Abstract
Titin is the largest known protein and a critical determinant of myofibril elasticity and sarcomere structure in striated muscle. Accumulating evidence that mRNA transcripts are post-transcriptionally regulated by specific motifs located in the flanking untranslated regions (UTRs) led us to consider the role of titin 5′-UTR in regulating its translational efficiency. Titin 5′-UTR is highly homologues between human, mouse, and rat, and sequence analysis revealed the presence of a stem-loop and two upstream AUG codons (uAUGs) converging on a shared in frame stop codon. We generated a mouse titin 5′-UTR luciferase reporter construct and targeted the stem-loop and each uAUG for mutation. The wild-type and mutated constructs were transfected into the cardiac HL-1 cell line and primary neonatal rat ventricular myocytes (NRVM). SV40 driven 5′-UTR luciferase activity was significantly suppressed by wild-type titin 5′-UTR (~70% in HL-1 cells and ~60% in NRVM). Mutating both uAUGs was found to alleviate titin 5′-UTR suppression, while eliminating the stem-loop had no effect. Treatment with various growth stimuli: pacing, PMA or neuregulin had no effect on titin 5′-UTR luciferase activity. Doxorubicin stress stimuli reduced titin 5′-UTR suppression, while H2O2 had no effect. A reported single nucleotide polymorphism (SNP) rs13422986 at position −4 of the uAUG2 was introduced and found to further repress titin 5′-UTR luciferase activity. We conclude that the uAUG motifs in titin 5′-UTR serve as translational repressors in the control of titin gene expression, and that mutations/SNPs of the uAUGs or doxorubicin stress could alter titin translational efficiency.
Keywords: Titin, Ttn, 5′-UTR, uORF, translation, cardiomyocytes
Introduction
The cardiac sarcomere is the basic contractile unit of the heart and is comprised of a large assembly of myofilament proteins that are responsible for force generation. At ~3–3.7mDa, titin (TTN) is the largest and third most abundant myofilament protein in striated muscle [1]. Titin serves as a molecular scaffold for contractile and many other sarcomeric proteins to organize around. Physiologically, titin accounts for the majority of passive tension during diastole and restoring force after systole [1]. While the majority of studies on titin have focused on genomic mutations, structural and mechanical roles in the sarcomere, the mechanisms regulating titin gene expression are virtually unknown.
Regulation of gene expression is a complex orchestration of events and is controlled at multiple levels including transcription, mRNA processing/decay, and protein translation [2,3]. Even though changes in mRNA transcript levels are commonly extrapolated to reveal functional differences at the protein level, the correlation between mRNA and protein expression is highly variable and this discrepancy may be due to post-transcriptional regulation [4,5]. Indeed, Schwanhäusser et.al. demonstrated in NIH3T3 fibroblasts that only 40% of cell protein levels were determined by mRNA levels and stressed the importance of translational control as the main regulator of protein abundance [6]. Cap-dependent translational initiation occurs when 43S ribosomal complex binds near the 5′-7-methylguanylate cap of the untranslated region (UTR) and scans in a 5′ to 3′ direction until it recognizes the AUG start codon of the main open reading frame (mORF) [7]. Upon recognition, the 60S ribosomal subunit joins and translational elongation occurs until a stop codon is read resulting in termination of translation [7]. It is increasingly being recognized that the 5′-UTR can contain cis-regulatory elements such as upstream AUGs (uAUGs), secondary structure, and upstream ORF (uORF) which are thought to repress translational efficiency[2,8]. uORFs are thought to reduce translational efficiency by a process called leaky scanning in which the 43S ribosomal complex either recognizes the uORF and initiates translation or scans past the uORF to the mORF. In the case of translating the uORF, the translating ribosome can either scan through and reinitiate at the mORF or stall which leads to mRNA degradation [2,8,9]. Overall, uORFs reduce translational initiation at the mORF. Calvo et al. reported 50% of human and mouse protein coding genes contain an uORF and further demonstrated that these uORF repress translational activity by 30–80% [9]. Secondary structures such as stem loops have been demonstrated to repress translational efficiency by reducing the 43S ribosomal complex’s ability to scan [7]. To examine whether titin is post-transcriptionally regulated by its 5′-UTR, we performed in silico analysis of titin 5′-UTR and identified a stem loop and two uORF that were highly conserved between human and mouse. We hypothesized that titin gene expression is post-transcriptionally regulated by cis-regulatory elements within its 5′-UTR.
