Abstract
Dynactin is the longest known cytoplasmic dynein regulator, with roles in dynein recruitment to subcellular cargo and in stimulating processive dynein movement. The latter function was thought to involve the N-terminal microtubule binding region of the major dynactin polypeptide p150Glued, though recent results disputed this. To understand how dynactin regulates dynein we generated recombinant fragments of the N-terminal half of p150Glued. We find that the dynein-binding coiled-coil α-helical domain CC1B is sufficient to stimulate dynein processivity, which it accomplishes by increasing average dynein step size and forward step frequency, while decreasing lateral stepping and microtubule detachment. In contrast, the immediate upstream coiled-coil domain, CC1A, activates a novel diffusive dynein state. CC1A interacts physically with CC1B and interferes with its effect on dynein processivity. We also identify a role for the N-terminal portion of p150Glued in coordinating these activities. Our results reveal an unexpected form of long-range allosteric control of dynein motor function by internal p150Glued sequences, and evidence for p150Glued auto regulation.
INTRODUCTION
A single major form of cytoplasmic dynein plays critical roles in many aspects of cell movement, including vesicular, virus, and nuclear transport, cell migration, nuclear import, and mitotic and meiotic chromosome movement. Dynein adapts to diverse functions via a number of regulatory factors, most notably dynactin, LIS1, NudE, and NudEL1. LIS1 increases dynein force output by prolonging stalling under load2 and also acts as a clutch to control dynein movement3. Dynactin is a megadalton-sized multi-subunit complex4 involved in dynein recruitment to subcellular cargo5, 6 and in promoting processive dynein travel along microtubules7–9. Despite the importance of the latter activity for neuronal viability and other aspects of basic cell physiology, its underlying mechanism remains unknown.
p150Glued is the largest polypeptide component of dynactin, and is thought to be the principal active subunit, containing both dynein and microtubule binding sites10–12 (Fig. 1A,B). The latter, near the p150Glued N-terminus, targets dynactin to growing microtubule ends13–15 and contributes to organization of the mitotic spindle16 and initiation of retrograde axonal transport17, 18. Antibody inhibition of the microtubule binding region was reported to diminish dynactin stimulation of dynein processivity7, suggesting that p150Glued might act by stabilizing and prolonging the dynein-microtubule interaction. However, removal of the p150Glued N-terminus had no effect on travel distance for individual dynein molecules in vitro or for vesicular cargo in vivo9, 15, 16. Sequential truncations of p150Glued through its coiled-coil domain CC1 produced a stepwise decrease in dynein processivity9. Despite a role for this region in dynein binding in vertebrate dyneins10–12, dynein binding persisted in the yeast dynactin complex9. Dynactin also contributes to coordinating kines in and dynein activities in vivo19, 20, though whether this effect is direct is unknown.
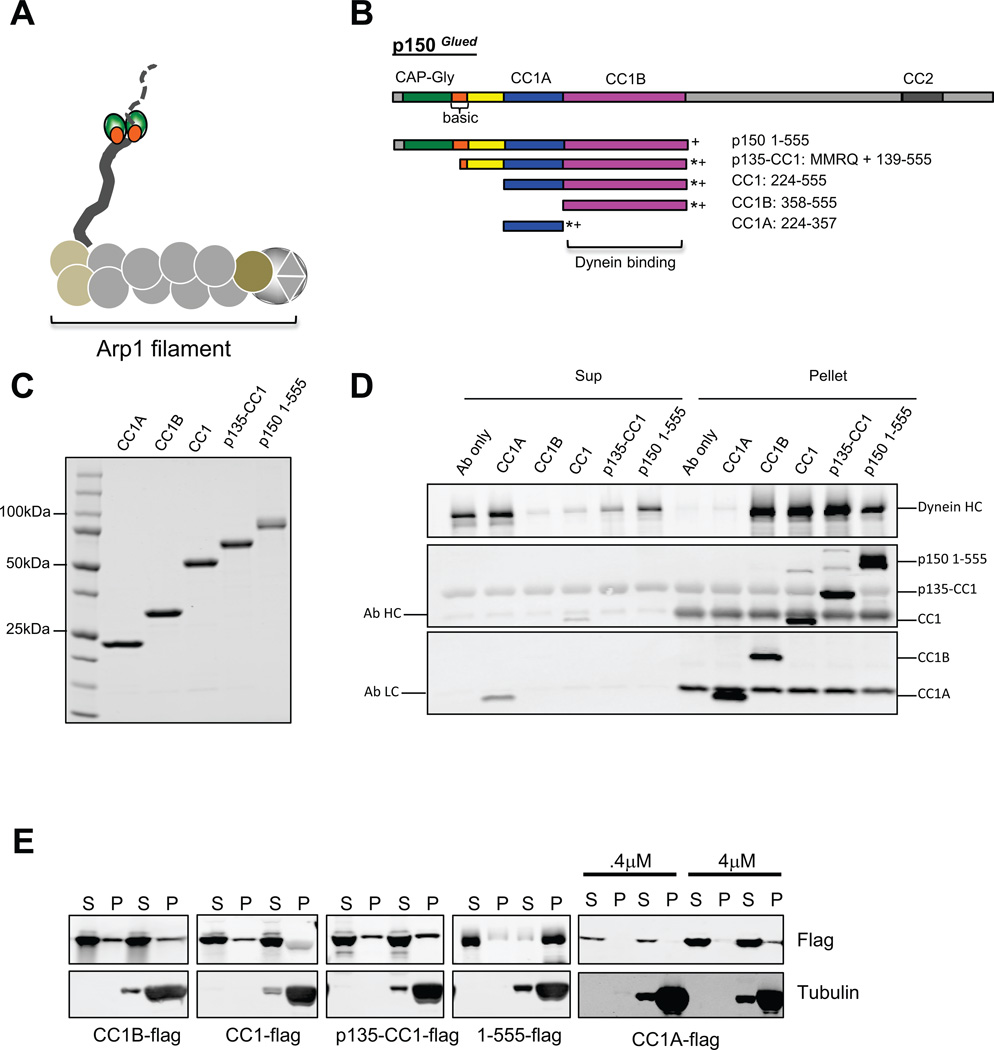
Figure 1. Characterization of dynactinp150Glued fragments.
(A) Diagram of the dynactin complex, an ~35 nm long filament of the actin-like protein Arp1 and associated factors. The p150Glued subunit is seen as a projecting arm at left with globular N-terminal microtubule binding CAP-Gly (green) and basic (orange) domains near the end. (B) Domain map of p150Glued and its subfragments used in this study, which are C-terminally flag (*) and His6 (+) tagged. CC1B and CC1A contain slightly different boundaries from those used in our previous study21. (C) Coomassie stained gel of the purified p150Glued fragments used in this study. (3 independent experiments) (D) Calf brain cytoplasmic dynein was tested for co immunoprecipitation with the Flag-tagged p150Glued fragments using anti-flag antibody. Bands were visualized by Western blotting with antibodies to dynein heavy chain and the flag tag. All fragments except CC1A bound dynein. (3 independent experiments) (E) Microtubule (MT) co sedimentation of p150Glued fragments. Fragments (0.1 uM, unless otherwise noted) were centrifuged in the absence or presence of taxol-stabilized MTs. Only p150 1-555, which alone contains the CAP-Gly and complete basic regions of p150Glued, showed substantial co-sedimentation with MTs. (2–3 independent experiments) CC: coiled-coil α-helix, Sup: supernatant, Ab: antibody.
We have now carried out detailed analysis of p150Glued fragments to reconstitute dynactin regulatory activity and understand the mechanisms by which dynactin regulates dynein. We find that the N-terminal half of p150Glued is sufficient to reconstitute stimulation of processive dynein travel along microtubules, as well as an additional form of behavior, dynein diffusion on microtubules. We identify specific processivity and diffusivity sub domains of p150Glued and test how dynein stepping behavior contributes to these functions. In the course of this work we also identify novel auto regulatory interactions between p150Glued sub domains, which for the first time reveals dynactin to be a highly complex regulatory machine.
