Summary
Autophagy is the primary catabolic process triggered in response to starvation. Although autophagic regulation within the cytosolic compartment is well established, it is becoming clear that nuclear events also regulate the induction or repression of autophagy. Nevertheless, a thorough understanding of the mechanisms by which sequence-specific transcription factors modulate expression of genes required for autophagy is lacking. Here, we identify Foxk proteins (Foxk1 and Foxk2) as transcriptional repressors of autophagy in muscle cells and fibroblasts. Interestingly, Foxk1/2 serve to counter-balance another forkhead transcription factor, Foxo3, which induces an overlapping set of autophagic and atrophic targets in muscle. Foxk1/2 specifically recruits Sin3A-HDAC complexes to restrict acetylation of histone H4 and expression of critical autophagy genes. Remarkably, mTOR promotes the transcriptional activity of Foxk1 by facilitating nuclear entry to specifically limit basal levels of autophagy in nutrient-rich conditions. Our study highlights an ancient, conserved mechanism whereby nutritional status is interpreted by mTOR to restrict autophagy by repressing essential autophagy genes via Foxk-Sin3-mediated transcriptional control.
Introduction
Macroautophagy (hereafter autophagy) is a well-conserved eukaryotic catabolic process that promotes cellular homeostasis and ensures cell survival. In response to stressors, such as starvation, cells form membrane-bound autophagosomes to engulf cytoplasmic proteins, lipids, and organelles1.These cargoes are then delivered for lysosomal degradation, which aids in the restructuring of cells during tissue development and differentiation and generates necessary metabolites to sustain energy demands in nutrient-limiting conditions.
Until recently, autophagy has been viewed largely as a cytoplasmic phenomenon that is exclusively regulated by cytoplasmic complexes. However, it is becoming clear that autophagy is sensitive to epigenetic and transcriptional changes2. For example, prolonged autophagy results in the genome-wide reduction of two histone modifications, tri-methylated H3K4 (H3K4me3) and acetylated H4K16 (H4K16ac)3. Furthermore, a number of DNA-binding transcription factors either positively or negatively regulate autophagy4. Two transcription factors, TFEB and Foxo3, rapidly respond to autophagy-inducing stimuli by translocating from the cytosol to the nucleus, leading to enhanced expression of autophagy genes5,6. Another factor, ZKSCAN3, negatively regulates autophagy, acting as the counterpart to TFEB to repress an overlapping set of autophagy genes7. Importantly, despite these recent observations, our understanding of the nuclear events associated with autophagy remains rudimentary, and it is unclear how autophagic stimuli specifically direct recruitment of chromatin-modifying enzymes to autophagy genes and how DNA-binding factors promote activation or repression of autophagy genes. Here, we describe a genome-wide transcriptional network directly linking two sequence-specific transcriptional repressors in the Foxk family to a chromatin remodeling complex, changes in histone modifications, and repression of the autophagic program.
Results
Foxk proteins are components of Sin3A, but not Sin3B, complexes
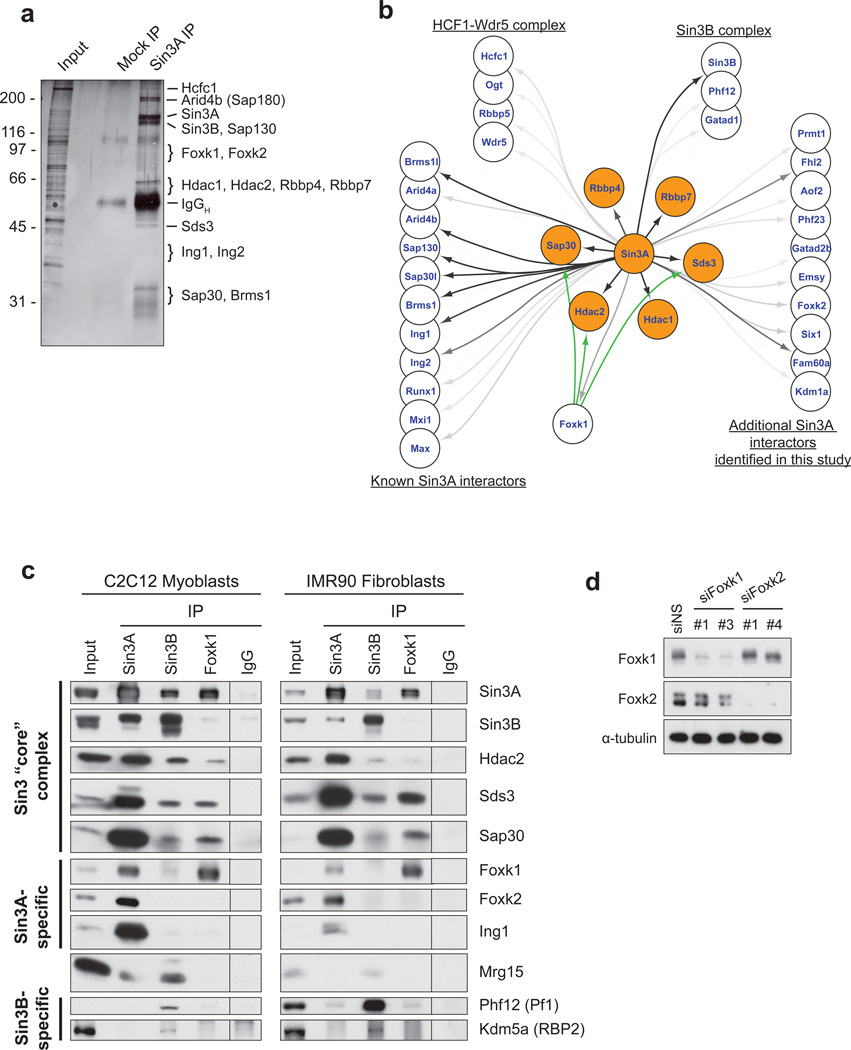