Methods
All experiments involving animals were approved by the Animal Care and Use Committee at Vanderbilt University Medical Center, and carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Primer sequences are provided in the online Supplement.
Cardiomyocyte Isolation and Cell Culture
Neonatal rat ventricular myocytes (NRVMs) were isolated from hearts of 1–2-day old Sprague-Dawley rat pups (Charles River Laboratory) as previously described [10]. Adult rat ventricular myocytes were isolated from hearts of adult male Sprague-Dawley rats (Charles River Laboratory) as previously described [10]. The HL-1 cardiac cell line was kindly provided by Dr. Claycomb and human embryonic kidney 293 (HEK-293) cell line purchased from ATTC.
Titin half-life
NRVM were treated with transcriptional inhibitor actinomycin D or dimethyl sulfoxide (DMSO) vehicle for 1hr, 6hr, 12hr, 24hr and 48hr. Total RNA was isolated using TRIzol® Reagent and DNAse treated according to manufacturer’s instructions (Life Technologies™). Total RNA was converted into cDNA and RT-PCR was performed using SYBR Green with specified Ttn and 18S primers. Titin mRNA expression was normalized to 18S RNA. Data was fitted into a nonlinear one phase exponential decay model (PRISM Software) to calculate the titin mRNA half-life.
Titin 5′-UTR fluorescence in situ hybridization
Cardiomyocyte mRNA in situ hybridization was performed according to the method of Cripe et al. [11]. Briefly, adult cardiomyocytes were fixed in a 1:9 formaldehyde/methanol (vol/vol) and hybridized with a digoxigenin labeled 222 nucleotide probe directed against the 5′-UTR of titin or a 265 nucleotide labeled probe against myosin heavy chain mRNA (5565–5830 nucleotides of X15938). The probes were detected using a rhodamine labeled antidigoxigenin conjugate.
5′ RACE and Plasmids
RNA was extracted from c57BL/6 mice cardiac and skeletal muscle tissue using Trizol. The titin 5′-UTR sequence was identified using 5′ Rapid Amplification of cDNA Ends kit (Ambion). The 5′-UTR of titin was subcloned downstream of an SV40 promoter and upstream of the firefly luciferase gene in a pGL3 reporter (Promega). All mutagenesis were performed using the QuikChange lightening mutagenesis kit (Agilent Technologies). Mutagenesis of the uORFs were performed by mutating uAUG1 and/or uAUG2 to AAG, rs13422986 SNP reporter construct was generated by mutating the G to A in the 5′-UTR.
Cell transfection and Stimulation
NRVMs were serum starved overnight (1% FBS/DMEM) followed by transient transfection with 200ng of pGL3 control or 5′-UTR pGL3 reporter constructs, along with 100ng of pRL-TK Renilla luciferase construct or pRL-null Renilla luciferase construct (Promega) via Lipofectamine 2000 delivery (Life Technologies). Following 24 h transfection, NRVM were either electrically paced at 2 Hz, 30V, 10ms using a Ion Optix myopacer, or treated with, 20ng/mL neuregulin, 200μM phorbol 12-myristate 13-acetateor (PMA), 0.5 μM doxorubicin, 50 μM H2O2, or 3 μM blebbistatin for 24hrs.
Luciferase Reporter Assay
Following treatment, cells were harvested and assayed for luciferase activities using a GloMax-Multi Detection system (Promega). The ratio of firefly:Renilla luciferase activities was calculated to normalize for differences in cell number and transfection efficiency. Each assay was run in triplicate and the ratio values were normalized to control to allow for between-experiment comparison.
Statistical Analysis
Data are reported as mean±SEM. Where appropriate, results were either analyzed by Student’s t-test or ANOVA with a Bonferroni’s multiple comparison post-hoc test. P<0.05 was considered statistically significant.
