasal (i.n.) route with 4,000 CFU (KIM6+/pCD1Ap i.n. LD50 = ~400 CFU) of each Y. pestis strain. Verification of proper inoculum concentration was ascertained by CFU determination on HIA plates. Experimental observation progressed for the course of four weeks. To discern more subtle changes in virulence, the experiment was replicated utilizing lower inoculum. Ten mice per group were challenged by the s.c. route with 200 CFU and 5 mice per group were challenged by the i.n. route with 2,000 CFU for each Y. pestis strain. The experiment was terminated at 16 days post-inoculation. Kaplan–Meier survival curves were generated, and statistical significance was determined by log-rank test (IBM SPSS Statistics v20).
All animal experiments were conducted in ABSL-3 facilities within the Galveston National Lab under a protocol approved by the Institutional Animal Care and Use Committee at UTMB. The experimental duration progressed to a humane endpoint in which case surviving mice were euthanized.
2.10 Crystal violet assay
Biofilm formation was quantified by crystal violet assay [28]. Bacterial strains were pre-grown as previously described for the growth kinetic experiments. 24-well polystyrene plates were inoculated to OD600 = 0.1 with either 0.75 ml of HIB or BCS supplemented with 0.2% K-gluconate and incubated at 26°C for 18–20 hours at 250 rpm. Media was aspirated, wells were washed with dH2O, and biofilms were dyed with 0.01% crystal violet for 20 min. Crystal violet dye was aspirated and wells were washed three times with dH2O. Bound crystal violet was solubilized with 1.5 ml of 33% acetic acid for 15 min whilst shaking at 150 rpm. Absorbance was measured utilizing a BioTek Synergy HT at 570 nm. Results reflect the average of 2 independent experiments, each consisting of 6 technical replicates. Statistical significance was determined by 2-tailed Student’s T-test with a threshold p-value <0.05. All strains were assessed for retention of the pigmentation locus containing the hmsHFRS genes essential for biofilm formation via Congo red phenotypic determination and PCR [29]. Preliminary findings reveal no difference in biofilm production amongst pCD1 positive and negative strains regardless of media composition and available primary/alternate carbon sources (Supplemental Fig S2). Therefore, pCD1-deficient strains were utilized for biofilm assessments.
3. RESULTS
3.01 Molecular basis of glycerol deficiency of biovar Orientalis isolates
The functional glpD gene of Y. pestis strain KIM6+ (biovar Medievalis) was replaced with the glpD’ allele containing the 93 bp in-frame deletion of Y. pestis strain CO92L (biovar Orientalis) via chromosomal allelic exchange. Alternatively, the reciprocal exchange was performed to replace the disrupted glpD’ of CO92L with the intact glpD of KIM6+. Phenotypic observation of glycerol fermentation on both MacConkey and fuchsin-infused indicator plates supplemented with 0.2% glycerol demonstrated the 93 bp in-frame deletion is sufficient to impair glycerol fermentation in KIM 6+ strain; however, expression of functional GlpD in CO92L did not restore glycerol fermentation (Table 2). Plasmid complementation of glpD from KIM6+ restored glycerol fermentation in the KIM6+ chromosomal allelic exchange mutant.
TABLE 2.
Characterization of Y. pestis Glycerol Fermentation
| Y. pestis strain / plasmid | Glycerol fermentation |
|---|---|
| KIM 6+ | Positive |
| KIM 6+ glpD’ | Negative |
| KIM 6+ glpD’ / pBGLPD5 | Positive |
| CO92L* | Negative |
| CO92L/pBGLPD5* | Negative |
| CO92L/pTpGLPFKX6* | Negative |
| CO92L/pBGLPD5, pTpGLPFKX6* | Positive |
| CO92L glpD + | Negative |
| CO92L glpD +/pBGLPD5 | Negative |
| CO92L glpD +/pTpGLPFKX6 | Positive |
Results consistent with 7 other biovar Orientalis strains used in this study.
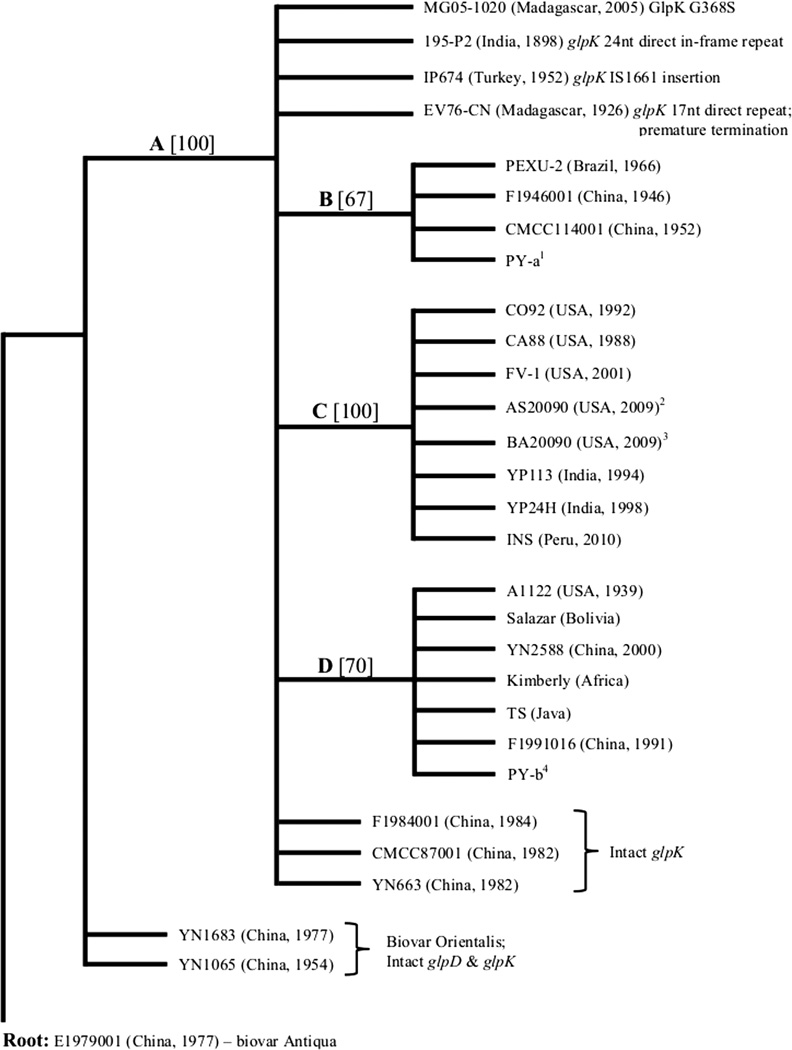