Mammalian cells express two isoforms of Sin3, Sin3A and Sin3B, and both have been shown to have non-redundant functions. Our previous studies prompted us to identify functionally distinct Sin3A complexes with important roles in skeletal muscle, since Sin3A is essential for viability and is specifically required for development of this tissue8–10. We focused on chromatin-associated complexes in mouse C2C12 myoblasts by immuno-purifying endogenous Sin3A complexes from solubilized chromatin11. This protocol enriched a cohort of sequence-specific transcription factors as well as proteins able to interact with, and modify, histones (Fig. 1a,b; Supplementary Table 1). Mass spectrometric analyses identified the “core” components of Sin3 complexes (e.g., HDAC1, HDAC2, Sap30, Sds3, Rbbp4, Rbbp7), as expected, together with a number of chromatin-associated proteins with DNA-binding, bromo-, chromo-, and PHD domains (Fig. 1b, Supplementary Table 1).
Figure 1. Identification of Foxk1 as a component of a Sin3A complex.
(a) Immuno-purification of Sin3A complexes from solubilized myoblast chromatin. (b) Sin3A interaction map showing protein-protein interactions connected by black and gray arrows. The intensity of the arrows indicates the number of peptides sequenced by mass spectrometry after Sin3A immuno-purification. Core components of Sin3 complexes are highlighted in orange. The green arrows signify additional interactors with Foxk1 as determined by IP-western in this study. (c) Nuclear Foxk1 and Foxk2 interact with Sin3A, but not Sin3B, complexes. The vertical line denotes a splice of the scanned image; immunoprecipitate from the non-specific IgG control was loaded on the same gel as all other samples. (d) siRNA-mediated depletion of Foxk1 and Foxk2. For each Foxk protein, two distinct targeting sequences yield similar knock-down efficiency. Western blotting documents the specificity of siRNAs and antibodies for the intended Foxk homologs.
Several sequence-specific transcription factors associated with Sin3A, including the closely related Foxk1 and Foxk2 proteins. In co-immunoprecipitation experiments in myoblasts and normal human (IMR90) fibroblasts, Foxk1 and Foxk2 (collectively termed “Foxk”) interacted strongly with Sin3A and several of the Sin3 core components independently of DNA (Fig. 1c; Supplementary Fig. 1), confirming their identity as components of Sin3 complexes. However, the Foxk proteins are not universal components of all Sin3A complexes, since the Foxk-Sin3A complexes were cleanly separated from other Sin3A complexes that contain Ing1 (Fig. 1c). Despite their extensive similarity, Foxk1 and Foxk2 antibodies specifically recognized their intended antigens, and Foxk1 and Foxk2 were independently enriched in separable Sin3A complexes (Fig. 1c, Supplementary Figs. 1). Furthermore, depletion of Foxk1 or Foxk2 with specific siRNAs led exclusively to reductions in their intended targets (Fig. 1d). Previous studies indicated that over-expressed Foxk1 interacts with both Sin3A and Sin3B, suggesting that Foxk1 has the potential to interact with the highly homologous PAH2 domain of either Sin3 isoform12–14. However, endogenous Foxk did not associate with Sin3B or any Sin3B-specific components (Phf12, Kdm5a/RBP2) in nuclear fractions (Fig. 1c and Supplemental Fig. 1). Likewise, Sin3B was not co-immunoprecipitated with anti-Foxk antibodies, indicating that Foxk proteins are components of Sin3A, but not Sin3B, complexes.