RESULTS
Analysis of p150GluedFragments
To elucidate the molecular basis for dynein regulation by dynactin we produced a series of p150Glued fragments spanning the N-terminal 555 a.a. residues, including the microtubule and dynein binding sites (Fig. 1 B, C). This region is thought to be highly elongated, as suggested by its extensive predicted α-helical coiled-coil content. Electron microscopy has also revealed a pair of small globular elements presumed to contain the microtubule binding domains toward the tip of a fine, projecting fiber, which may contain the predicted CC1 α-helical coiled-coil4 (Fig. 1A,B). This structure is broken into two sub regions, one of which, CC1B21, 22 is responsible for binding to the dynein intermediate chains located within the tail portion of the dynein complex (Fig. 1 A–D).
Each of our p150Glued fragments was well-behaved as judged by physicochemical analysis (Supplementary Fig 1, Supplementary Table 1). Circular dichroism spectroscopy showed substantial α-helical structure, suggested to be organized into a coiled-coil by a ratio of θ220/θ208 >123 (Supplementary Table 1). The fraction of α-helix in each fragment showed an approximate correspondence to the predicted coiled-coil content (Supplementary Table 1). CC1, CC1A and CC1B consisted largely of reversible, temperature-sensitive α-helical structure (Supplementary Fig. 1), as recently reported for similar fragments with somewhat different boundaries22.
Our largest p150Glued fragment, p150 1-555, expressed using baculovirus, showed substantial microtubule binding, in contrast to the shorter fragments (Fig. 1E), consistent with a role for the N-terminal CAP-Gly and nearby basic domain ofp150Glued in microtubule binding8, 10, 12. All fragments containing CC1B pulled down purified calf brain cytoplasmic dynein, whereas CC1A did not (Fig. 1D).
Effects of p150Glued Fragments on Single-molecule Dynein Behavior
We used a laser trap bead assay to permit simultaneous analysis of both dynein force generation and transport along microtubules. Beads adsorbed with the p150 1-555 fragmental one bound to microtubules and exhibited prolonged bidirectional motility (τ = 54.3 ± 11 sec), determined to be diffusional (D0=0.069 ± 1E-4 µm2/sec; Table 1) by MSD analysis (Supplementary Fig. 2D). This behavior is reminiscent of that for some previously characterized p150Glued fragments8 as well as for brain dynein-dynactin mixtures24. Beads adsorbed with the other p150Glued fragments did not interact with microtubules.
Table 1.
Summary of single molecule data.
| p150 1-555 |
Dynein | Dynein + p150-1-555 |
Dynein + p135-CC1 |
Dynein* + CC1 |
Dynein+ CC1B |
Dynein+ CC1A |
Dynein*+ CC1A† (1:7000) |
Dynein + CC1A+ CC1B |
|
|---|---|---|---|---|---|---|---|---|---|
| Binding fraction | N/A | N/A | Unaffected | Unaffected | Decreased (70% → 30%) | Unaffected | Unaffected | Decreased (34% →17%) | Decreased (37% →23%) |
| Processive beads | 0% | 83 ± 4% | 70 ± 6 % | 50 ± 5% | 30 ± 4% | 83 ± 4% | 83 ± 6% | 0% | 48 ± 6% |
| Run length (µm) | N/A | 0.92 ± 0.09 | 1.84 ± 0.24 | 1.94 ± 0.17 | 0.86 ± 0.05 | 1.99 ± 0.16 | 0.76 ± 0.17 | N/A | 0.65 ± 0.05 |
| Velocity (µm/sec) | N/A | 0.41 ± 0.04 | 0.24 ± 0.02 | 0.33 ± 0.03 | 0.24 ± 0.05 | 0.33 ± 0.03 | 0.28 ± 0.03 | N/A | 0.31 ± 0.04 |
| Force (pN) | N/A† | 1.18 ± 0.03 | 1.03 ± 0.02 | 1.09 ± 0.02 | 1.13 ± .02 | 1.2 ± .03 | 1.1 ± .02 | N/A | 1.05 ± 0.01 |
| MT binding time of processive beads (sec) | N/A | 2.7 ± 0.4 | 6.6 ± 0.8 | 5.4 ± 0.6 | 2.8± 0.4 | 5.2 ± 1 | 2.7 ± 0.6 | N/A | 2.8± 05 |