Independent expression of either the glpD gene or the glpFKX operon derived from KIM6+ in separate plasmid vectors did not facilitate glycerol fermentation in CO92L. However, glycerol fermentation was repaired when both glpD and glpFKX genes were introduced simultaneously either on compatible plasmids or when plasmid with glpFKX operon was present in the CO92L derivative expressing the chromosomally restored glpD gene. Similarly, we found that the ability to ferment glycerol in seven other biovar Orientalis isolates of Y. pestis isolated from distinct geospatial origins can be restored only by complementation with both glpD and glpFKX genes (Tables 1 and 2; Supplemental Fig. S3). Analysis of Y. pestis genome sequences deposited to date in the GenBank database revealed that all biovar Orientalis isolates, with the exception of strains YN1065 and YN1683, contained identical 93 bp in-frame deletion in glpD gene. In addition to the deletion in glpD, all biovar Orientalis strains aside from Chinese isolates F1984001, CMCC87001, and YN663 displayed a variety of disruptions in the glpFKX operon. Strain CO92L possessed an extensive 941 bp deletion that disrupted both glpK and glpX genes. Mutations in other isolates consisted of nonsynonymous substitutions, single and multiple nucleotide insertions or deletions, as well as point mutation, resulting in amino acid change. In all cases, the various mutations affected glpK (Fig. 1). Overall, the 93 bp in-frame deletion in glpD gene evident in biovar Orientalis isolates is sufficient to cause the deficiency in glycerol fermentation; however, the inability to ferment glycerol is often ensured by additional impairments of the glpFKX operon (all of which notably disrupt the glpK gene).
FIG. 1.
Maximum parsimony tree comparing glpD, glpK, and glpX amongst biovar Orientalis isolates rooted to biovar Antiqua strain E1979001. Branch bootstrap values calculated from 200 replicates. A. Acquisition of the 93 bp in-frame glpD deletion and GlpD missense mutation (A125V); B. glpK poly(A) region insertion; C. glpKX pseudogene; D. glpK poly(A) region deletion. 1 PY strains 01–05; 2 AS20090 strains 1156, 1434, 1509, 1539, 2147; 3 BA20090 strains 1703, 1799, 1990, 2009; 4 PY strains 19, 71, 72, 76.
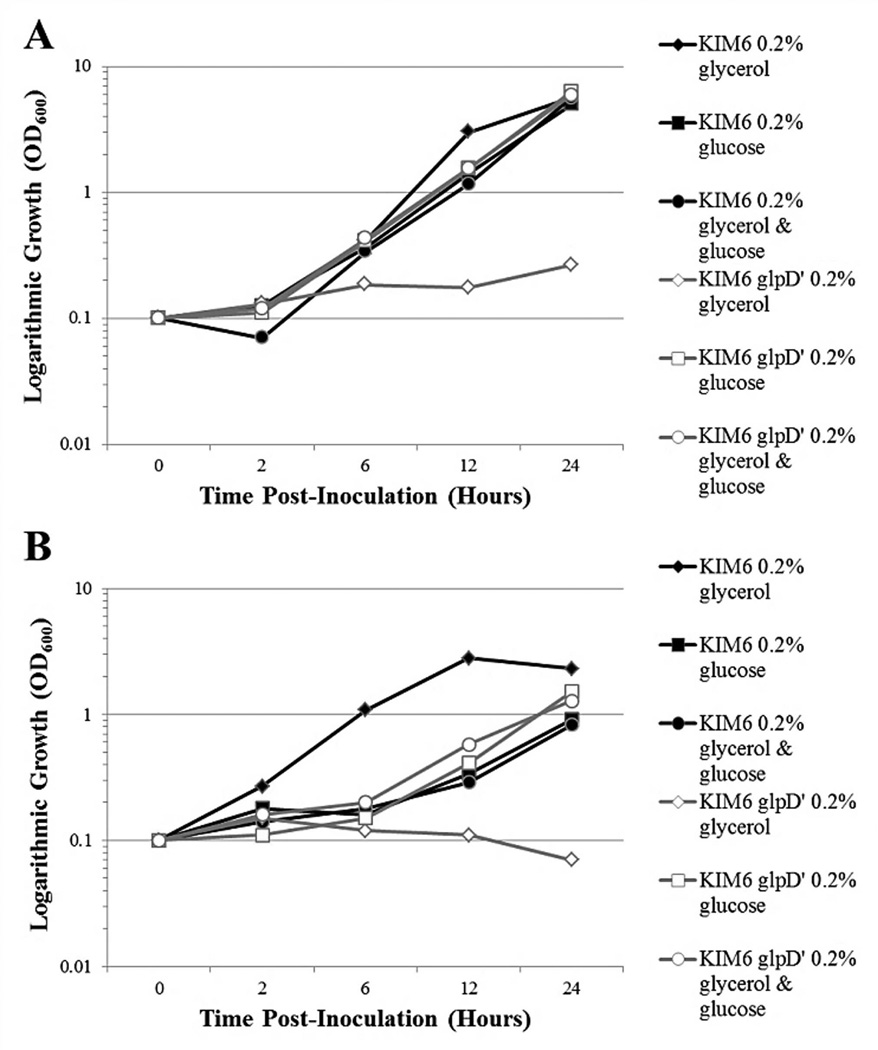
3.02 Disruption of aerobic glycerol metabolism does not hinder the rate of growth
Growth kinetics of the KIM6+ glpD’ allelic exchange mutant were comparable to the parent strain during growth at 26°C (Fig. 2A) and 37°C (Fig. 2B) when cultured in BCS chemically defined medium utilizing glucose as the sole carbon source. As expected, BCS media containing glycerol as the sole carbon source did not facilitate growth of the glpD deficient mutant. Notably, the 37°C growth rate of KIM6+ in BCS medium supplemented with glycerol alone was higher than that in BCS medium supplemented with glucose, or both glucose and glycerol. These observations indicate that i) Y. pestis KIM 6+ can either utilize glycerol faster than glucose at this temperature, or the presence of glucose has some inhibitory effect on growth at 37°C, and ii) Y. pestis KIM 6+ bacteria preferably use glucose when both glycerol and glucose are present in the medium suggesting typical catabolite repression phenomenon. The latter is confirmed by the fact that simultaneous addition of both glucose and glycerol resulted in nearly identical growth of both the KIM6+ and the glpD-deficient mutant in comparison with that provided by supplementation of the glucose alone. Moreover, the 37°C growth rates of KIM6+ and glpD deficient mutant were comparable when utilizing non-phosphoenolpyruvate (PEP) : carbohydrate phosphotransferase system (PTS) carbon sources other than glycerol, such as K-gluconate, as well as in peptide-rich HIB medium (data not shown).