Foxk1 localizes to gene promoters and enhancers
Our results were notable because Foxk1 and Sin3A have been shown to play important roles in muscle biology: Sin3A knock-out mice exhibit severe muscle defects and early post-natal lethality, and Foxk1 loss severely impairs the proliferation of muscle stem cells (satellite cells) and muscle regeneration10,15,16. Given its role in muscle biology, we focused primarily on Foxk1 in subsequent studies. First, we used chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIP-seq) in myoblasts, which led to the identification of ~7700 genomic Foxk1 binding sites. We confirmed a subset of these data with ChIP and quantitative PCR (qChIP) and showed that depletion of Foxk1 with two different siRNAs significantly reduced its enrichment at identified targets (Supplementary Fig. 2a).
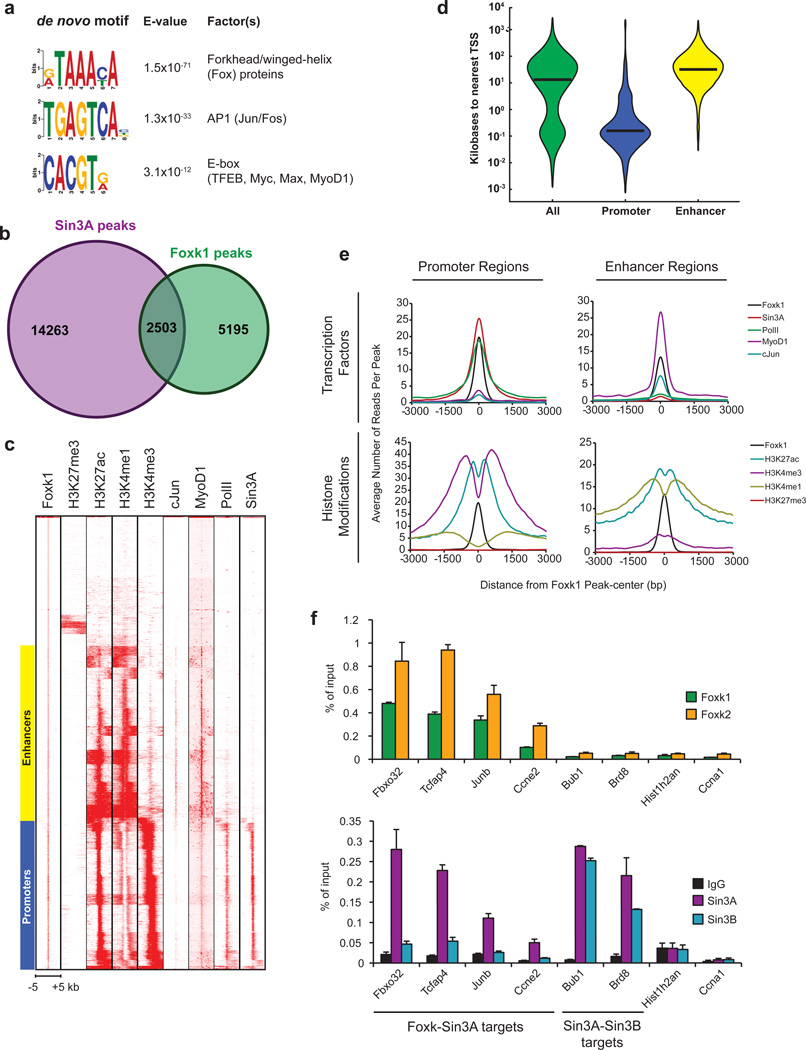
Using our ChIP-seq data, we interrogated genomic Foxk1 binding sites using a de novo computational approach, which identified the forkhead/winged-helix motif as the most significantly enriched DNA sequence (Fig. 2a). Other motifs were also strongly enriched within a 100 bp window (Fig. 2a; Supplementary Fig. 2b), indicating that additional factors may co-regulate Foxk1 targets. Next, we used genome-wide sequence comparisons with a mouse strain (C57/BL6) distinct from the C2C12 parental strain (C3H) to identify homozygous polymorphisms in this motif on a subset of genes. We found that each polymorphism completely abrogated Foxk1 binding to chromatin (Supplementary Fig. 2c), confirming that Foxk1 binds directly to this motif.