| MT binding time of diffusive beads (sec) | 54.3 ± 11 | 12.9 ±2.07 | 29.8 ± 7.8 | 41.2 ± 9.4 | 8.2 ± 2 | N/A | N/A | 78.4 ± 13.6 | 3.5 ± 0.6 |
| Diffusion coefficient of diffusive beads (µm2/sec) | 0.07 ± .0001 | 0.017 ± .0002 | 0.04 ± 0.0015 | 0.018 ± .0003 | 0.0198 ± .0002 | N/A | N/A | 0.018 ± .000013 | 0.025 ± .00053 |
p150Glued 1-555 alone or diffusive dynein motors do not generate force, as shown in Fig. S3 (and see Supplemental Discussion).
Microtubule-associated transport of beads to which p150Glued 1-555 alone, cytoplasmic dynein alone, or dynein with p150Glued fragments were adsorbed. To test the effects of p150Glued fragments on dynein, the motor protein was adsorbed at single molecule concentrations, the beads blocked to further protein binding, exposed to a 150-fold molar excess of fragment, centrifuged, and examined for motile behavior in an optical trap.
CC1 and high concentrations of CC1A required increased dynein to maintain a 30% bead-microtubule binding fraction. Error is SEM.
CC1A was used at a 7000-fold molar excess over dynein and remained present during motility analysis.
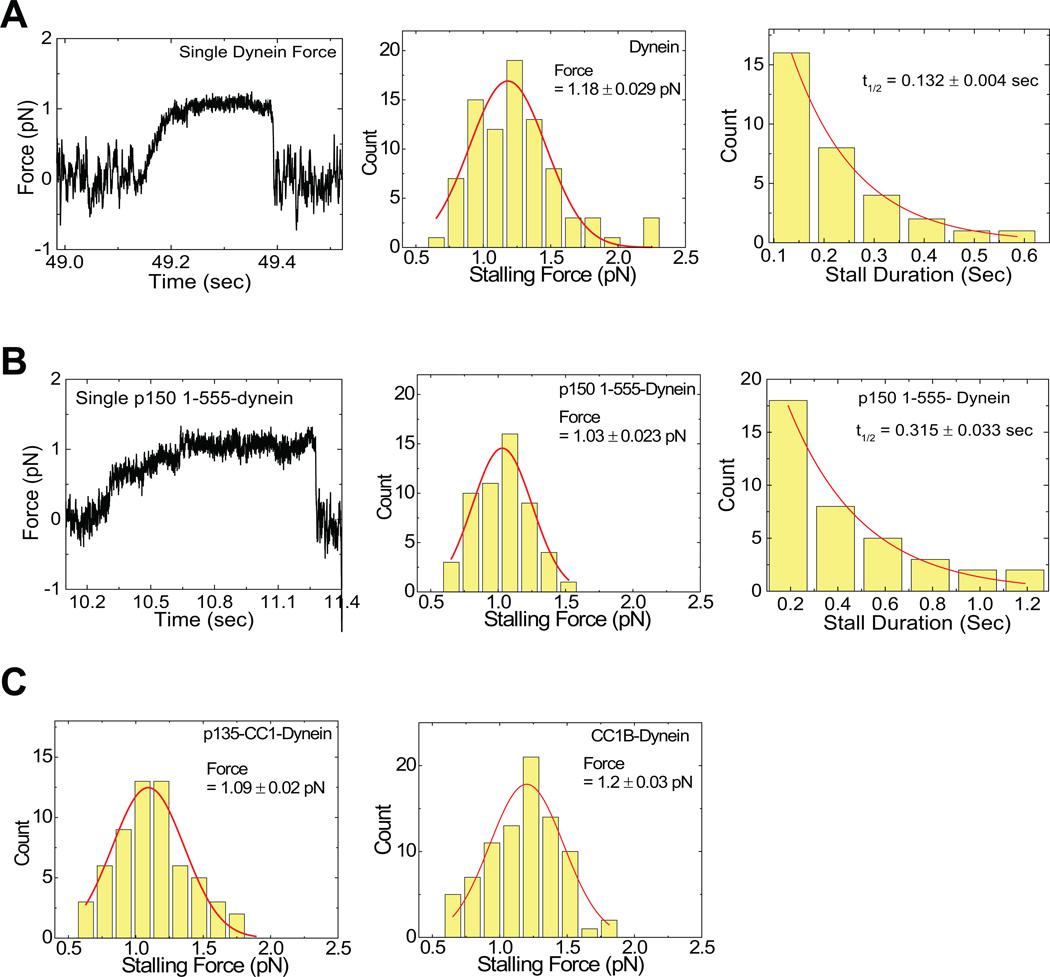
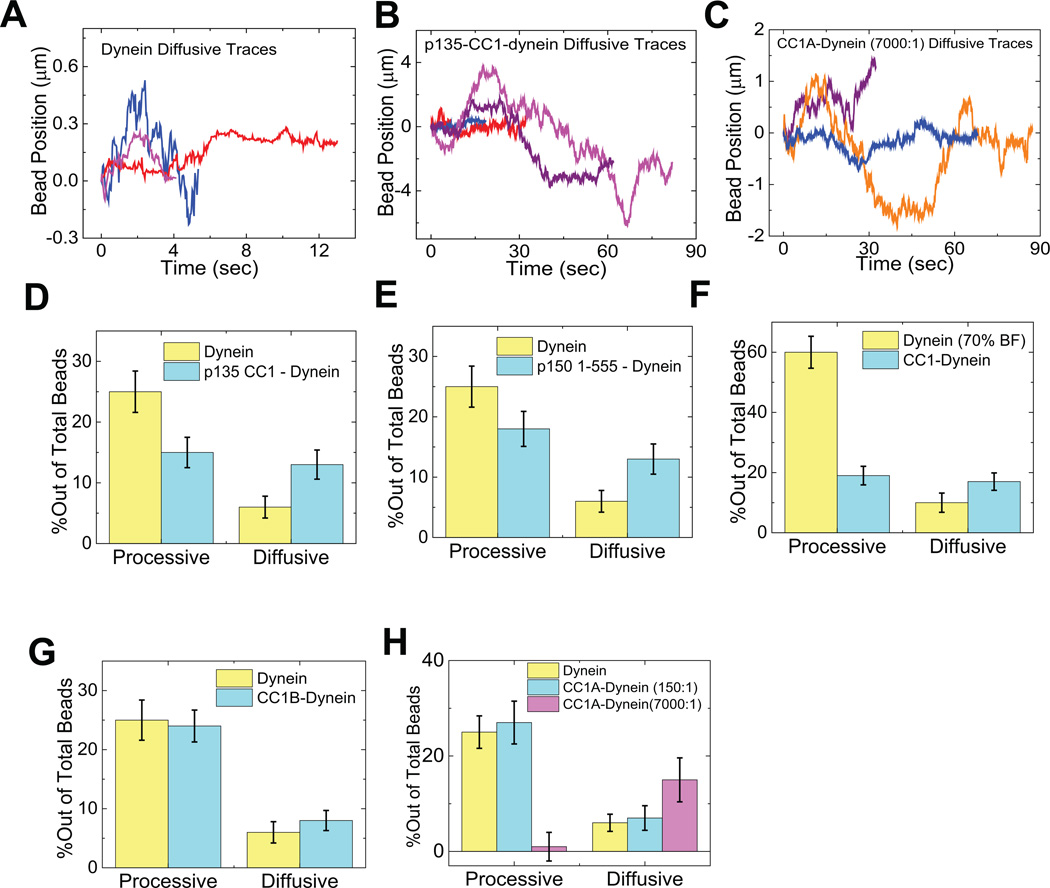
To examine dynein behavior, we adsorbed the motor protein to beads at single-molecule concentrations, blocked the beads with casein to prevent further protein recruitment, exposed themtoa 150-foldmolar excess of dynactin fragment, and washed them by centrifugation and re-suspension in motility buffer. We then captured individual beads using a laser trap, applied them to microtubules, determined stall force for the bound dynein, and then allowed the bead to travel freely along the microtubule (Supplementary Fig. 2A). Dynein alone showed predominantly processive behavior (Fig. 2A, Table 1) associated with an average stall force of approximately 1.2 pN, corresponding to a single molecule (Fig. 4 A,B)2, 25. This result is consistent with our earlier analysis25 supporting processive movement for individual dynein molecules based on Poisson analysis of processivity as a function of motor dilution. As previously reported by us and others25, 26 some events were diffusive (Fig. 3A–C), as indicated by a linear MSDplot (not shown, and Suppl. Fig 2 D–G). The ratio of processive to diffusive runs was generally high for our calf brain dynein (Fig. 3 D, E, G), but varied among preparations and appeared to decrease with preparation age (see Methods)
Figure 2. Effects of dynactin fragments on dynein single molecule processivity.
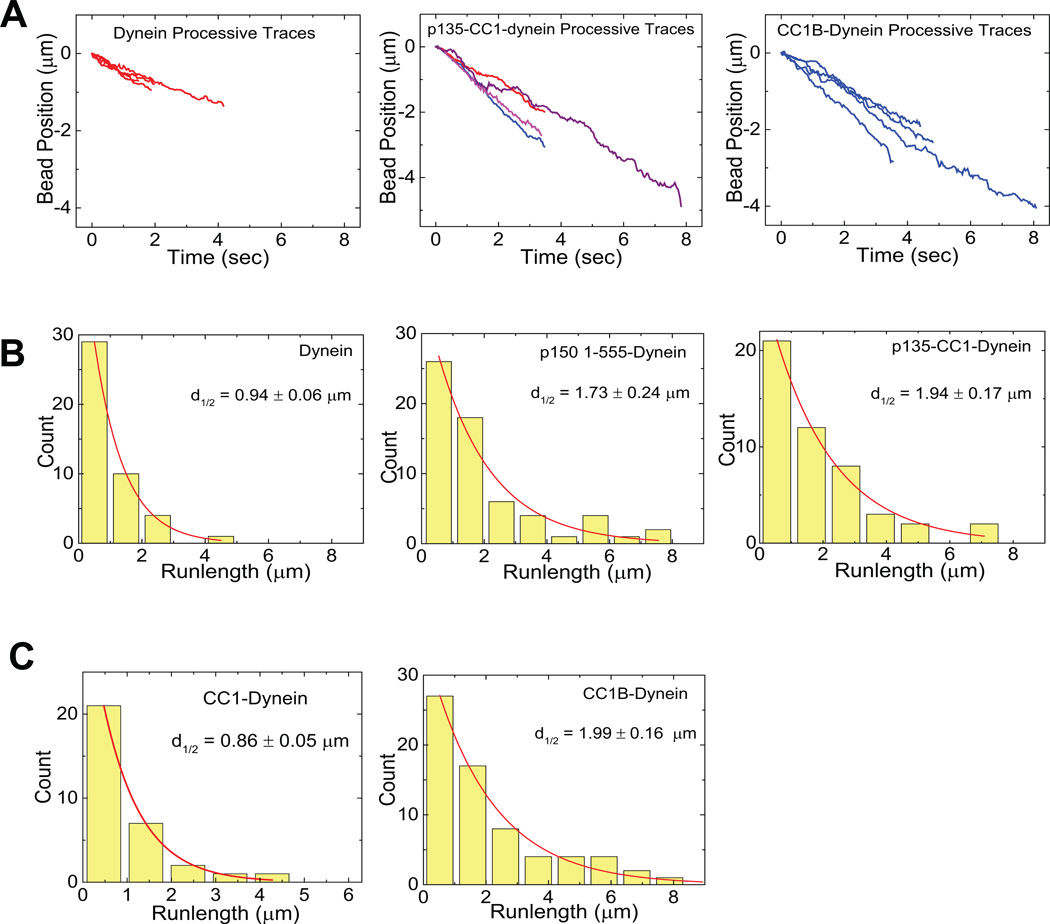
(A) Sample traces (4 each) showing processive motion of beads with dynein alone (left) dynein plus p135-CC1 (middle) and dynein plus CC1B (right). (B) Bead run length distributions for dynein alone (left), dynein with P150 (middle), or dynein with P135 (right). (C) Run length histograms for dynein with CC1 (left) and CC1B (right). In each case, data were well described by a single decaying exponential (chi-squared test) with mean travel for dynein plus the p150Glued 1-555, p135-CC1, and CC1B fragments approximately double that for dynein alone (see also Table 1). All processivity measurements were done at a bead binding fraction of ~30%.
Figure 4. Effects of dynactin fragments on dynein single molecule force production.
(A) reflects dynein alone, and (B) reflects dynein with P150. Shown are example force traces (A, B, Left), distributions of stalling forces (100 force events) (A, B, middle) and distributions of durations of force producing events (40 stalling events) (A,B, Right). C) shows distributions of stalling forces for dynein with P135 (left) and CC1B (right).