Results
Titin mRNA half-life and localization in cardiomyocytes
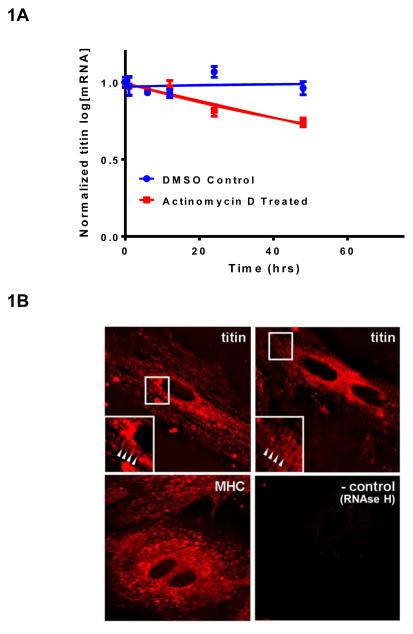
To assess the half-life of titin mRNA, NRVMs were treated with transcriptional inhibitor actinomycin D and steady state titin mRNA levels measured at various time points. Fitting the data to a one phase exponential decay model, titin mRNA half-life was estimated at 66hr (Figure 1A). Subcellular mRNA targeting provides a mechanism to control gene expression[12]. We used fluorescent in situ hybridization (FISH) and observed that titin mRNA was localized to the sarcomere (based on the periodic staining pattern) and in the cytoplasm (Figure 1B, top panels). This striation pattern appeared to be specific for titin as the mRNA for another myofilament protein, myosin heavy chain, was non-sarcomeric and diffusely distributed (Figure 1B, bottom left panel). The relatively long half-life of titin mRNA and its sarcomeric localization is presumed to place greater importance on post-transcriptional control as a means of regulating protein expression.
Figure 1.
Titin mRNA half-life and subcellular localization in cardiomyocytes. A: NRVM were treated with Actinomycin D or DMSO control for 0,1,6,12,24, and 48hrs. Titin gene expression was analyzed by qRT-PCR and normalized to 18S ribosomal RNA. Normalized ct values were normalized to a standard curve of titin gene expression. Data was fitted to a one-phase exponential decay model to calculate titin mRNA half-life. Data is presented as mean ± SEM, (n=3). B: Titin mRNA is localized to the sarcomere and cytoplasm. Top panels: Striated appearance of titin mRNA staining (see arrowheads in inset) characteristic of muscle sarcomeres. Bottom panels: MHC mRNA staining was diffuse with no detectable periodicity. A sample subjected to Rnase treatment served as a negative control.
Identification of mouse titin 5′-UTR
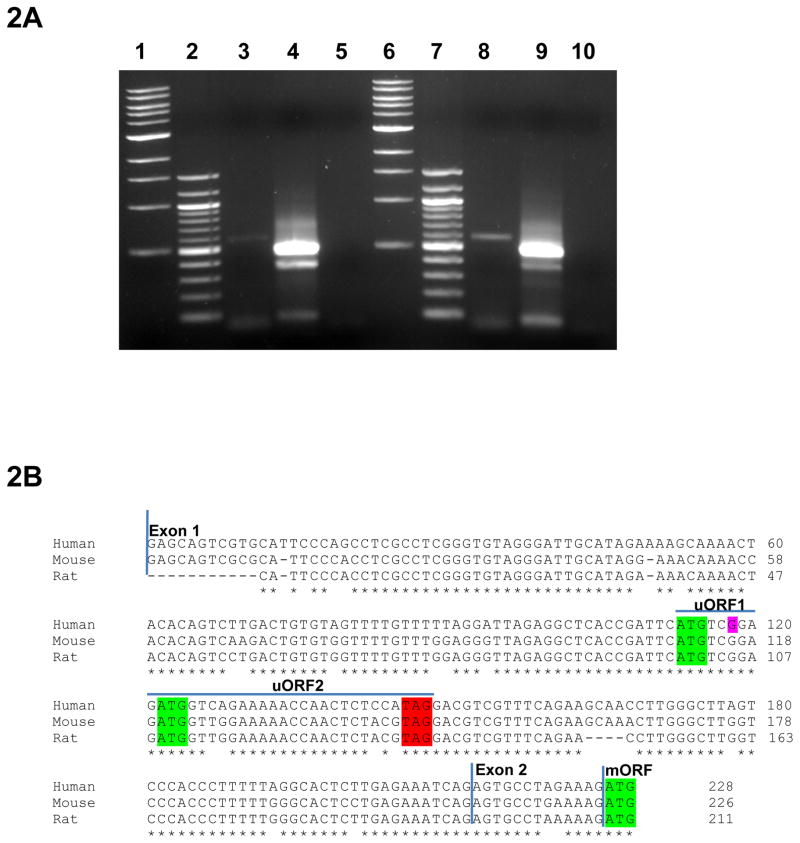
The UTRs of mRNA transcripts are known to regulate protein translation; in this study we specifically investigated the role of titin 5′-UTR on regulating translational efficiency. To identify titin 5′-UTR we performed a 5′ RACE and amplified two RACE PCR products from mouse cardiac and skeletal muscle total RNA (Figure 2A). The expected 508 bp product contained the 223 nt canonical titin 5′-UTR as determined by sanger sequencing. The faint 400 bp product was a PCR artifact in both cardiac and skeletal muscle. The 5′-UTR sequence was entirely consistent with Genebank’s mouse annotation, and sequence alignment shows high homology (>87%) between mouse, human and rat (Figure 2B). We performed in silico analysis of titin 5-’UTR and were able to identify two uAUGs (uAUG1 and uAUG2) that were both in frame with the same stop codon which we will denote as uORF1 and uORF2. We used the Mfold computer program to predict the secondary structure of the 5′-UTR sequence and were able to identify a highly stable stem loop structure (Figure S1A). Importantly, these motifs were conserved in human, mouse, and rat suggesting regions of greater regulatory importance.