Figure 2. Genome-wide identification of Foxk1 binding sites at promoters and enhancers.
(a) Genome-wide discovery of Foxk1 binding motif and co-enrichment of additional transcription factor motifs. (b) Venn diagram depicting the number of Sin3A, Foxk1, and Sin3A-Foxk1 co-localized peaks deduced from our ChIP-seq data. The p value for co-localization of Foxk1 and Sin3A peaks (p<10−7869) was determined as outlined in the Methods section. (c) Supervised k-means clustering of Foxk1 ChIP-seq data revealed Foxk1 binding sites at promoters and enhancers. Each row represents a single 10 kb region surrounding the center of a Foxk1 enriched region (peak). (d) Promoter and enhancer regions predicted by k-means clustering are depicted as violin plots showing the distance of Foxk1 binding sites from the nearest TSS. Median distances are represented by black lines. (e) Tag densities of histone modifications and factors centered on Foxk1 peaks at promoters and enhancers. (f) qChIP analysis of Sin3 isoforms at Foxk1 and Foxk2 binding sites. Data represent n = three independent biological replicates from separate dishes and lysate preparations and are presented as mean+SEM.
We compared our Foxk1 and previously established Sin3A binding sites, which revealed extensive and significant overlap (p < 10−7868) in myoblasts (Fig. 2b, c; Supplementary Table 2). Supervised k-means clustering of Foxk1 and Sin3A data with previously published histone modification (H3K27me3, H3K27ac, H3K4me1, H3K4me3) and transcription factor (c-Jun, MyoD1, RNA PolII) occupancy (ChIP-seq) data revealed that Foxk1 is recruited to both promoters and distal enhancers (Fig. 2c–e). Further examination of these clusters indicated that nearly all promoters were associated with an active chromatin signature typified by high levels of H3K27ac, H3K4me3, and PolII and low levels of H3K27me3 and H3K4me1 (Fig. 2e). In contrast, putative enhancer sites were marked by high levels of H3K27ac, H3K4me1, c-Jun, and MyoD1 and low levels of H3K27me3, H3K4me3, and PolII, as expected for distal regulatory elements17.
We observed strong enrichment of Sin3A with Foxk1 at promoters but not at enhancers, consistent with previous studies10 (Fig. 2c, e; Supplementary Fig. 2d). Confirmatory qChIP showed that many sites strongly bound by Foxk1 and Foxk2 displayed a corresponding enrichment of Sin3A and negligible levels of Sin3B, reinforcing the observation that the Foxk proteins preferentially recruit Sin3A complexes to chromatin (Fig. 2f). Further, Foxk2 recruitment to each of the Foxk1 targets that we examined suggests that Foxk proteins are recruited to a highly overlapping set of genes (Fig. 2f), consistent with the fact that their DNA-binding domains are 91% identical (and 96% similar).