FIG. 2.
Growth curves of Y. pestis strain KIM6+ and the isogenic KIM6+ glpD’ dysfunctional mutant in chemically defined BCS media containing 4 mM CaCl2 supplemented with either 0.2% glycerol; 0.2% glucose; or both 0.2% glycerol and 0.2% glucose. A. 26°C; B. 37°C. Bacteria were pre-grown with aeration in HIB at 26°C for 24 hours with two transfers, harvested in log phase, and washed in 0.033 M K-phosphate buffer pH 7.0. The initial inoculum was approximately OD600 = 0.1. Optical density readings were taken at 0, 2, 6, 12, and 24 hours post-inoculation.
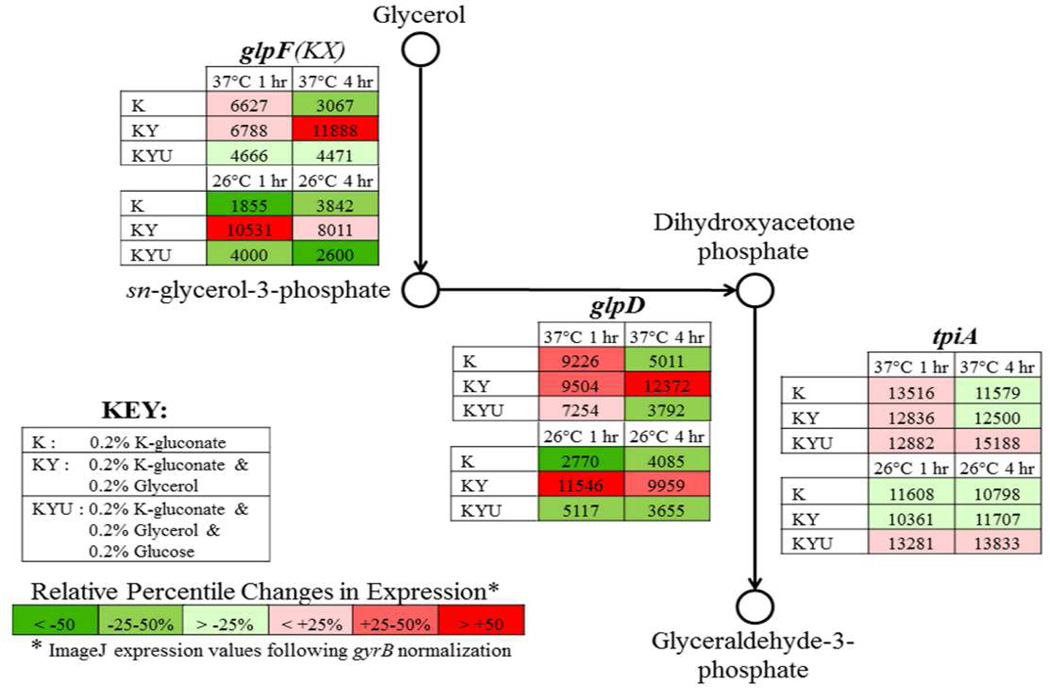
3.03 Temperature shift from 26°C to 37°C induces transient enhanced expression of aerobic glycerol metabolism
Expression patterns of the aerobic glycerol metabolic pathway during transition from 26°C to 37°C in the presence of different carbon sources were evaluated via semi-quantitative RT-PCR (Fig. 3, Supplemental Figs. S4 and S5). Y. pestis KIM 5 was assessed during growth at both 26°C and following temperature shift to 37°C in chemically defined BCS medium containing 4 mM CaCl2 supplemented with either 0.2% K-gluconate alone (BCS-K), both 0.2% K-gluconate and 0.2% glycerol (BCS-KY), or 0.2% K-gluconate, 0.2% glycerol and 0.2% glucose (BCS-KYU). The transition from 26°C to 37°C in BCS-K induced the expression of the aerobic glycerol metabolic pathway glpFKX and glpD genes at 1 hour time point followed by reduction in their transcription at 4 hours to the level seen at 26°C. The expression of the tpiA gene encoding the triose phosphate isomerase which facilitates the glycolytic metabolism of glycerol remained constant. The presence of glycerol in BCS-KY resulted in a steady expression of the aerobic glycerol pathway at both temperatures and both time points. The presence of glucose in BCS-KYU led to the suppression of the transcription of glpFKX and glpD genes at both temperatures.