Figure 2.
5′ RACE identifies single titin 5′ UTR in mouse cardiac and skeletal tissue. A: 5′ RACE assay was performed using mouse cardiac and skeletal tissue. 5′ RACE assay identifies single expected 514 bp RACE product. Lanes 1 and 6 are 1 kb ladder. Lanes 2 and 7 are 100bp ladder. Lanes 3, 4 and 5 are cardiac outer PCR reaction, cardiac inner PCR reaction and cardiac TAP treated negative control, respectively. Lanes 8, 9 and 10 are skeletal outer PCR reaction, skeletal inner PCR reaction and skeletal TAP treated negative control respectfully. B: Clustal Omega multiple sequence alignment of titin 5′-UTR between human, mouse and rat. Titin 5′-UTR sequence spans from exon 1 to part of exon 2. Human, mouse and rat 5′-UTR sequences are >87% homologous. uAUG1, uAUG2 and the mORF between human, mouse and rat (shown in green) are conserved. The in frame TAG stop codon for uAUG1 and uAUG2 is shown in red. Overline blue depicts the uORF reading frames. SNP rs13422986 is shown in purple.
Characterization of functional cis regulatory elements within mouse titin 5′-UTR
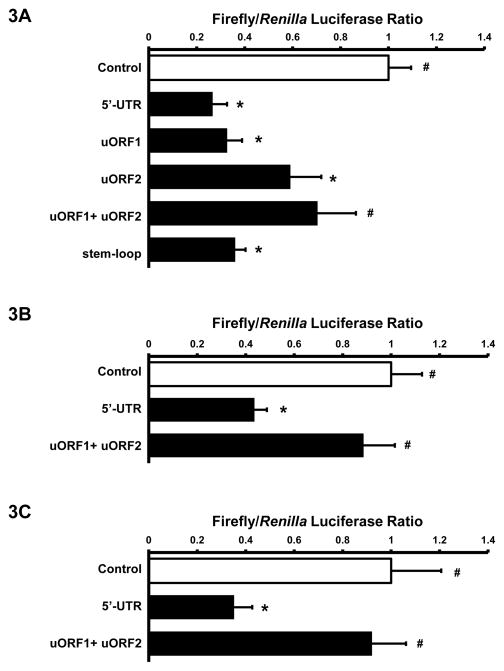
To determine if titin 5′-UTR regulates titin translation, we subcloned the 223 nucleotide 5′-UTR into the pGL3 firefly luciferase reporter vector. The wild-type titin 5′-UTR reporter activity showed significant repression in transiently transfected HL-1 cells (Figure 3A). To determine the functional relevance of the 5′-UTR stem-loop, we mutated 3 sites in the stem-loop that according to Mfold resulted in complete disruption of the stem-loop [13]. Dual luciferase reporter assays in HL-1 cells showed that the mutated stem-loop had no effect on titin 5′-UTR suppression of luciferase activity (Figure 3A). To investigate the role of the two uORFs we mutated the uAUG of each uORF alone (uORF1 and uORF2) and in combination (uORF1+uORF2). The results demonstrated that mutated uORF1 had no effect, while mutated uORF2 increased luciferase activity when compared to 5′-UTR reporter activity. Mutated uORF1+uORF2 reporter generally restored firefly luciferase activity to pGL3 control reporter levels (Figure 3A). These results were confirmed in NRVM showing that titin 5′-UTR resulted in 57% repression compared to pGL3 control luciferase activity while mutated uORF1+uORF2 resulted in near complete de-repression of titin 5′-UTR activity (Figure 3B). Addition of the mouse titin 3′-UTR did not significantly reduce luciferase activity in NRVM (Figure S1.B), thus we conclude that under basal conditions titin 5′-UTR is the main regulatory component controlling titin translation.