Foxk1 regulates atrophy and autophagy genes
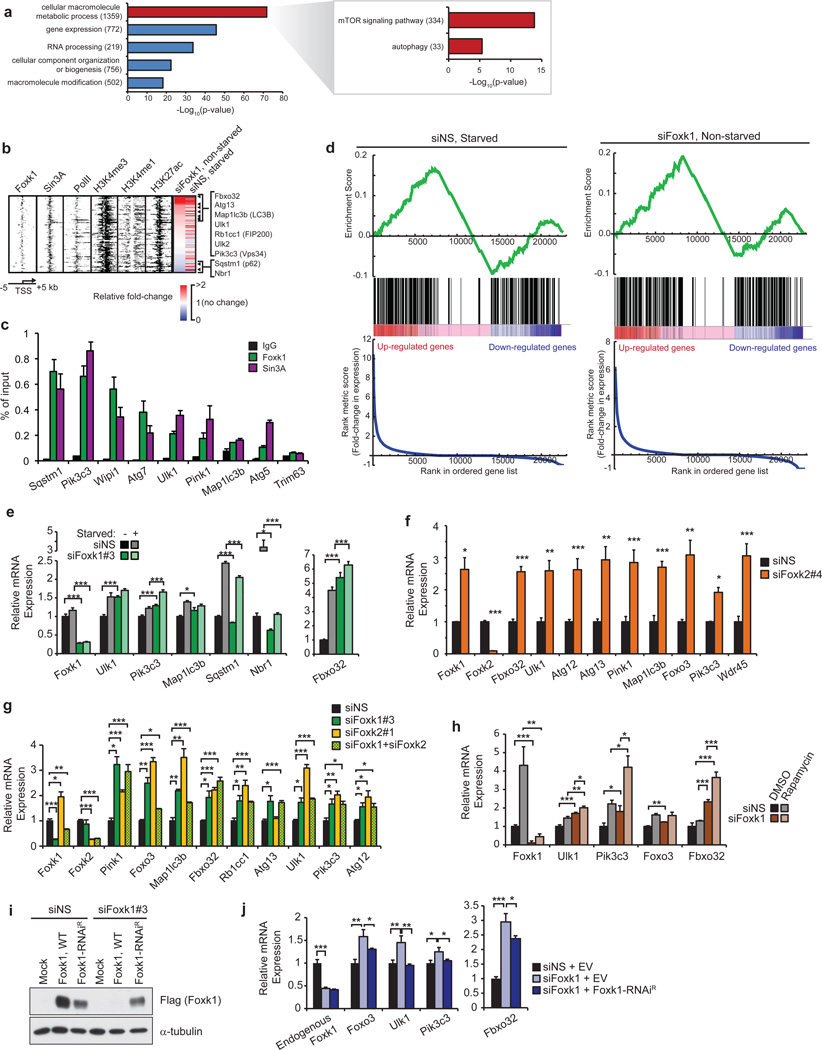
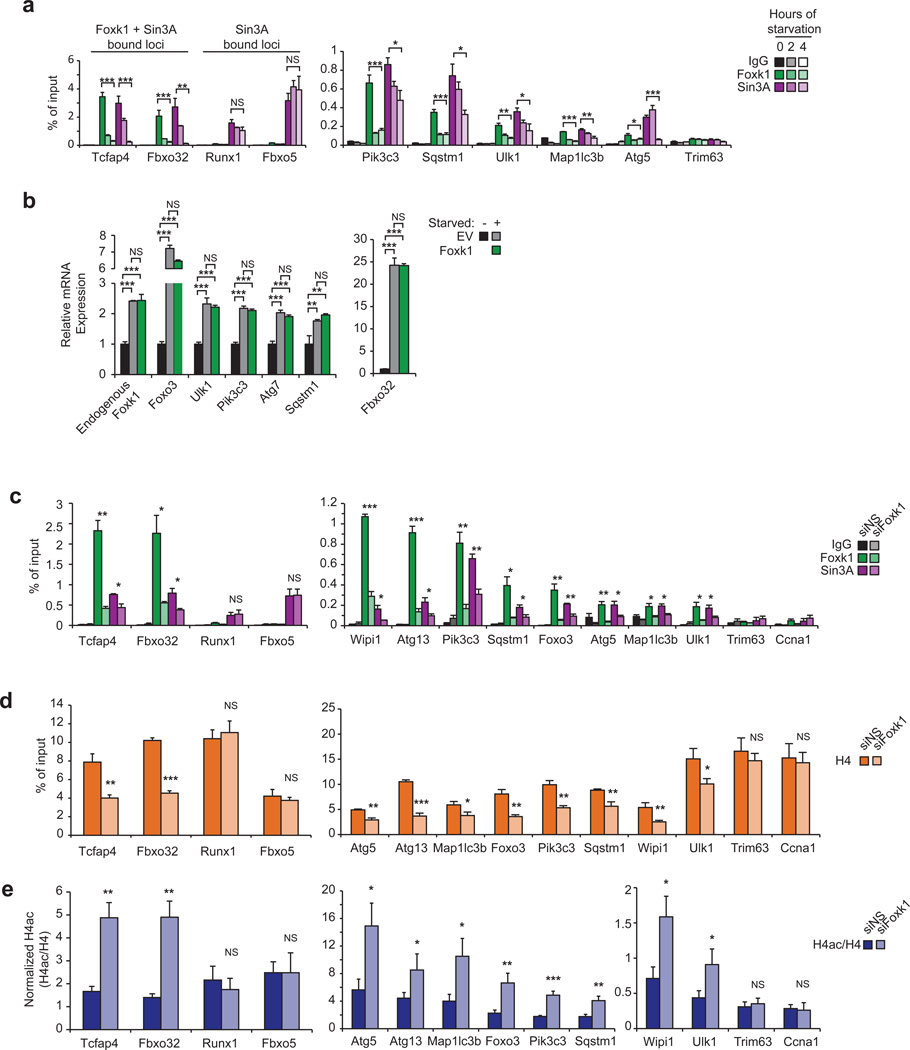
Using gene ontology (GO) analysis, we found that Foxk1 binding was strongly associated with genes involved in regulating metabolic processes (Fig. 3a). More specifically, Foxk1 was significantly enriched on genes that play roles in catabolic processes, including autophagic pathways and mTOR signaling. Since autophagy is associated with muscular atrophy6, Foxk recruitment to the pro-atrophy gene, Fbxo32 (also known as atrogin-1 or MAFbx), was notable (Fig. 2f). Fbxo32, an E3 ubiquitin-ligase that is dramatically up-regulated in response to starvation, cancer, disuse, denervation, or diabetes, is a key contributor to muscle atrophy18–22. We examined whether Foxk1 could directly regulate atrophy-associated genes via direct binding to promoter regions. Using a comprehensive list of 110 previously defined atrophy genes20 and 247 known autophagy-associated genes, we found that Foxk1 co-localized with Sin3A at the promoters of 108 of these genes (χ2 test, p = 2.5×10−27), 73% of which were autophagy genes (79 genes, χ2 test, p = 5.3×10−10)(Fig. 3b; Supplementary Table 3). We confirmed recruitment of Foxk proteins and Sin3A to a subset of these genes (Fig. 2f, 3c and Supplementary Fig. 3a).
Figure 3. Foxk1 represses transcription of genes associated with induction of atrophy and autophagy initiation.
(a) GO analysis of Foxk1 targets showed enrichment of genes involved in mTOR signaling and autophagy. All indicated p-values are Bonferroni-corrected. Numbers in parentheses indicate the number of Foxk1-bound genes in the respective categories. (b) Heatmap depicting ChIP-seq data for Foxk1, Sin3A, RNA PolII, H3K27ac, H3K4me3, and H3K4me1 at 108 Foxk1-bound autophagy genes significantly deregulated upon Foxk1 knockdown in non-starved conditions. RNA-seq expression, relative to non-starved