FIG. 3.
Results of semi-quantitative RT-PCR of the aerobic glycerol metabolic pathway. Y. pestis KIM5 were pre-grown with aeration in BCS medium supplemented with 0.2% K-gluconate at 26°C for 24 hours with two transfers. At OD600 = 0.3 (0 hours time point) additional carbon sources were added and temperature from 26°C to 37°C was changed in half of the samples. Approximately 1×108 bacteria were taken at 1 hour and 4 hours post-temperature shift from both 26°C and 37°C cultures grown in the presence of either 0.2% K-gluconate alone (K); both 0.2% K-gluconate and 0.2% glycerol (KY); or 0.2% K-gluconate, 0.2% glycerol, and 0.2% glucose (KYU). RT-PCR conditions consisted of 25 ng total RNA for 25 cycles of amplification. Expression values were derived from ImageJ software analyses and normalized to gyrB. Color scale reflects relative percentile changes in gene expression from the mean.
3.04 Mutants deficient in glpD do not accumulate enhanced concentrations of glycerol-3-phosphate
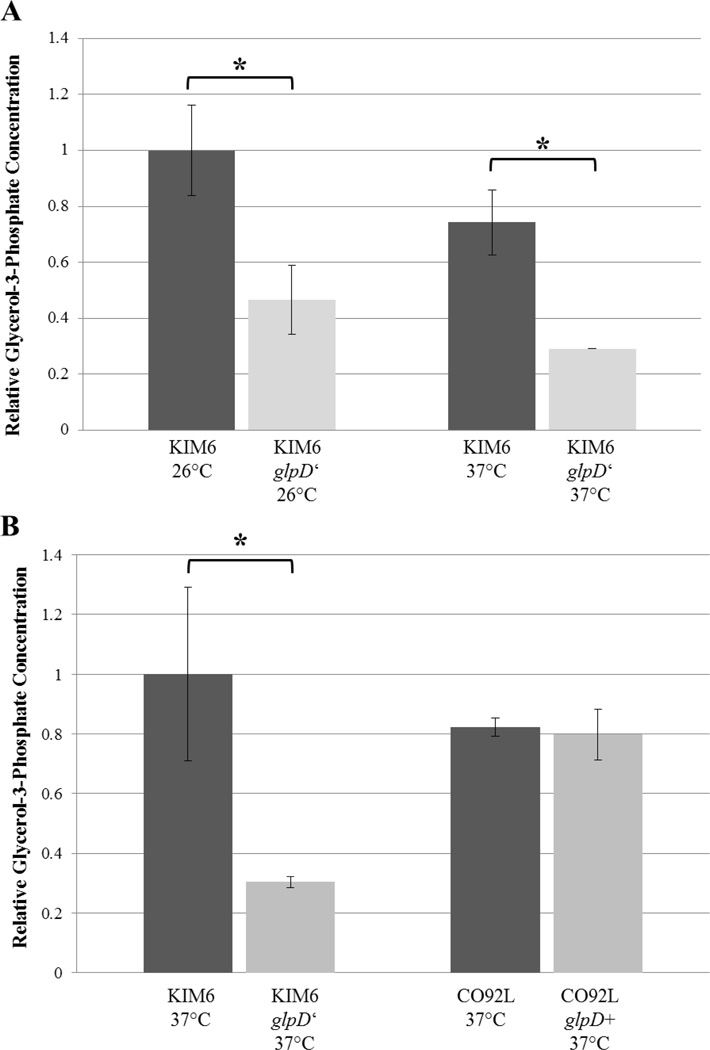
Glycerol-3-phosphate (G3P), the product of GlpK, is capable of repressing CRP activation in E. coli, and therefore regulates catabolite repression. Thus, the degree of G3P accumulation in Y. pestis glpD allelic exchange mutants was quantified via colorimetric assay (Biovision). The relative concentrations of G3P were inferior in the glpD deficient KIM6 mutant in respect to the control strain when incubated at either 26°C or 37°C. KIM6+ Y. pestis grown to exponential phase prepared by methanol/chloroform extraction demonstrate approximate 2.2-fold and 1.9-fold enhanced concentrations of G3P relative to KIM6+ glpD’ samples when incubated at 26°C and 37°C, respectively (Fig. 4A). Moreover, KIM6+ direct cell lysates indicated an approximate 3.3-fold increase in G3P relative to the glpD-deficient mutant. (Fig. 4B). No change in relative levels of G3P was detected amongst the CO92L and CO92L glpD+ mutant (Fig. 4B). Significant changes in expression amongst the glpD allelic exchange mutants and the respective controls were determined by Student’s T test (p-value < 0.05).
FIG. 4.
Colorimetric glycerol-3-phosphate determination (Biovision) of glpD allelic exchange mutants. A. KIM6 and KIM6 glpD’ were grown to logarithmic phase in HIB media supplemented with 0.2% glycerol at both 26°C and 37°C. Approximately 2×109 total cells as determined by optical density were pelleted, lysed, and processed by methanol/chloroform extraction. B. Intracellular concentrations of glycerol-3-phosphate quantification of KIM6, KIM6 glpD’, CO92L, and CO92L glpD+ direct lysates of 2×109 37°C logarithmic phase cultures. Relative glycerol-3-phosphate concentrations were determined from 50 µl, 25 µl, 12.5 µl aqueous phase extract (A) or cellular homogenate supernatant (B) per sample. Error bars reflect standard deviation from the mean. * P-value < 0.05 as determined by Student’s T test.