Figure 3. Effects of dynactin fragments on dynein single molecule diffusivity.
Sample traces (3–4) showing diffusive motion of beads with dynein alone (A), dynein with p135-CC1 (B) and dynein with high amounts of CC1A (C). (D–H) Quantitation of the effects of different dynactin fragments on the overall amount of diffusive vs processive binding events. Because the percentage of dynein-only diffusion could change between experiments, each graph reports the results of the control (dynein-alone) performed at the same time as the fragment experiment. The sum of the diffusive and processive components reflects the total bead binding fraction (e.g. in D, the total bead binding fraction for dynein alone was 30%). P135 (D) and P150 (E) decreased the gross number of processive events, and increased the number of diffusive events. CC1 (F) decreased the gross number of processive events, but did not increase diffusive events, and CC1B (G) did not alter the number of either class of events. CC1A (H) had no effect at low concentrations, but induced diffusion and suppressed processive motion at high concentrations. These data reflect numerous experiments. Error bars (Fig 3 D–H) were obtained using equation , where p refers to bead binding fraction and N refers to the total number of bead tested. For dynein alone, dynein-p135 CC1, dynein-p150 1-555, dynein-CC1B, dynein-CC1 (70% BF), dynein-CC1A (150:1), and for dynein-CC1A (7000:1) the respective number of beads checked was N = 165, 195, 180, 253, 161, 70, and 70. For high dynein (70% BF), N = 85. Each experiment was reproduced in the lab at least 3 times. The single dynein alone control experiment was repeated before each measurement of dynein with a dynactin fragment.
The addition of the p150Glued fragment p150 1-555 had two effects: it increased the frequency of diffusive behavior and the duration of both diffusive and processive behavior (Fig. 2B, Fig. 3E, Table 1). Because the absolute bead-binding fraction was unchanged by the fragment (Table 1), the change in relative diffusive vs processive frequencies reflects a conversion from one form of behavior to the other. Importantly, the average length for the processive runs was increased ~2.0-fold relative to that for dynein alone (Fig. 2B, middle; Table 1), very similar to the effect reported for the complete dynactin complex7–9. Stall forces for processively moving dynein beads exposed to p150 1-555 were again close to 1 pN (Fig. 4B, sup fig 2), and stall duration was prolonged (Fig. 4, A,B, right).
In addition to their greater relative frequency, (Fig. 3E), the duration of diffusive dynein events was prolonged by ~3-fold. (Table 1) The diffusion coefficient was smaller than that for p150Glued1-555 alone (Suppl. Fig. 2F), but greater than that for dynein, suggesting that at least a component of the observed diffusion is associated with the dynein-microtubule interaction. The diffusing beads produced minimal force (see Supplementary Fig. 3 and methods), with small force peaks at 0± 0.35–0.5 pN, behavior which was not seen for trapped beads diffusing in the trap without a motor (Supp. Fig 3, methods). The minimal motor effects likely reflect random binding to microtubules and release, rather than directed motion, because their magnitude is consistent with thermal noise without the motor (Supp. Fig 3A, right), and because individual trajectories (Supp Fig 3, F,I) show sudden decreases in thermal motion, rather than obvious binding and subsequent directed transport.. The observation of genuine diffusive and processive states, and the dynactin-induced changes in their relative frequency and properties, suggested that the dynein regulatory activity of the complete dynactin complex resides substantially within the N-terminal half of p150Glued, encouraging us to search further for specific regulatory loci.
The p135-CC1 fragment was designed to correspond to the N-terminus of a naturally occurring p150Glued splice variant lacking the CAP-Glydomain and most of the basic region27. Remarkably, when combined with dynein, p135-CC1 had effects similar to those of p1501-555, again increasing the frequency of diffusive events (Fig. 3D), as well as the duration of both processive and diffusive microtubule interactions (Fig 2, 3; Table 1). Because p135-CC1 doesn’t show significant MT binding (Fig. 1E), its effect on diffusion must result from changes to dynein behavior (Suppl. Fig 2G). The absence of the microtubule binding CAP-Gly and basic regions in p135-CC1 reveals further that these regions are dispensable for regulating dynein processivity as well as diffusion. For the diffusive beads, force production was again minimal (Supplementary Fig. 3 H–J). Dynein in this state could easily be displaced along microtubules at the lowest optical trap setting (< 0.4 pN), though lateral detachment required higher forces (data not shown). Thus, these data demonstrate that dynactin can actually turn off dynein force production, a novel regulatory function, while allowing dynein to retain its interaction with microtubules.
In contrast to these results, the CC1 fragment severely inhibited the dynein-microtubule interaction, requiring a 2.3-fold increase in dynein concentration to achieve an equivalent number of microtubule binding events. CC1 inhibition reflected a selective decrease in the frequency of processive events (Fig. 3F), as the absolute number of diffusive microtubule interactions was unchanged. These results together revealed CC1 to have a potent inhibitory effect on processive dynein motion), despite the presence in this fragment of the dynein-binding portion of p150Glued. This observation has relevance for the long-standing use of CC1 in cell expression studies as a potent dynein inhibitor28. While part of its effect seems due to its ability to compete with both dynactin and NudE-LIS1 for dynein binding21, 29, our current results identify an additional direct toxic effect on dynein function.
Distinct p150Glued Processivity and Diffusivity-enhancing Domains
To identify subdomains responsible for dynactin regulation of dynein and the unexpected inhibitory effects of CC1, we examined smaller p150Glued fragments. Strikingly, CC1B induced a clear 2.2-fold increase in dynein run-length (Fig. 2A right, 2C, middle; table 1) but did not change the frequency of either processive or diffusive events (Fig. 3G). The magnitude of the effect on run lengths was similar to values observed for the entire dynactin complex7, 9 and to that for the longer p150Glued fragments characterized in this study (Table 1; Fig. 2). They support a role for the CC1 region reported in yeast9 though our analysis of CC1 (above) and CC1A (below) reveals clear differences from the yeast work. Dynein force production was normal (Fig. 4), confirming single motor behavior. These results identified CC1B as the processivity-stimulating domain of p150Glued.
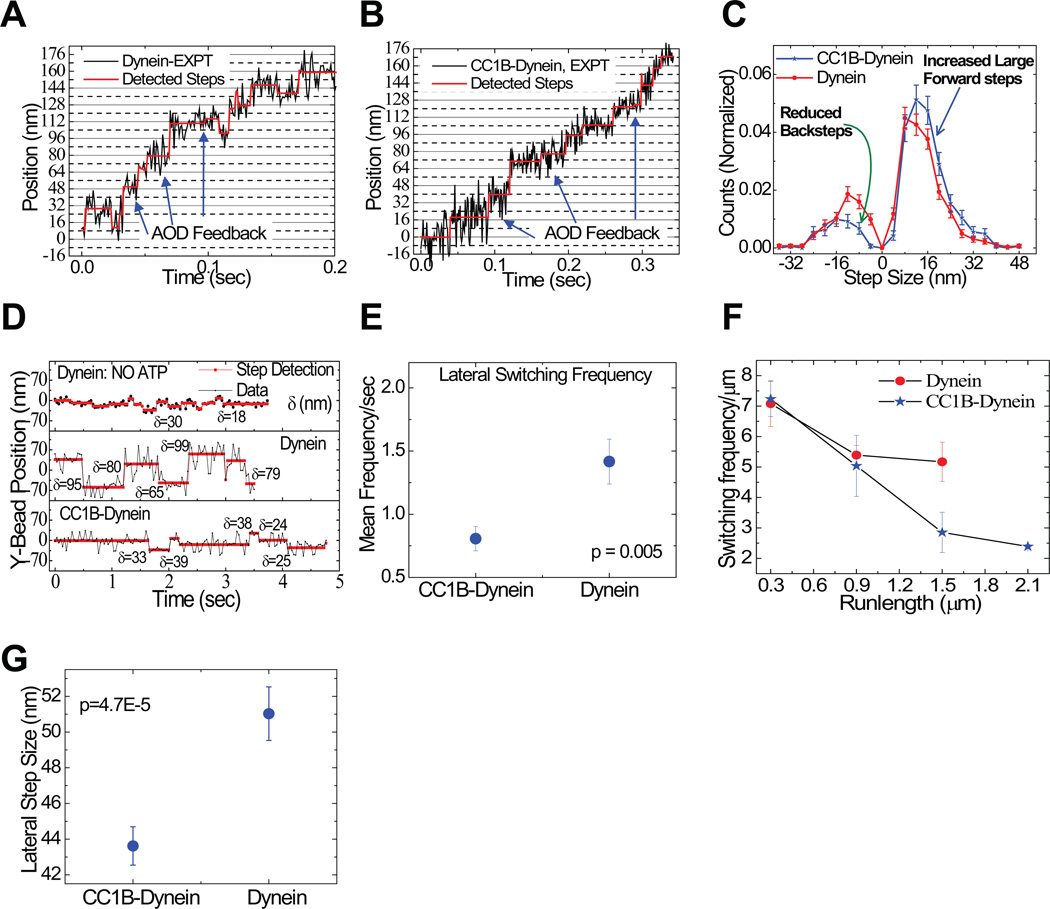
To gain insight into the mechanism responsible for this activity we evaluated dynein stepping along microtubules, combining a force-feedback optical trap with a novel step-detection approach (see Methods; Supplementary Fig. 4; Supplementary Table 2,). The motor protein alone exhibited a mix of step sizes. Forward 8 nm steps predominated (Fig. 5 A–C; Supplementary Table 2), though reverse and lateral steps were also observed, as previously reported30–32 (Fig. 5 C–G). CC1B altered each of these behaviors, causing a higher proportion of forward to reverse steps, an increase in forward step size and a decreased frequency and size of lateral steps (Fig. 5; Supplementary Figs. 4 and 5; Supplementary Table 2, methods). The average step size increased from 6.28 to 11.22 nm (see methods), corresponding to a predicted 1.8-fold increase in travel distance, slightly less than the experimentally observed 2-fold increase. The remaining increase reflects a decreased probability of detachment per step (average of 177 steps with CC1B vs 146 for dynein alone). Our observations, thus, identify a mechanism for dynein processivity regulation independent of the N-terminal p150Glued microtubule-binding region, and involving changes to stepping behavior.