control conditions, after either starvation or Foxk1 knockdown is displayed to the right. Rows are sorted by fold-change in expression upon Foxk1 knockdown, where red indicates increased, and blue indicates decreased, gene expression. Genes displayed to the right are those known to play important roles in atrophy and autophagy processes. (c) qChIP of Foxk1 and Sin3A at the promoters of autophagy genes. The promoter of Trim63 is a negative control devoid of Foxk1 and Sin3A. (d) Gene set enrichment analysis (GSEA) of atrophy and autophagy genes in starved (left) or Foxk1-depleted (right) myoblasts. Genes are ranked by fold-change in expression relative to control, non-starved cells. Atrophy and autophagy-associated genes are denoted by black bars in the middle of the panels. A normalized enrichment score of 3.5 and 4.1 for starved and Foxk1-depleted cells, respectively, were calculated from the GSEA. Both enrichment scores have a nominal p-value < 0.0001. (e) Quantitative reverse-transcriptase PCR (qRT-PCR) of siRNA-depleted Foxk1 with or without starvation. Foxk1 depletion de-repressed a subset of autophagy genes and Fbxo32 under basal conditions, and starvation further increased expression of some genes beyond levels seen with starvation or knock-down alone. (f) Foxk2 depletion leads to de-repression of a panel of autophagy genes and Fbxo32. (g) Depletion of Foxk1 and Foxk2 alone or together leads to de-repression of autophagy genes and Fbxo32. (h) qRT-PCR of siRNA-depleted Foxk1 with or without rapamycin treatment. (i) Ectopic production of Flag-tagged RNAi-resistant Foxk1 (Foxk1-RNAiR). (j) Transfection with Flag-tagged Foxk1-RNAiR, but not empty vector (EV), restored repression of target genes. All qChIP and qRT-PCR data represent n = three independent biological replicates from separate dishes and lysate preparations, and are presented as mean+SEM. * p <0.05, ** p<0.01, *** p<0.001 (two-tailed t-test).