3.05 Y. pestis glycerol metabolism does not affect persistence in macrophages
Y. pestis aerobic glycerol metabolism has been shown to be simulated during conditions representative of the macrophage intraphagosomal environment [17]. Therefore, the potential contribution of glycerol metabolism upon Y. pestis survival and persistence in macrophages was assessed. We found that both Y. pestis uptake (0 hours time point post-infection) and intracellular viability (18 hours time point) were not influenced by either inactivation or restoration of glpD in the respective KIM6+ and CO92L backgrounds (Table 3). Furthermore, to circumvent unintentional shunting and enhanced induction of the anaerobic glycerol metabolic pathway, the glpABC system was also impaired in KIM6+ and KIM6+ glpD’ strains. Deletion of the glpABC operon had no impact upon Y. pestis persistence in macrophages.
TABLE 3.
Log10 CFU/mL Intracellular Viability of Y. pestis in RAW 264.7 Murine Macrophage-like Cells
| Y. pestis strain | 0 hrs PI | 18 hrs PI |
|---|---|---|
| CO92L | 5.33 ± 0.09 | 4.58 ± 0.29 |
| CO92L glpD + | 5.31 ± 0.14 | 4.82 ± 0.12 |
| KIM 6+/pCD1Ap | 5.64 ± 0.14 | 4.67 ± 0.24 |
| KIM 6+ glpD’/pCD1Ap | 5.55 ± 0.19 | 4.82 ± 0.06 |
| KIM 6+ ΔglpABC/pCD1Ap | 5.55 ± 0.2 | 4.74 ± 0.11 |
| KIM 6+ glpD’ΔglpABC/pCD1Ap | 5.67 ± 0.25 | 5.03 ± 0.34 |
3.06 No impact of glycerol metabolism upon Y. pestis in vivo infection
In order to ascertain any involvement amongst glycerol metabolism and virulence during infection of a mammalian host, Swiss Webster mice were challenged by subcutaneous and intranasal routes. The glpD dysfunctional mutants constructed in the KIM6+ background did not reveal any alterations in survival when challenged with either 2,000 CFU or 4,000 CFU by the i.n. route (Fig. 5A, 5C). Furthermore, no significant differences in survival were observed for mice challenged by the s.c. route with either 200 CFU or 500 CFU of the glpD dysfunctional mutant in respect to the virulent KIM6+ control (Fig. 5B, 5D). No change in survival was observed between mice challenged with the glpABC or the control strain. Inoculum as determined by plate counts were comparable amongst all strains.
FIG. 5.
Survival curves of murine challenge with KIM6+ and mutants deficient in glpD and/or the glpABC operon. A. Intranasal inoculation with 4000 CFU (~6 LD50 KIM6+/pCD1Ap) n = 5 mice per group. B. Subcutaneous inoculation 200 CFU (~20 LD50 KIM6+/pCD1Ap) n = 10 mice per group. C. Intranasal inoculation with 4000 CFU (~10 LD50 KIM6+/pCD1Ap) n = 5 mice per group D. Subcutaneous inoculation 500 CFU (~50 LD50 KIM6+/pCD1Ap) n = 5 mice per group. Verification of proper inoculum was determined by CFU enumeration. P-values were calculated by log-rank test.
3.07 Biofilm formation is enhanced by functional glpD
Despite displaying a comparable growth rate to the isotype control, the KIM6+ glpD-deficient mutant demonstrated a slight, yet significant, defect in biofilm formation as determined by crystal violet assay during growth in both HIB (Fig. 6A) as well as BCS chemically defined media utilizing 0.2% K-gluconate as the sole carbon source (Fig. 6B). Plasmid-based complementation of functional glpD restored wild-type biofilm formation in the KIM6+ glpD-deficient mutant. Conversely, the CO92L glpD+ allelic exchange mutant complemented with glpFKX derived from KIM6+ also conferred a significant increase in biofilm formation during growth in HIB (Fig. 6C). Both the CO92L chromosomal allelic exchange mutant expressing functional glpD derived KIM6+ as well as the CO92L glpD+ mutant complemented with glpFKX demonstrated a significant increase in biofilm formation when grown in BCS chemically defined media utilizing 0.2% K-gluconate as the sole carbon source (Fig. 6D). Biofilm formation of plasmid complemented glpD in CO92L resulted in comparable biofilm formation to the allelic exchange mutant.
FIG. 6.
Crystal violet assays of two independent experiments, each consisting of 6 technical replicates. Initial inoculum of OD600 = 0.1 Y. pestis aerobic glycerol metabolism mutants were incubated at 26°C for 24 hours incubation whilst shaking at 250 rpm. Error bars represent standard deviation from the mean. A. KIM6+ mutants cultured in HIB media. B. KIM6+ mutants grown in BCS-MOPS media supplemented with 0.2% K-gluconate. C. CO92 mutants grown in HIB media. D. CO92 mutants cultured in BCS-MOPS supplemented with 0.2% K-gluconate. * P-value < 0.05 as calculated by 2-tailed Student’s T test.
4. DISCUSSION
Phenotypic determination of glycerol fermentation amongst KIM6+ (biovar Medievalis) and CO92L (biovar Orientalis) glpD allelic exchange mutants provides formal proof that the 93 bp in-frame deletion within the glpD gene of Y. pestis biovar Orientalis isolates is sufficient to impair glycerol metabolism. In order to oxidize G3P to DHAP, GlpD requires a concurrent reduction of flavin adenine dinucleotide (FAD) to FADH. The deletion in the biovar Orientalis glpD gene (amino acids 7 to 37) overlaps with a characterized flavin-binding domain (amino acids 5 to 34) in E. coli GlpD [30].