Figure 3.
Titin 5′-UTR suppresses pGL3 reporter activity. A: HL-1 cells were transiently transfected with 100ng RTK and 200ng of either pGL3 control, wild-type Ttn 5′UTR, or mutant ttn 5′UTR reporter constructs (uORF1, uORF2, uORF1+ uORF2, stem-loop). Cells were lysed and assayed for firefly and Renilla luciferase activities (n=5–6 for each group). B: NRVM were transfected with pGL3 control, wild-type, or mutant uORF1+uORF2 ttn 5′UTR reporter construct and assayed for firefly and Renilla luciferase activity (n=3–4 for each group). C: HEK 293 cells were transfected with pGL3 control, wild-type 5′UTR, or mutant uORF1+uORF2 Ttn 5′UTR reporter construct and assayed for firefly and Renilla luciferase actvity (n=4–6 for each group). All data displayed as Mean±SEM, * p<0.05 vs. pGL3 control; # p<0.05 vs. 5′ UTR; groups compared by ANOVA.
To determine if the repressive effect of titin 5′-UTR is cardiac specific, we transfected the titin 5′-UTR reporter plasmids into non-muscle HEK293 cells. Similar to the results observed in cardiac cells, 5′-UTR luciferase activity was significantly suppressed in HEK 293 cells (Figure 3C), and by and large the suppression stems from the uORFs.
Taken together, these data suggests that the presence of uORF motifs in titin 5′-UTR results in innate suppression of translational efficiency.
Titin 5′-UTR Stimulation
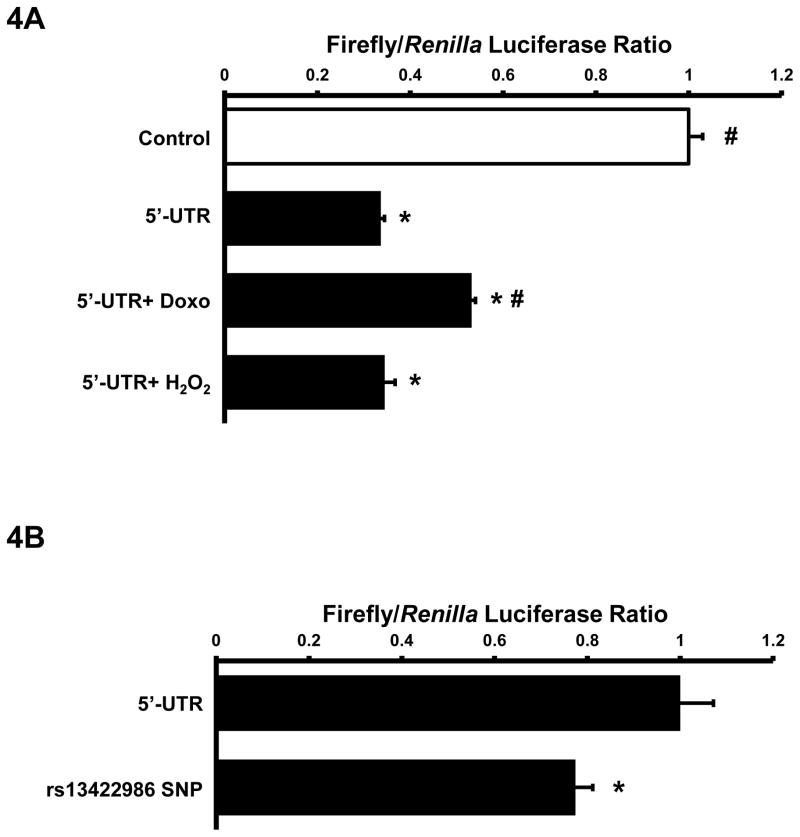
To test whether cardiac hypertrophic stimuli could enhance titin translational efficiency via 5′-UTR de-repression, we transiently transfected NRVM with the titin 5′-UTR reporter and electrically paced cells at 2 Hz or treated cells with neuregulin or PMA. Neuregulin and PMA stimulation did not alter titin promoter activity in NRVM (data not shown), suggesting that these growth stimuli did not alter titin mRNA transcripts. Neither electrical stimulation nor neuregulin and PMA altered titin 5′-UTR luciferase activity (Figure S2.A–C). Since NRVM spontaneously contract in culture we also mechanically arrested NRVM with blebbistatin, but again found no change in titin 5′-UTR luciferase activity. Stress stimuli, such as doxorubicin and oxidative stress enhance overall protein degradation which has been shown to stimulate de novo protein synthesis. NRVMs treated with doxorubicin significantly enhanced titin 5′-UTR luciferase activity compared to the untreated controls (Figure 4A). H2O2, on the other hand, had no effect titin 5′-UTR luciferase activity.