These analyses suggested a potential link between Foxk1 function and nutrient sensing or restriction. To determine how Foxk1 functionally regulates this cohort of atrophy and autophagy genes, we analyzed genome-wide expression profiles in cells grown under nutrient-rich or starved (serum and amino acid-depleted) conditions and in cells depleted of Foxk1 (Supplementary Table 4). A gene set enrichment analysis (GSEA) of atrophy and autophagy-associated genes was used to assess differential gene expression in starved versus control cells (Fig. 3d, left panels). Starvation showed a mixture of up- and down-regulated atrophy and autophagy genes, but there was a strong positive enrichment of these genes among the most highly up-regulated genes. GSEA of non-starved, Foxk1-depleted (siFoxk1) versus control cells showed a strikingly similar enrichment of the same atrophy and autophagy genes (Fig. 3d, right panels), suggesting that Foxk1 ablation instigates changes in gene expression that closely mirror those induced by starvation.
Next, we examined the 108 genes directly bound by Foxk1 and Sin3A (Supplementary Table 3). Interestingly, the group of genes up-regulated by either Foxk1-depletion or starvation alone included Fbxo32 and critical components of the Ulk1 and Vps34 complexes (Ulk1, Ulk2, Pik3c3/Vps34, Ambra1, Rb1cc1/FIP200, and Atg13; Fig. 3e; Supplementary Fig. 4a), which comprise the machinery necessary to initiate autophagy and to nucleate vesicles that become autophagosomes23–27. Similar results were obtained with a second, distinct siRNA (Supplementary Fig. 4b). Likewise, Foxk2 depletion using two distinct siRNAs significantly up-regulated the same group of genes (Fig. 3f–g), and co-depletion of Foxk1 and Foxk2 did not further de-repress these genes (Fig. 3g). Interestingly, we noted that Foxk2 depletion led to de-repression of Foxk1 expression, suggesting that transcription of Foxk family members is tightly linked through a negative feedback mechanism. These results indicate that Foxk1 and Foxk2 act redundantly, but independently, to repress the expression of atrophy and autophagy genes, but the loss of either Foxk protein is sufficient to up-regulate their target genes.
Enhanced expression of Fbxo32 and autophagy-initiation genes in Foxk-depleted cells suggests that Foxk-mediated recruitment of the Sin3A complex results in the silencing of autophagic and atrophic responses (Fig. 3b, e–g). Moreover, induction of Fbxo32 and autophagy genes after Foxk1 depletion mimicked rapamycin-induced starvation, and expression of many genes was further enhanced upon combined rapamycin treatment and Foxk1 depletion (Fig. 3h). The cumulative effect of mTOR inhibition (through rapamycin treatment) and Foxk1 depletion was more than additive, suggesting that mTOR and Foxk1 could modulate both overlapping and distinct pathways. In reciprocal experiments, we ablated Foxk1 and expressed an RNAi-resistant cDNA (Fig. 3i). Importantly, over-expression of Foxk1 restored gene repression, confirming that (1) de-repression of Foxk1 target genes was not due to an off-target effect and (2) Foxk1 silences expression of autophagy genes (Fig. 3j).