Our findings also demonstrate the inability to ferment glycerol by biovar Orientalis strains is almost exclusively insured by additional impairments of the glpFKX operon. Upon expression of functional glpD in biovar Orientalis strains solely dysfunctional in glpK (PEXU-2, 195-P2, TS, and Kimberly), glycerol fermentation was not restored. However, glycerol fermentation was afforded upon supplemental expression of functional glpFKX. These findings indicate that y0876, annotated as a putative glycerol kinase, is insufficient to support glycerol fermentation [6]; whereas, glpK is essential for aerobic glycerol fermentation. Phylogenetic analyses suggest the diverse glpFKX disruptions in biovar Orientalis isolates were acquired subsequent to the shared deletion in glpD. We postulate the acquisition of mutations in glpK may reflect negative selection against GlpK activity in the presence of dysfunctional GlpD. In the absence of selective pressure, one could expect a random rate of mutation acquisition resulting in comparable degree of disruptions in glpF as observed in glpK. However, there are no alterations in the glpF gene amongst any biovar Orientalis isolates, Moreover, BLAST analysis of strains available in GenBank revealed that glpK is intact in all other Y. pestis biovars, suggestive of glpK evolutionary bias in the absence of functional glpD.
Nevertheless, three Orientalis isolates from China (F1984001, CMCC87001, and YN663) containing the characteristic 93 -bp deletion in glpD gene appear to have intact glpFKX operon (Fig. 1). Taking into account that the Orientalis biovar likely evolved in China, these type of strains circulating amongst the original endemic plague reservoirs of this region may reflect ancestors of the clone which spread over the globe during the 3rd plague pandemic [22]. Interestingly, two Chinese isolates YN1683 and YN1065 (respective whole genome shotgun sequence project accession numbers ADTD01000001.1 and ADTC00000000.1) are annotated as Orientalis biovar, despite possessing intact glpD and glpFKX genes (Fig. 1) [31]. Likely, the phenotypic inability to ferment glycerol resulted in the formal classification of both isolates as Orientalis biovar. The existence of such atypical glycerol non-fermenting isolates in the plague natural reservoirs where the vast majority of isolates are capable to ferment glycerol is described in the literature. Through subsequent culture of such glycerol non-fermenting isolates, it is possible to select colonies displaying a positive glycerol fermentation phenotype. For example, a well- known Russian bivalent live plague vaccine 1–17 consisted of the glycerol non-fermenting strain #1 and fermenting strain #17 [32]. Nevertheless, it was later found that the glycerol-negative phenotype of the strain #1 can revert to glycerol-positive one after several passages in liquid culture with extensive aeration [33].
In this study, we confirmed prior observations demonstrating enhanced expression of the aerobic glycerol metabolic pathway during early phase temperature shift from 26°C to 37°C via semi-quantitative RT-PCR [16]. When utilizing alternate carbon sources, there is a short-lived up-regulation of the aerobic glycerol metabolic pathway when incubated at 37°C. Thereafter, the expression normalizes to values consistent with growth at 26°C. Interestingly, expression of the tpiA gene encoding the triose phosphate isomerase remained constant throughout both temporal and incubation conditions. These results suggest that the short-term increase in glycerol metabolism observed during incubation at 37°C may have no implications upon energy metabolism. This notion is supported by 37°C growth kinetics which are comparable amongst the glpD dysfunctional mutant and the KIM6+ control. The transient early phase enhanced expression of the glycerol metabolic pathway upon temperature shift from 26°C to 37°C may be a reflection of temperature sensitive enzyme kinetics (such as the CRP positive regulator, or ArcB/A & GlpR negative regulators) resulting in a temporary relaxation of carbon catabolite regulation.
In E. coli, G3P has been shown to mediate catabolite repression through inhibition of adenylate cyclase, thus decreasing cAMP and preventing activation of the cAMP receptor protein, CRP [11]. We sought to address whether functional GlpK in the presence of inactive GlpD stimulates intracellular accumulation of G3P in Y. pestis. To the contrary, our findings reveal that Y. pestis KIM6+ mutants expressing dysfunctional glpD whilst retaining functional glpK have decreased concentrations of G3P in respect to glycerol fermentation positive controls. Sole complementation of glpD in the CO92L background had no impact upon G3P concentration. Taken together, deletion of glpD in the KIM6 background (retaining functional glpFKX) results in decreased concentrations of G3P; whereas, complementation of glpD in CO92L (dysfunctional glpKX) had no impact upon G3P levels. Therefore, GlpD activity may impart regulatory feedback upon GlpK and/or GlpX.
Pseudomonas aeruginosa infection in cystic fibrosis patients is primarily associated with lipid and amino acid metabolism. The most abundant lung surfactant lipid, phosphatidylcholine, is readily degraded into fatty acids and glycerol by P. aeruginosa [34]. Components of the aerobic glycerol metabolic pathway are notably up-regulated in patients infected with cystic fibrosis and are suggested to play a central role in the nutrient acquisition and pathogenesis of P. aeruginosa [35]. Y. pestis Psa fimbriae have been characterized to interact with phosphatidylcholine on alveolar epithelial cells and pulmonary surfactant [36]. Moreover, bioinformatics predictions suggest Y. pestis is capable of catabolizing phosphatidylcholine via the glycerol metabolic pathway. In our study, no change in the mean time to death was observed amongst the glpD dysfunctional mutants and the virulent controls when challenged by the intranasal route. Therefore, glycerol metabolism does not appear to play a role in Y. pestis pneumonic infection, highlighting a core difference in pathogenesis amongst chronic P. aeruginosa infection and the acute infectious process of Y. pestis. [37].
The glpB gene has been shown to be up-regulated in the rat bubo relative to in vitro growth at both 21°C and 37°C [38]. Moreover, virulence screening suggests that glpABC anaerobic respiration contributes to plague [39]. In our study, the anaerobic glpABC operon was deleted from KIM6+ as well as the KIM6+ glpD deficient mutant. In both cases, deletion of the glpABC operon did not result in an appreciable change in virulence for either intranasal or subcutaneous routes of inoculation. This discrepancy may result from two confounding variables. Foremost, the glpABC knock-out mutants utilized in our study were constructed in a biovar Medievalis background; whereas, biovar Orientalis strains were used in the aforemention virulence screening assessments. The evolution of Y. pestis is characterized by reductionist gene loss and inactivation [40]. Biovar Orientalis, a more recent evolutionary divergent, may have lost additional pathways which operate in a compensatory manner upon deletion of the glpABC operon in biovar Medievalis. Alternately, unique host physiology may exist amongst our virulence studies performed a murine model; whereas, a rat model system was utilized by other investigators.