Figure 5. Effects of processivity-enhancing fragment CC1B on dynein stepping behavior.
Sample dynein bead traces with detected steps (in red) for single dynein alone (A) and with CC1B (B). Arrows indicate points of force feedback (see Methods). (C) Distribution of step-sizes for single dyneins with (blue) or without (red) CC1B (428 stepping events). CC1B reduces back-stepping (green arrow) and increases large forward steps (blue arrow). (D) Sample traces showing lateral bead position on microtubule vs. time for stationary dynein (without ATP - top); and dynein alone (middle) or with CC1B (bottom) moving on a microtubule in the presence of saturating ATP. Step detection identifies changes in lateral bead position (red lines); δ indicates size of detected lateral step in nm. (E) Lateral dynein bead switching frequency (95 switching events) on the microtubule surface, and (F) as function of run length (94 run lengths); p value from t-test. Longer CC1B-dynein runs correlate with decreased lateral switching frequency. (G) Magnitude of lateral step size (428 detected steps). CC1B decreases the frequency and magnitude of lateral dynein steps. Error bars are SEM.
We next examined CC1A, which, at low concentrations, had a minimal effect on dynein behavior (Fig. 3H, Table 1). At a very high ratio to dynein (7000:1), CC1A somewhat reduced dynein interactions with microtubules (Fig 3H, Table 1). Strikingly, all motile events were diffusive (Fig. 3C, H, Suppl. Fig 2D), with minimal force production. CC1A, thus, promotes the diffusional dynein state exclusively. CC1A binding to dynein cannot be readily detected by biochemical means (Fig. 1), and must, therefore, involve a weak, transient interaction, as discussed below. We note that the diffusion coefficient for dynein in the presence of CC1A or p135-CC1 is comparable to that for dynein alone (Table 1), suggesting dynein has the same interaction with the microtubules in each case, though the duration of diffusive events is increased by thesep150Glued fragments. CC1A and larger CC1A-containing fragments showed little evidence of microtubule binding (Fig. 1E), suggesting that the CC1A region may directly stabilize dynein in an inherent diffusive conformation.
Interaction between CC1A and CC1B
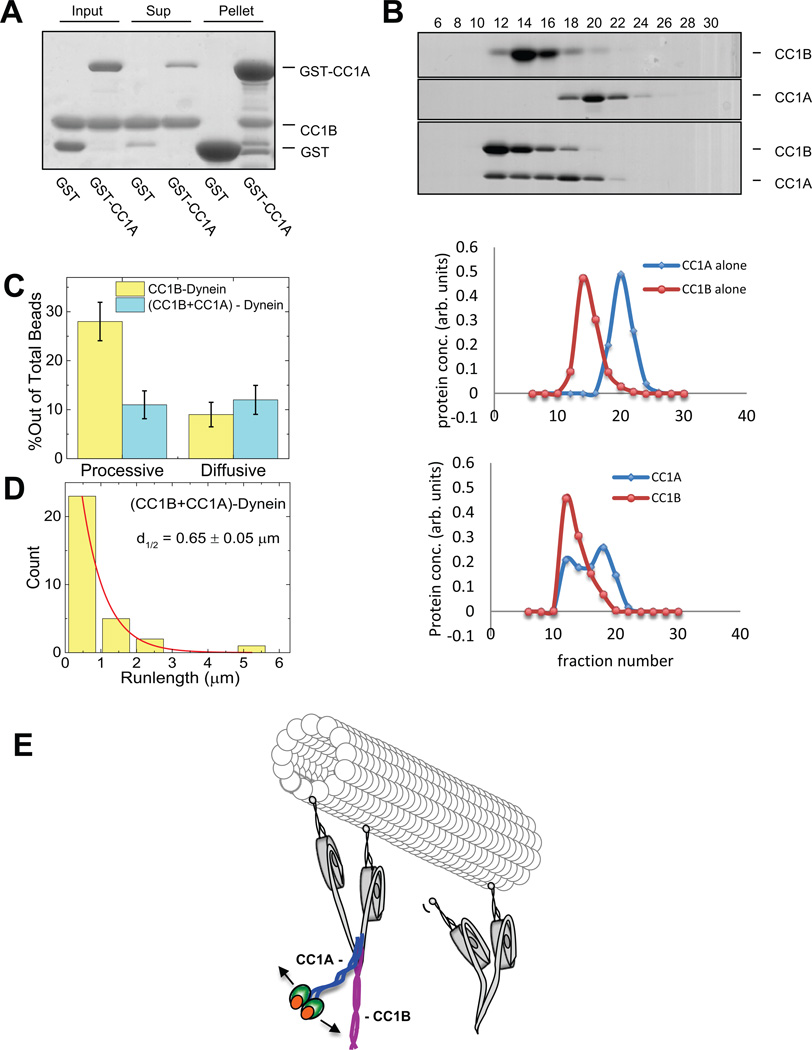
Dynein inhibition by CC1 differs from the effects of its CC1A and CC1B sub fragments alone, suggesting a potential cooperative interaction between them. Indeed, we observed clear evidence that GST-CC1A binds CC1B in pull-down assays (Fig. 6A). We also tested for co fractionation of CC1A and CC1B by size exclusion FPLC, and found clear evidence for a shift to a greater hydrodynamic radius for the combined fragments relative to each alone (Fig. 6B). To test for an effect on motor behavior we exposed beads sequentially to dynein, CC1B, and CC1A, the latter at a 150-fold molar excess relative to dynein. At this ratio CC1A alone has minimal effect on dynein (Table I). However, it clearly suppressed CC1B enhancement, decreasing the overall frequency of processive motion (Fig 6C) and mean travel distance (Fig. 6D).
Figure 6. Interaction between coiled-coil p150Glued fragments CC1A and CC1B.