Figure 4.
Regulation of titin translational efficiency. A: NRVM were transiently transfected with 100ng RTK and 200ng pGL3 Control or Ttn 5′ UTR via lipofectamine delivery for 24 hours. NRVM were either treated with vehicle, 0.5μM doxorubicin or 50μM H2O2 for 24hrs. Cells were lysed and assayed for luciferase activities (n=3). B: NRVM were transiently transfected with 100ng RTK and 200ng Ttn 5′-UTR or mutated rs13422986 5′-UTR for 24hrs. Cells were lysed and assayed for luciferase activities (n=4). All data presented as Mean±SEM, *p<0.05 vs 5′-UTR. Groups were analyzed by ANOVA or student’s t-test.
These data suggest that growth stimuli do not, but stress stimuli such as doxorubicin could modulate titin translational efficiency via its 5′-UTR.
SNPs in titin 5′-UTR
We scanned the NIH SNP database and found several SNPs throughout the 5′-UTR titin; however, none modulated the generation or loss of the uORFs. We did find a SNP in the −4 position of uAUG2 Kozak sequence. Strong Kozak sequences contain conserved −3 and +4 sites (underlined) of the GCCA/GCCAUGG which promotes efficient 43S ribomosomal complex recognition [14,15]. Given that uORF2 accounts for the majority of suppression of titin 5′-UTR translational efficiency, we hypothesized that SNP at the −4 position of uAUG2 could alter titin reporter function. We generated the SNP 5′-UTR reporter and were able to show that the rs13422986 SNP significantly decreased titin 5′-UTR luciferase activity by 17% in NRVM (Figure 4B).
Discussion
Titin is the largest known protein (~3–3.7 mDa) and presumably the costliest to synthesize. In this report, we found that titin mRNA is localized at the sarcomere and has a relatively long half-life of ~3 days which would favor control of titin protein expression at the post-transcriptional level. We provide evidence that titin 5′-UTR represses its translation efficiency through a conserved uORF. Mutations/SNP in the titin 5′-UTR or stress stimuli could alter titin translational efficiency and ultimately affect cardiac development and growth.
Schwanhäusser et.al. analyzed the global mRNA half-lives in mammalian NIH3T3 cells via pulse labelling mRNA followed by deep sequencing and found that median mRNA half-life is 9 hours [6]. We found that titin mRNA is remarkably stable with a half-life of ~3 days, which is similar to the reported protein half-life of titin in skeletal muscle cells in culture [16]. Housekeeping genes are abundantly expressed and energetic constraints placed on the cell may explain why these genes tend to have stable mRNAs and proteins [6]. We speculate that stable titin mRNAs and proteins may have evolved in part to minimize the energetic cost of transcription, synthesis and transport of large proteins in a cell. Our in situ hybridization study together with previous reports showing that titin mRNA is striated and co-localized with titin protein as well as sarcomeric 60S ribosomal subunit, suggests that titin mRNA is targeted to the sarcomere directly at the site of protein localization and translation [17,18]. This close spatial proximity between titin translation and sarcomere incorporation may facilitate local feedback regulation of mRNA post-transcriptional control.