Components of the glycerol metabolic pathway have been shown to be induced during infection of the flea vector in addition to flowcell biofilm formation [18]. Our findings reveal that dysfunctional glpD in the presence of functional glpK in a KIM6+ background results in a slight, yet significant, decrease in biofilm formation when cultured in both peptide rich HIB as well as chemically defined BCS media supplemented with both 0.2% glucose and 0.2% glycerol. Moreover, expression of functional glpD in CO92L results in a substantial increase in biofilm formation in the presence of both functional as well as dysfunctional glpK. However, sole complementation of glpK in CO92L did not enhance biofilm formation. The stimulation of biofilm formation upon the metabolism of non-PEP : PTS carbon sources corresponds with aspects of the Y. pestis infectious cycle. Mammalian blood and digestive byproducts found in the flea midgut contain relatively little carbohydrates [2]. Furthermore, Y. pestis carbohydrate metabolism is known to be repressed during infection of the flea [18]. However, the molecular mechanism defining the interplay amongst Y. pestis glycerol metabolism and biofilm formation has not been discerned.
Glycerol metabolism has also been shown to impair transcription of the lsr operon in E. coli [41]. The protein products of the lsr operon are essential for the import and processing of the autoinducer AI-2 quorum-sensing molecule. AI-2 quorum sensing has been shown to differentially modulate biofilm formation in a variety of pathogens. In E. coli, Salmonella enterica serovar Typhimurium, and Streptococcus intermedius, AI-2 quorum sensing enhances biofilm formation [42–44]. Glycerol negatively regulates E. coli AI-2 quorum sensing through two modes of actions: Foremost, glpD-deficient E. coli have enhanced intracellular concentrations of G3P which directly inhibits CRP-dependent activation of lsr transcription. Alternately, E. coli mutants deficient in glpD accumulate DHAP which is suggested to impair lsr transcription by a cAMP-CAP-independent mechanism involving LsrR, the lsr operon repressor. Interestingly, our findings suggest that Y. pestis glycerol metabolism does not regulate biofilm formation in a manner consistent with AI-2 regulation in E. coli considering Y. pestis mutants deficient in glpD do not accumulate G3P.
Contrary to E. coli, Staphylococcus aureus biofilm formation has been shown to be impaired by AI-2 [45]. Moreover, S. aureus Em-Mu disruption of glpD resulted in a defect in biofilm formation, potentially arising from a partial reduction in the β-1,6-linked N-acetylglucosamine polysaccharide intercellular adhesin (PIA) [46]. However, the impact of AI-2 upon Y. pestis biofilm formation is remains unclear considering biofilm phenotypes of Y. pestis AI-2 deficient mutants were assessed in combination with acyl homoserine lactone knockouts [47, 48]. In conclusion, Y. pestis biofilm regulation may transpire through a manner analogous to that of S. aureus in which AI-2, potentially subject to glycerol-mediated inhibition, may impair biofilm formation.
Supplementary Material
Highlights.
Biovar Orientalis glpD inactivation impairs glycerol fermentation
The inability to ferment glycerol is often insured by glpFKX mutations
Inactivation of GlpD in the presence of functional GlpK had no impact on virulence
Yersinia pestis aerobic glycerol metabolism promotes biofilm production
ACKNOWLDGEMENTS
Studies conducted for this manuscript were supported by the NIH/NIAID award Y1-AI-8401. S.P.W. was supported by the NIH/NIAID T32 pre-doctoral training grant on Tropical and Emerging Infectious Diseases (T32AI007526; PD: A.D.T. Barrett, Ph.D.). We would like to extend gratitude to Dr. Robert D. Perry for kindly providing the pCD1Ap construct. Also, we would like to thank Dr. Florent Sebbane for sharing his unpublished observations regarding the glycerol anaerobic metabolism of Y. pestis in a rat model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chouikha I, Hinnebusch BJ. Yersinia--flea interactions and the evolution of the arthropodborne transmission route of plague. Curr Opin Microbiol. 2012;15:239–246. doi: 10.1016/j.mib.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devignat R. [Varieties of Pasteurella pestis; new hypothesis] Bull World Health Organ. 1951;4:247–263. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou D, Tong Z, Song Y, Han Y, Pei D, Pang X, et al. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J Bacteriol. 2004;186:5147–5152. doi: 10.1128/JB.186.15.5147-5152.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charusanti P, Chauhan S, McAteer K, Lerman JA, Hyduke DR, Motin VL, et al. An experimentally-supported genome-scale metabolic network reconstruction for Yersinia pestis CO92. BMC Syst Biol. 2011;5:163. doi: 10.1186/1752-0509-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar JR, Bell RM. Biosynthesis in Escherichia coli of sn-glycerol 3-phosphate, a precursor of phospholipid. Kinetic characterization of wild type and feedback-resistant forms of the biosynthetic sn-glycerol-3-phosphate dehydrogenase. J Biol Chem. 1978;253:6354–6363. [PubMed] [Google Scholar]
- 8.Lin EC. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 9.Anderson A, Cooper RA. Gluconeogenesis in Escherichia coli The role of triose phosphate isomerase. FEBS Lett. 1969;4:19–20. doi: 10.1016/0014-5793(69)80184-5. [DOI] [PubMed] [Google Scholar]