(A) Pull-down of bacterially-expressed CC1B with GST-CC1A or GST alone shows specific binding of CC1B to CC1A using Coomassie Blue staining. (3 independent experiments) (B) Size exclusion FPLC of CC1A and CC1B using Coomassie Blue Staining. (3 independent experiments) When in combination the two fragments elute in a common, higher molecular size peak. (C, D) Single-molecule bead assays reveal that CC1A, at a level which has no effect on its own on dynein (150:1 CC1A:dynein), strongly suppresses the effect of CC1B on dynein (see Figure legend 3 for number of beads checked). The number of processive dynein events and their average run-length are reduced relative to values for dynein alone, suggesting that, as for CC1, the combined fragments actively inhibit dynein function. Error is SEM. E) Schematic representation of dynein regulation by dynactin. (Left) Dimer of p150Glued N-terminus (aa 1-555) binds via its CC1B coiled-coil domain with the dynein intermediate chains situated within the dynein tail. CC1B alone is sufficient to increase the length of processive dynein movement along microtubules. CC1A is shown interacting physically with CC1B. As the two domains are covalently linked in the intact p150Glued and p135 polypeptides, we propose that CC1 must bend as shown. In this conformation, the N-terminal globular portion of p150Glued and p135 might be able to interact with the C-terminal portion of CC1B. Thus, we envision a series of intramolecular interactions within p150Glued or p135, ultimately modulating dynein behavior through its tail domain. We propose that the ultimate target of regulation are the dynein motor domains, the behavior of which seem more clearly coordinated in the presence of the dynactin fragments, as evidenced by more efficient stepping along microtubules and longer runs.
These results thus identify a direct interaction between CC1A and CC1B, and reveal that CC1A inhibits the functional effects of the CC1B-dynein interaction (Fig 6D). The net outcome is similar to the effects of CC1, suggesting that its subdomains are capable of interacting to regulate each other. The observation that CC1 binds dynein, but inhibits its motility, suggests that its CC1B sub domain remains associated with dynein when dynactin is in either a stimulatory or inhibitory conformational state.
DISCUSSION
These results indicate that the subdomains of CC1 have the remarkable ability to modulate processive and diffusive dynein behavior. However, in either covalent or non covalent combination with CC1A, the effect of CC1B on dynein processivity is suppressed. Thus, CC1 alone cannot completely account for the dynein regulatory properties of dynactin. Instead, our results argue for further intramolecular regulation by the N-terminal globular region of p150Glued or p135. Fragments including the globular domain stimulate both processivity and diffusivity, and mimic the effects of the complete dynactin complex. Thus, the globular domain must suppress the inhibitory effect of CC1A, further modulating its behavior. Although the structural organization of p150Glued within the complete dynactin complex remains incompletely understood, we speculate that CC1 must be capable of folding upon itself in order to allow CC1A and CC1B to interact (Fig 6E). Because CC1A is substantially shorter than CC1B, this arrangement would allow the N-terminal globular domain of p150Glued or p135 to contact the C-terminal portion of CC1B, and, perhaps, further modulate its behavior. The globular N-terminal region is known to bind to microtubules and other interactors33. Based on our current data we speculate that the N-terminal regions ofp150Glued and of p135 may, in turn, modulate interactions between CC1A and CC1B, and between the latter and dynein.
The only known site for this interaction is within dynein in the N-terminal 44 residues of the intermediate chain (IC)21, 22, 29, which is a major component of the dynein tail. Our data, therefore, suggest that dynactin regulation involves very long-range communication between the dynein tail and motor domains, where CC1B helps keep the motor domains close to each other, allowing improved coordination and interactions. A related mechanism but with an opposite effect on motor activity, was identified by analysis of dynein purified from the Loa mutant mouse30. We found in this case that a mutation in the tail portion of the dynein heavy chain (HC) specifically inhibits processivity, with an increase in wandering behavior on the microtubule surface. Recent results in the yeast S. cerevisiae showed further that deletion of the dynein light chain (LC) subunit weakens the IC-HC interaction, as was seen in Loa dynein, again decreasing processivity34. Together, these results suggest a common regulatory mechanism inhibited by the Loa mutation but stimulated by dynactin, likely mediated through components of the dynein IC-LC complex. Our results support findings that, within the cytoplasmic dynein dimer, the motor domains are inefficiently coordinated31, 32 and suggest that dynactin may act to increase motor coordination through a long-range allosteric mechanism. The effects on motor behavior are manifested in increasing average forward step size, suppressing backward steps, and decreasing overall probability of motor detachment from the microtubule per step.
Our data also identify mechanisms for switching dynein between processive and diffusive states. Such behavior has recently been reported for individual fluorescently-tagged dynein molecules in the yeast S. pombe. Dynein exhibited one-dimensional diffusion along cytoplasmic microtubules to reach Num1 cortical anchorage sites, after which the dynein exhibited directed, processive movement35. Expression of the dynein-binding portion of Num1, which interacts with the base of the complex36, converted dynein from diffusive to processive behavior. Our results identify dynactin as a physiological dynein regulator capable of inducing what may be equivalent states. We speculate that in different contexts, the ability of dynactin to turn off dynein force production could playa role in minimizing tug-of-war interactions between kinesin and dynein19, 20 in the cell.
Recent studies have identified a family of RDD (Rab-dynein-dynactin) binding adapters24, 37 capable of strongly stimulating dynactin-mediated cytoplasmic dynein processivity38, 39. Whether these factors act simply as scaffolds to facilitate the weak dynein-dynactin interaction40, or, instead, up-regulate the mechanism discovered in our current study remain important questions for future investigation. We note that dynein used in the recent studies38, 39 exhibited very limited processivity. The use of beads as in the current work not only allows dynein force production to be determined, but also enables higher resolution spatial tracking than is currently possible with fluorescently tagged motor, and has revealed mammalian dynein to be clearly processive25, 30. We note that the length of runs detected in the current in vitro assays are also more representative of the behavior of physiological cargo in vivo41, 42. Thus, the generality of RDD adapter function among cargo forms, and how their behavior may be modulated, remain additional questions for further research.
MATERIALS AND METHODS
Protein Purification and Characterization
Cloning and Protein Expression and Purification
All p150Glued constructs for recombinant protein expression were cloned from full-length rat cDNA (EDL91133.1). p150Glued CC1, CC1B, CC1A and p135-CC1 were cloned with C-terminal Flag and 6X His tags into pGEX 6P-1 (Amersham Biosciences), which encodes an N-terminal GST tag (Fig. 1). Proteins were expressed in BL21-CodonPlus RIPL competent cells (Agilent Technologies, #230280) with 0.5 mM IPTG for 4–6 hours at 20°C. Bacterial pellets were lysed in PBS with 1 mM DTT and 1:500 protease inhibitor cocktail (Sigma, P8340) by sonication and centrifuged at 4°C for 30 min at 150,000g with a final concentration of 1% Triton-X. Proteins were purified with glutathione beads (GE, 17-0756-01) for 1 hour at 4°C. Washed beads were incubated overnight at 4°C with Precission Protease (GE, 27-0843-01) in cleavage buffer (50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA) supplemented with 1 mM DTT to remove the GST tag. Proteins were supplemented with 5% glycerol, flash frozen and stored at −80°C. p150 1-555 was expressed using BaculoDirect™ System (Invitrogen). p150 1-555-Flag-6X His was cloned into entry vector pENTR1A (Invitrogen) and recombined with linear BaculoDirect™. SF9 insect cells (Invitrogen, 11496-015) were infected with the recombination product a single population of virus was isolated by plaque assay. SF9 cells were infected for 48 hrs, and collected in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM Imidazole) with 1:100 protease inhibitor cocktail and 1% IGEPAL CA-630 (Sigma I-3021). Cleared lysate was incubated with NTA-Nickel beads (Qiagen, 1018611) for 1 hour at 4°C, washed with lysis buffer containing 40 mM Imidazole and protein was eluted in lysis buffer containing 250 mM Imidazole. Buffer was exchanged to cleavage buffer using NAP columns (GE Healthcare) and protein was supplemented with 5% glycerol, flash frozen and stored at −80°C. Full length Drosophila kinesin was purified from wild type Drosophila as in43. Bovine brain cytoplasmic dynein was purified as described44 except that whole brains were flash frozen and stored at −80°C before purification.