The control of gene expression via the 5′-UTR represents one important mechanism of control within the complex network of gene regulation. Identification of cis-regulatory elements such as secondary structure, uAUGs, uORFs have been shown to negatively regulate translational efficiency [8,9,19]. Mutations in the 5′-UTR of protein coding genes have been associated with diseases [8]. Moreover, alternative transcriptional start sites for a gene when expressed in different tissues or in disease have been reported that have altered 5′-UTR control of translational efficiency [20,21]. We identified a single 223 nt titin 5′-UTR transcript via 5′RACE assay in wild-type mouse cardiac and skeletal muscle. The absence of any additional bands signifies that titin 5′-UTR does not stem from alternative promoters, transcriptional initiation sites, and RNA processing events under normal conditions. Whether alternative transcriptional start sites exist for titin with muscle development or disease remains to be determined.
The −3 and +4 nucleotides of the consensus Kozak sequence are key nucleotides involved in binding eIF2α of the mammalian 43S subunit and are present in uAUG2 but absent in uAUG1 of titin 5′UTR [15]. Thus, uAUG1 does not have a strong Kozak sequence and may be skipped over by the ribosome in a process called leaky scanning [8]. Our results are consistent with this notion as uORF2 accounted for most of titin 5′-UTR translational repression, while uORF1 had a negligible effect. Short hairpin motifs in the 5′-UTR have been shown to repress translational efficiency by impeding ribosomal scanning [7]. We found a conserved short hairpin motif in titin 5′-UTR, however, mutation of the short hairpin motif within the 5′-UTR did not relieve repression, suggesting that this motif does not play a role in titin translational regulation. It is possible that our mutagenesis may not have sufficed to fully disrupt the hairpin, regardless our data suggest that the titin 5′-UTR translational repression comes primarily from uORF2.
A further question relates to the degree of titin 5′-UTR translational control following treatment with growth factors. We compared growth stimuli neuregulin (PI-3-kinase and p70S6K activation) and PMA (ERK activation) on titin 5′-UTR translational efficiency. We also investigated the effect of electrical pacing which was previously shown in feline cardiomyocytes to increase 5′-UTR translational efficiency of c-jun 5′-UTR reporter [22]. On the other hand, mechanical inactivation of spontaneously beating NRVM has been shown to depress alpha myosin heavy chain mRNA translation and result in cellular atrophy [23]. Neither growth stimuli, electrical pacing, nor mechanical arrest in cardiomyocytes altered titin 5′-UTR reporter activity, thus it appears that stimuli modulating cardiac growth do not affect titin protein synthesis at the 5′-UTR level.
Our data shows that doxorubicin stress was able to increase 5′-UTR activity. This observation is interesting because doxorubicin has been shown to induce calpain mediated titin degradation [10]. Increasing titin protein synthesis via enhanced 5′-UTR translational efficiency could act as a compensatory feedback mechanism under conditions of stress-induced titin degradation. Support for stress induced translational upregulation stems from a study in which endoplasmic reticulum stress in kidney cells bypassed the inhibitory uORF in the 5′-UTR of PKD2 mRNA thereby increasing it translation [24].
Calvo et al. reported that SNPs generated or deleted uORFs in the 5′-UTRs of at least 507 human genes and showed that all SNPs resulted in a 30–60% decrease in reporter levels when compared to control reporters [9]. Currently there are 24 disease conditions associated with SNPs altering uORF function [8]. A SNP at the −6 position of the Kozak sequence of cardiac transcription factor GATA4 was shown to co-segregate with atrial septal defect [25]; in vitro experiments showed that this SNP resulted in decreased translational initiation and protein expression of GATA4. We demonstrated that the titin rs13422986 T/C SNP at the uAUG2 Kozak sequence decreases titin translational efficiency. This SNP has a minor allele count of T=0.015/32 in 1000 genomes project. We therefore speculate that the population harboring the rs13422986 SNP is synthesizing less titin which may potentially affect cardiac growth.
It should be noted that the 3′-UTR of titin mRNA may be another important control mechanism at the post-transcriptional level [26]; we are currently investigating this notion. Nevertheless, our result showing that addition of the titin 3′-UTR to the 5′-UTR reporter did not alter luciferase activity suggests that the 5′-UTR is the main regulatory component of basal titin mRNA translational efficiency.
In summary, our study found that titin mRNA transcripts have a long half-life and are localized at the sarcomere. We provide evidence that titin translational efficiency is controlled in part by its 5′-UTR mainly through a cis-regulatory uORF. Given that titin synthesis is energetically costly, we believe that the uORF could play a role in fine tuning titin mRNA translation by serving as a passive brake to prevent overproduction of titin and hence wasting of cellular resources.
Supplementary Material
Highlights.