- 10.Motin VL, Georgescu AM, Elliott JM, Hu P, Worsham PL, Ott LL, et al. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD) J Bacteriol. 2002;184:1019–1027. doi: 10.1128/jb.184.4.1019-1027.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppler T, Postma P, Schütz A, Völker U, Boos W. Glycerol-3-phosphate-induced catabolite repression in Escherichia coli. J Bacteriol. 2002;184:3044–3052. doi: 10.1128/JB.184.11.3044-3052.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 13.Kim TJ, Chauhan S, Motin VL, Goh EB, Igo MM, Young GM. Direct transcriptional control of the plasminogen activator gene of Yersinia pestis by the cyclic AMP receptor protein. J Bacteriol. 2007;189:8890–8900. doi: 10.1128/JB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan L, Han Y, Yang L, Geng J, Li Y, Gao H, et al. The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar microtus. Infect Immun. 2008;76:5028–5037. doi: 10.1128/IAI.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W, Roland KL, Kuang X, Branger CG, Curtiss R. Yersinia pestis with regulated delayed attenuation as a vaccine candidate to induce protective immunity against plague. Infect Immun. 2010;78:1304–1313. doi: 10.1128/IAI.01122-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, et al. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J Bacteriol. 2004;186:6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou D, Han Y, Qiu J, Qin L, Guo Z, Wang X, et al. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microbes Infect. 2006;8:2669–2678. doi: 10.1016/j.micinf.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog. 2010;6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holman WL. The Use of Decolorized Acid Fuchsin as an Acid Indicator in Carbohydrate Fermentation Tests with Some Remarks on Acid Production by Bacteria. The Journal of Infectious Diseases. 1914;15:7. [Google Scholar]
- 21.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 22.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler JM, Brubaker RR. Physiological basis of the low calcium response in Yersinia pestis. Infect Immun. 1994;62:5234–5241. doi: 10.1128/iai.62.12.5234-5241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J Cancer. 2004;41:47. [PubMed] [Google Scholar]
- 25.Cathelyn JS, Crosby SD, Lathem WW, Goldman WE, Miller VL. RovA, a global regulator of Yersinia pestis specifically required for bubonic plague. Proc Natl Acad Sci U S A. 2006;103:13514–13519. doi: 10.1073/pnas.0603456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JB, Telepnev MV, Zudina IV, Bouyer D, Montenieri JA, Bearden SW, et al. Evaluation of a Yersinia pestis mutant impaired in a thermoregulated type VI-like secretion system in flea, macrophage and murine models. Microb Pathog. 2009;47:243–251. doi: 10.1016/j.micpath.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heroven AK, Sest M, Pisano F, Scheb-Wetzel M, Steinmann R, Böhme K, et al. Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front Cell Infect Microbiol. 2012;2:158. doi: 10.3389/fcimb.2012.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YC, Hinnebusch BJ, Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A. 2008;105:8097–8101. doi: 10.1073/pnas.0803525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Surgalla MJ, Beesley ED. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin D, Larson TJ. Nucleotide sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1991;173:101–107. doi: 10.1128/jb.173.1.101-107.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci U S A. 2013;110:577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feodorova V, Motin V. Plague vaccines. In: Feodorova VA, Motin VL, editors. Vaccines against bacterial biothreat pathogens. Kerala, India: Research Signpost; 2011. pp. 176–233. [Google Scholar]
- 33.Korobkova EI, Pavlova LP. Influence of repeated passages at conditions of aeration on properties of vaccine strains 1, EV, 17. In: Nikolaev NI, editor. Specific prophylaxis of particularly dangerous infections. Medicine, Moscow, Russia: 1964. pp. 237–246. [Google Scholar]
- 34.Bernhard W, Wang JY, Tschernig T, Tümmler B, Hedrich HJ, von der Hardt H. Lung surfactant in a cystic fibrosis animal model: increased alveolar phospholipid pool size without altered composition and surface tension function in cftrm1HGU/m1HGU mice. Thorax. 1997;52:723–730. doi: 10.1136/thx.52.8.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Son MS, Matthews WJ, Kang Y, Nguyen DT, Hoang TT. In vivoevidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galván EM, Chen H, Schifferli DM. The Psa fimbriae of Yersinia pestis interact with phosphatidylcholine on alveolar epithelial cells and pulmonary surfactant. Infect Immun. 2007;75:1272–1279. doi: 10.1128/IAI.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finegold MJ. Pneumonic plague in monkeys. An electron microscopic study. Am J Pathol. 1969;54:167–185. [PMC free article] [PubMed] [Google Scholar]
- 38.Sebbane F, Lemaître N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, et al. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pradel E, Lemaître N, Merchez M, Ricard I, Reboul A, Dewitte A, et al. New Insights into How Yersinia pestis Adapts to Its Mammalian Host during Bubonic Plague. PLoS Pathog. 2014;10:e1004029. doi: 10.1371/journal.ppat.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jesudhasan PR, Cepeda ML, Widmer K, Dowd SE, Soni KA, Hume ME, et al. Transcriptome analysis of genes controlled by luxS/autoinducer-2 in Salmonella enterica serovar Typhimurium. Foodborne Pathog Dis. 2010;7:399–410. doi: 10.1089/fpd.2009.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed NA, Petersen FC, Scheie AA. AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob Agents Chemother. 2009;53:4258–4263. doi: 10.1128/AAC.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu D, Zhao L, Xue T, Sun B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 2012;12:288. doi: 10.1186/1471-2180-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, et al. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun. 2007;75:1079–1088. doi: 10.1128/IAI.01143-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 48.Bobrov AG, Bearden SW, Fetherston JD, Khweek AA, Parrish KD, Perry RD. Functional quorum sensing systems affect biofilm formation and protein expression in Yersinia pestis. Adv Exp Med Biol. 2007;603:178–191. doi: 10.1007/978-0-387-72124-8_15. [DOI] [PubMed] [Google Scholar]
- 49.Doll JM, Zeitz PS, Ettestad P, Bucholtz AL, Davis T, Gage K. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am J Trop Med Hyg. 1994;51:109–114. doi: 10.4269/ajtmh.1994.51.109. [DOI] [PubMed] [Google Scholar]
- 50.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.