Protein Interaction Analysis
p150Glued fragments were tested for dynein binding by immunoprecipitation with anti-flag antibody in PEM-35 (35 mM PIPES, 5 mM MgSO4, 1 mM EGTA, .5 mM EDTA, pH 7.0) supplemented with .05–1 ug/ul BSA + 1 mM DTT + .1% Tween for 1–2 hrs at 4°C with protein A beads (Invitrogen). MT sedimentation assays were performed as follows. Purified bovine dynein (8 nM) mixed with 10X p150 fragment and 2.5 uM taxol-stabilized MTs (Cytoskeleton Inc, TL238) with or without 10 mM ATP (Sigma, A9187) in BRB80 (80 mM K-Pipes pH 6.9, 1 mM MgCl2, 1 mM EGTA) supplemented with 20 uM taxol, 1 mM DTT, and .05 ug/ul BSA was incubated for 30 min at room temperature, and centrifuged for 45 min at 35,000 × g. Supernatants and pellets were analyzed by western blotting. Bacterially-expressed CC1B was tested for an interaction with CC1A by pull-down with either GST-CC1A or GST alone in 20 mM Tris-HCL, pH 7.0 plus 40 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 1 mM DTT, and protease inhibitor cocktail (Sigma, St. Louis, MO) at 4°C. Size exclusion FPLC was performed using a 24 mL Superose 6 column pre equilibrated with 20 mM Tris-HCL, pH 7.0, containing 1 mM EDTA at 4°C). IP and/or blotting antibodies used are: anti-alpha tubulin (Sigma, T9026 used at 1:10,000 for blotting) 74.1 anti-dynein intermediate chain (gift of K.P fister used at 1:5000 for blotting), anti-Flag (Sigma, F1804 M2 used at 1:10,000 for blotting) anti-DDDDK (Abcam ab1162 used at 1:40 for immunoprecipitation), and anti-dynein heavy chain (used at 1:1000 for blotting)45.
Circular Dichroism
All proteins were dialyzed overnight into 50 mM sodium phosphate buffer, pH 7.0, and CD measurements were taken on a Jasco-J815 spectrapolimeter. Fixed temperature measurements were collected in 0.1 mm cuvettes at 185–260 nm wave lengths, 0.1 nm data pitch, continuous scanning mode, at standard sensitivity, with scanning speed of 50 nm/min, response of 8 sec., and bandwidth of 1 nm. For each sample 3 data sets were accumulated per run. Melting curve data were accumulated at 222 nm from 5–85°C (for CC1) and 5–60°C (for CC1B) in a 1 mm cuvette with data pitch of 0.5°C, a 10 sec. delay, a temperature slope of 40°C/hr, standard sensitivity, 8 sec. response, and bandwidth of 1 nm. The cooling curve was collected in the same manner reversing the temperature 15 sec. after reaching the maximum. Molar ellipticities were calculated as described46–47. Secondary structure content predictions were made using the online server DICHROWEB (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). For comparison, the online server COILS (http://embnet.vital-it.ch/software/COILS_form.html) was used to predict coiled-coil structure from the amino acid sequences.
In vitro Optical Bead Assay
Optical-trapping motility assays, data recording, particle tracking and stalling-force analysis were performed as previously described in48, with the exception of the new implementation of force-feedback as described below.
For single molecule dynein assays, a 489 nm diameter carboxylate polystyrene bead (Poly-sciences, catalog # 09836-15), with non specifically attached motors, was positioned in a flow chamber above a taxol-stabilized microtubule for 30 sec to allow for binding. The single motor range was attained when the percentage of beads binding is smaller than or equal to 30%49. For run length measurements of single dynein, a run was defined as the distance travelled from initial binding until the bead detached (with optical trap turned off). The distribution of run lengths was fit to a single exponential decay to obtain mean run length and associated uncertainty. For processivity measurements, we made measurements on at least 40–50 beads with active motors. For each run, velocity was obtained by dividing run length by the duration of the run. The mean velocity and associated uncertainty was calculated from a statistical average of all velocities. We define the stall force (Fs) as the mean value of the load force at which the motor stops moving. Stall force measurements of single dynein beads were made with a trap stiffness of 1.5 pN per 100 nm. An event was classified as a stall when a single motor bead moved away from trap center and held its plateau position with a velocity of < 10 nm/sec for ≥ 100 ms before detachment. The mean stall force (and S.E.M.) were calculated from the Gaussian peak position (and uncertainty) of the stall force distribution. For force measurements, we made measurements with at least 40–50 beads with active motors. The obtained decay constant and uncertainty presented in each plot represents the average detachment time and S.E.M. respectively. Statistical significance was determined using the Student’s t-test. To determine the overall frequency of diffusive vs processive motion (e.g. in experiments summarized in Fig 3, we ultimately looked at 50–60 independent beads which had MT-binding activity. Since the binding fraction was ~30%, this required experiments on roughly 150–200 independent beads chosen randomly in solution. All summarized bead experiments were replicated at least 3 times, but usually more.
Single Molecule Bead Assay of Dynein with p150Glued Fragments
Dynein was adsorbed onto carboxylate beads, which were then blocked with casein (10 mM) and subsequently exposed to a 150-fold molar excess of p150Glued fragment. Excess dynein and fragments were removed by gentle centrifugation) Optical trap bead assays were performed in dynein motility buffer (35 mM PIPES pH 7.0, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA) in the presence of 1 mM ATP and an oxygen scavenging system (250 µg/ml glucose oxidase, 30 µg/ml catalase, 4.6 mg/ml glucose50.
Quantitation of diffusive vs processive dynein
We (see e.g. Fig 6 in51) and others24,26 have in the past reported that dynein can be in both diffusive and processive states. The likelihood of diffusive events decrease when more motors are present on the bead51. Experiments are done on MT-purified dynein (before experiments, a MT-pelleting/release step selects for active dynein. At the single-molecule level the percentage of processive versus diffusive beads can vary between different purifications (though not among multiple aliquots from the same original purification): typically ~25% of tested beads show processive motion, and 5% show diffusive motion (Fig. 3D). Thus, roughly 83% of binding events are processive, and 17% diffusive. If the dynein is aged (on ice, 4C, for 2–3 days) the diffusive fraction increases to ~40%. Some purifications show significantly more diffusive events from the start; we typically do not use those. The studies in this paper reflect the use of bovine dynein from 3–4 independent purifications. In the experiments to assess effects of dynactin fragments on dynein’s diffusion, we always do the experiment and the control on the same day, using the same dynein aliquot, with experiments interspersed.
Determination of force generation for diffusive beads
To determine whether there was any remaining force production by diffusive beads, we compared motion of beads diffusing due to the p150Glued 1-555 fragment alone (lacking motors) to the other diffusing dynein-dynactin fragment beads, in each case monitoring the position of the bead in the optical trap, originally sampled at 4 KHz, and subsequently averaged to 1 KHz to decrease noise. These traces were then analyzed using Kerssemakers’ step detection algorithm. The detected steps were restricted with a condition of waiting time ≥ 50 ms, and the distribution of forces at which the bead survives with this condition was obtained.
Force-Feedback for Step Size Distribution
To carry out step-size measurements at saturating ATP concentrations we used an optical trap with force feedback to suppress thermal noise; the trap moved to follow the bead and minimize opposition to motion. High temporal and spatial resolution measurements were made using a laser to detect the bead’s location.
The experimental set-up and calibration was done as in52 augmented by an Acousto-Optic Deflector (AOD) to allow force-feedback. Video tracking of a trapped bead in two dimensions was used to obtain the conversion parameters from AOD drive frequency (MHz) to position (nm). Trap stiffness was calibrated using Quadrant Photo Diode (QPD) signals with the power-spectrum method. With the AOD, the trap stiffness perpendicular to the microtubule (Y-direction) is < 20% smaller than the trap stiffness parallel to the microtubule (x-direction). The calibration of trap stiffness was confirmed by measuring kinesin stall forces with and without the AOD. Since trap stiffness along the microtubule was constant up to 2 micron from the center, all step size measurements were made within ±1.5 um from the center to maintain constant trap stiffness. The trap stiffness was 1.5 pN/100 nm, so the maximum force the bead stepped against was lower, and dependent on step size. For example a 16 nm step attempt, starting at 24 nm from the trap center and ending at 40 nm, would experience 0.36 pN of load. Using this system we measured rapidly stepping single motors attached to beads undergoing a low force opposing motion. The QPD signal was obtained at scanning rate of 4 KHz and AOD feedback occurred every 40 nm (the largest dynein step size possible) of bead displacement from the center of the trap, to maintain the bead at the center of the laser. This signal was averaged to 1 KHz, and analyzed for steps within each 40 nm stationary period using Kerssemakers’53 step detection algorithm.
Theoretical Simulation for Step Size Analysis
Rapid sequential steps of potentially varying size can be ‘fused’ together by the step detection procedure, resulting in “detection” of excessive large steps54. To address this confounding issue, we developed a partly synthetic data approach, as done by the Odde group in another context55.