Ttn 5′-UTR contains a conserved uORF that represses mRNA translational efficiency.
Growth stimuli does not affect Ttn 5′UTR translational repression.
Doxorubicin stress stimuli attenuates Ttn 5′UTR translational repression.
A SNP in the Kozak sequence of uORF exacerbates Ttn 5′UTR translational repression.
Acknowledgments
Funding
This study was funded in part by NIH grant RO1HL095813 (CCL), a Graduate Research Assistant Supplement RO1HL095813-S1 (AGC), and Vanderbilt University Stahlman (CCL).
We thank Douglas B. Sawyer for stimulating discussions and valuable feedback.
Footnotes
Conflict of Interest
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Cox B, Kislinger T, Emili A. Integrating gene and protein expression data: pattern analysis and profile mining. Methods. 2005;35:303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 7.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbosa C, Peixeiro I, Romao L. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet. 2013;9:e1003529. doi: 10.1371/journal.pgen.1003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim CC, Zuppinger C, Guo X, Kuster GM, Helmes M, Eppenberger HM, Suter TM, Liao R, Sawyer DB. Anthracyclines induce calpain-dependent titin proteolysis and necrosis in cardiomyocytes. J Biol Chem. 2004;279:8290–8299. doi: 10.1074/jbc.M308033200. [DOI] [PubMed] [Google Scholar]
- 11.Cripe L, Morris E, Fulton AB. Vimentin mRNA location changes during muscle development. Proc Natl Acad Sci U S A. 1993;90:2724–2728. doi: 10.1073/pnas.90.7.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 15.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs WB, Kim IS, Struve A, Fulton AB. Biosynthesis of titin in cultured skeletal muscle cells. J Cell Biol. 1989;109:2189–2195. doi: 10.1083/jcb.109.5.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen TH, Saetersdal T. Translocation of 60S Ribosomal Subunit in Spreading Cardiac Myocytes. Journal of Histochemistry & Cytochemistry. 1998;46:963–969. doi: 10.1177/002215549804600810. [DOI] [PubMed] [Google Scholar]
- 18.Fulton AB, Alftine C. Organization of protein and mRNA for titin and other myofibril components during myofibrillogenesis in cultured chicken skeletal muscle. Cell Struct Funct. 1997;22:51–58. doi: 10.1247/csf.22.51. [DOI] [PubMed] [Google Scholar]
- 19.Capell A, Fellerer K, Haass C. Progranulin transcripts with short and long 5′-untranslated regions (UTR) are differentially expressed via post transcriptional and translational repression. J Biol Chem. 2014 doi: 10.1074/jbc.M114.560128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegrzyn JL, Drudge TM, Valafar F, Hook V. Bioinformatic analyses of mammalian 5′-UTR sequence properties of mRNAs predicts alternative translation initiation sites. BMC Bioinformatics. 2008;9:232. doi: 10.1186/1471-2105-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Zalen S, Nijenhuis M, Jonkman MF, Pas HH. Two major 5′-untranslated regions for type XVII collagen mRNA. J Dermatol Sci. 2006;43:11–19. doi: 10.1016/j.jdermsci.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Spruill LS, McDermott PJ. Role of the 5′-untranslated region in regulating translational efficiency of specific mRNAs in adult cardiocytes. FASEB J. 2009;23:2879–2887. doi: 10.1096/fj.08-128447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikcevic G, Heidkamp MC, Perhonen M, Russell B. Mechanical activity in heart regulates translation of alpha-myosin heavy chain mRNA but not its localization. Am J Physiol. 1999;276:H2013–2019. doi: 10.1152/ajpheart.1999.276.6.H2013. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Zheng W, Wang Q, Lara C, Hussein S, Chen XZ. Translational up-regulation of polycystic kidney disease protein PKD2 by endoplasmic reticulum stress. FASEB J. 2013;27:4998–5009. doi: 10.1096/fj.13-236075. [DOI] [PubMed] [Google Scholar]
- 25.Mohan RA, van Engelen K, Stefanovic S, Barnett P, Ilgun A, Baars MJ, Bouma BJ, Mulder BJ, Christoffels VM, Postma AV. A mutation in the Kozak sequence of GATA4 hampers translation in a family with atrial septal defects. Am J Med Genet A. 2014 doi: 10.1002/ajmg.a.36703. [DOI] [PubMed] [Google Scholar]
- 26.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.