Theoretically simulated motion was combined with experimentally measured noise to generate simulated tracks that were then analyzed for stepping. Programs to generate simulated tracks for step detection were developed using self-written Matlab codes. The tracks comprised a known percentage of steps with given step sizes, with a mean dwell time between steps derived from a decaying exponential distribution. The mean dwell time was chosen to match experimentally determined velocities, taking into account the distribution of step sizes and the percentage of forward and backward steps. The tracks were generated at 4 KHz as measured in the experiment. Experimentally measured noise, generated from beads tethered to microtubules via non-stepping dynein (in the absence of ATP) with the trap feedback engaged, was added to the simulated tracks, which were then averaged to 1 KHz and analyzed using step detection. The steps-size distributions were normalized to the total number of steps (to avoid the effect of data size) and compared to the experimental step size distribution by calculating the residual, which is the difference between the two normalized step-size distributions. Smaller residuals indicated better agreement between the two. Simulated distributions were adjusted to minimize the residual and determine the correct experimental distribution.
We confirmed this method by measuring kinesin’s step distribution, which is comprised of predominately 8 nm plus-end directed steps with occasional back steps56 (Fig. S4 A–E). For dynein, experimental trajectories were derived from beads driven by single dyneins with or without CC1B moving at ~400 nm/sec with saturating (1 mm) ATP.
Procedure for Lateral Motion/Lateral Step-Size Determination
We used video enhanced DIC microscopy, coupled with sub-pixel resolution particle tracking57 to analyze lateral motion. As a control, kinesin was first tested as follows. The distributions of motion for 200 nm diameter carboxylate beads with single molecule concentrations of kinesin either tethered to the microtubule (in the presence of AMP-PNP) or moving freely along the microtubule (with 1 mM ATP) were measured (Fig S5 C, D). We projected the bead’s motion along the microtubule onto the best-fit line trajectory, and determined the Y-bead position (perpendicular to the microtubule). Protofilament switching events were detected by analyzing traces using step detection. Experiments were done at ~30% binding fraction to insure single kinesin or dynein activity, with the trap turned off to allow free lateral motion.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Richard McKenney for help in initiating this project and Dr. Alexandre Baffet for helpful comments on the manuscript. Supp. by GM102347 to RBV and GM070676 to SPG.
Footnotes
AUTHOR CONTRIBUTIONS:
RV, SPG, SJW, and SKT designed experiments; SKT, SJW, and CC performed the experiments; PA provided kinesin; RV, SPG, SKT, CC, and SJW wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- 2.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Roberts Anthony J, Leschziner Andres E, Reck-Peterson Samara L. Lis1 Acts as a “Clutch” between the ATPase and Microtubule-Binding Domains of the Dynein Motor. Cell. 150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroer TA. Dynactin. Annu Rev Cell Dev Bi. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 5.Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. The Journal of Cell Biology. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holleran EA, Tokito MK, Karki S, Holzbaur EL. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. The Journal of Cell Biology. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 8.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 9.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. P Natl Acad Sci USA. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holzbaur ELF, et al. Homology of a 150K cytoplasmic dynein-associated polypeptide with the Drosophila gene Glued. Nature. 1991;351:579–583. doi: 10.1038/351579a0. [DOI] [PubMed] [Google Scholar]
- 11.Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. The Journal of Cell Biology. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc Natl Acad Sci U S A. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson P, Stephens DJ. Microtubule plus-end loading of p150Glued is mediated by EB1 and CLIP-70 but is not required for intracellular membrane traffic in mammalian cells. J. Cell Sci. 2006;119:2758–2767. doi: 10.1242/jcs.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit R, Levy JR, Tokito M, Ligon LA, Holzbaur ELF. Regulation of Dynactin through the Differential Expression of p150(Glued) Isoforms. J. Biol. Chem. 2008;283:33611–33619. doi: 10.1074/jbc.M804840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, et al. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. Journal of Cell Biology. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moughamian AJ, Holzbaur EL. Dynactin is required for transport initiation from the distal axon. Neuron. 2012;74:331–343. doi: 10.1016/j.neuron.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd TE, et al. The p150(Glued) CAP-Gly domain regulates initiation of retrograde transport at synaptic termini. Neuron. 2012;74:344–360. doi: 10.1016/j.neuron.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross SP, Welte MA, Block SM, Wieschaus EF. Coordination of opposite-polarity microtubule motors. J Cell Biol. 2002;156:715–724. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haghnia M, et al. Dynactin is required for coordinated bidirectional motility, but not for dynein membrane attachment. Molecular Biology of the Cell. 2007;18:2081–2089. doi: 10.1091/mbc.E06-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siglin AE, et al. Dynein and dynactin leverage their bivalent character to form a high-affinity interaction. Plos One. 2013;8:e59453. doi: 10.1371/journal.pone.0059453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau SY, Taneja AK, Hodges RS. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J Biol Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 24.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur ELF. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 25.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokito MK, Howland DS, Lee VM, Holzbaur EL. Functionally distinct isoforms of dynactin are expressed in human neurons. Mol Biol Cell. 1996;7:1167–1180. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintyne NJ, et al. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan JL, Song Y, Barbar E. Structural dynamics and multiregion interactions in dynein-dynactin recognition. J Biol Chem. 2011;286:39349–39359. doi: 10.1074/jbc.M111.296277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat Cell Biol. 2010;12:1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic Dynein Moves Through Uncoordinated Stepping of the AAA+ Ring Domains. Science. 2012;335:221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisbrich A, et al. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 34.Rao L, et al. The yeast dynein Dyn2-Pac11 complex is a dynein dimerization/processivity factor: structural and single-molecule characterization. Molecular Biology of the Cell. 2013;24:2362–2377. doi: 10.1091/mbc.E13-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ananthanarayanan V, et al. Dynein Motion Switches from Diffusive to Directed upon Cortical Anchoring. Cell. 153:1526–1536. doi: 10.1016/j.cell.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Tang X, Germain BS, Lee W-L. A novel patch assembly domain in Num1 mediates dynein anchoring at the cortex during spindle positioning. The Journal of Cell Biology. 2012;196:743–756. doi: 10.1083/jcb.201112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai AK, Rai A, Ramaiya AJ, Jha R, Mallik R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell. 2013;152:172–182. doi: 10.1016/j.cell.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paschal BM, et al. Characterization of a 50-kDa polypeptide in cytoplasmic dynein preparations reveals a complex with p150GLUED and a novel actin. J Biol Chem. 1993;268:15318–15323. [PubMed] [Google Scholar]
- 41.Yi JY, et al. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bremner KH, et al. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009;6:523–535. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigua R, Tripathy S, Anand P, Gross SP. Isolation and purification of kinesin from Drosophila embryos. J Vis Exp. 2012 doi: 10.3791/3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paschal BM, Shpetner HS, Vallee RB. Purification of brain cytoplasmic dynein and characterization of its in vitro properties. Methods Enzymol. 1991;196:181–191. doi: 10.1016/0076-6879(91)96018-m. [DOI] [PubMed] [Google Scholar]
- 45.Mikami A, et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 46.Bohm KJ, Stracke R, Unger E. Speeding up kinesin-driven microtubule gliding in vitro by variation of cofactor composition and physicochemical parameters. Cell Biol Int. 2000;24:335–341. doi: 10.1006/cbir.1999.0515. [DOI] [PubMed] [Google Scholar]
- 47.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Kunwar A, et al. Mechanical stochastic tug-of-war models cannot explain bidirectional lipid-droplet transport. Proc Natl Acad Sci U S A. 2011;108:18960–18965. doi: 10.1073/pnas.1107841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 50.Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proc Natl Acad Sci U S A. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mallik R, Petrov D, Lex SA, King SJ, Gross SP. Building complexity: an in vitro study of cytoplasmic dynein with in vivo implications. Curr Biol. 2005;15:2075–2085. doi: 10.1016/j.cub.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 52.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerssemakers JW, et al. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 54.Carter BC, Vershinin M, Gross SP. A comparison of step-detection methods: how well can you do? Biophys J. 2008;94:306–319. doi: 10.1529/biophysj.107.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearson CG, et al. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol Biol Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 57.Carter BC, Shubeita GT, Gross SP. Tracking single particles: a user-friendly quantitative evaluation. Phys Biol. 2005;2:60–72. doi: 10.1088/1478-3967/2/1/008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.