Starvation leads to re-localization of Foxk1 from the nucleus to the cytoplasm
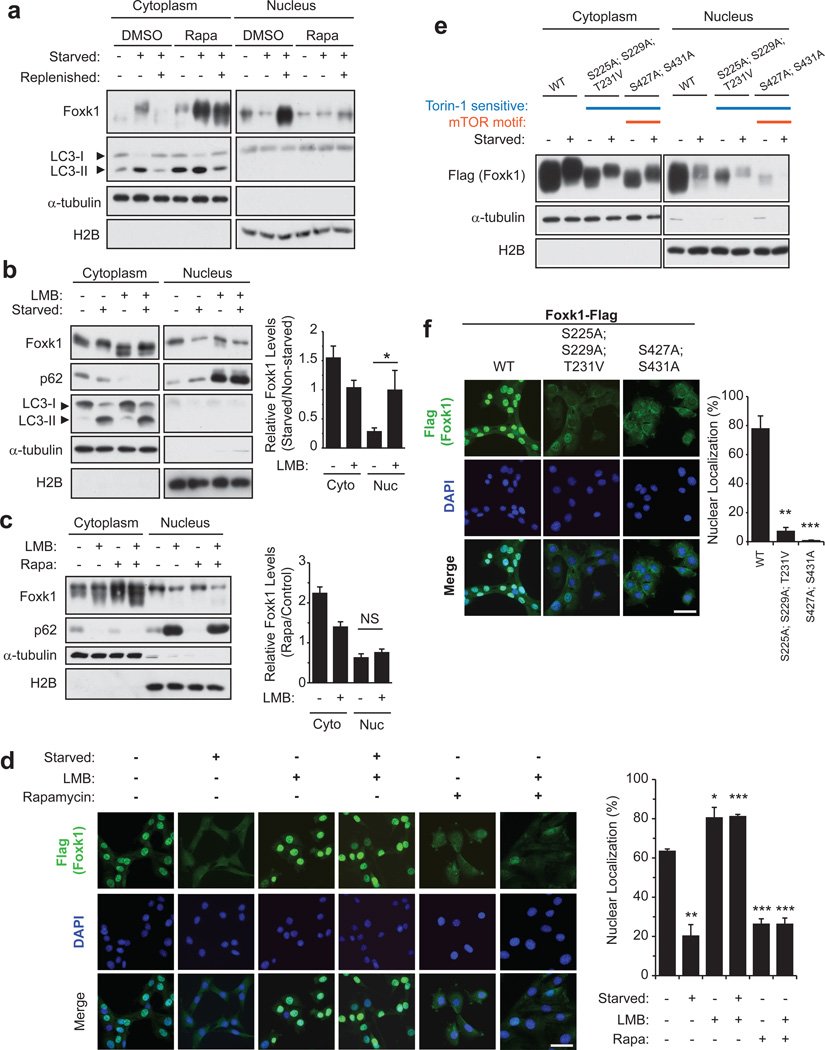
The transcriptional activity of another forkhead family member, Foxo3, is dictated by nutritional status, which regulates its nuclear localization28. We tested whether starvation signals could similarly affect the subcellular localization of Foxk1 and thereby impact gene expression. Upon starvation, Foxk1 was robustly transported from the nucleus into the cytoplasm (Fig. 4a, d), although total Foxk1 levels were not significantly altered during prolonged starvation (Supplementary Fig. 5a). Intriguingly, Foxk1 exported to the cytoplasm exhibited a clearly reduced mobility (Fig. 4a), and phosphatase treatment of Foxk1 immunoprecipitates confirmed that this was due to phosphorylation (Supplementary Fig. 5b). We assessed the lipidation of LC3-I (to form LC3-II), an indicator of autophagosome formation, as a marker to confirm that starvation had induced autophagy.
Figure 4. Nuclear import and export of Foxk1 is mTOR- and CRM1-dependent, respectively.
(a) Foxk1 is transported from the nucleus to the cytoplasm during autophagy, and nuclear localization was mTOR-dependent. Cells were incubated in nutrient rich medium with or without 100 nM rapamycin for 16 h prior to starvation for 4h, which was then followed by replenishment with nutrient-rich medium for 2 h. Where indicated, cells were continuously incubated in the presence of 100 nM rapamycin. (b) Cells were treated with 1 ng/μL leptomycin B (LMB) 1 hour before and during starvation. Control cells were treated with ethanol vehicle (EtOH). Quantification of n = 4 independent experiments is shown to the right. (c) Non-starved cells were incubated with rapamycin for 16 h prior to the addition of 1 ng/μL LMB for 5 hours. Quantification (n = 3 independent experiments) is shown to the right. (d) Cells stably over-expressing Flag-tagged Foxk1were starved, treated with LMB, and/or incubated with rapamycin as in panels a-c. Quantification of at least 88 cells viewed in 5–8 random fields from n = 3 independent experiments is shown to the right. Bar, 50 µm. (e) Myoblasts were transfected with the indicated Flag-tagged Foxk1 cDNAs. WT, wild-type; S225A;S229A;T231A, Torin1-sensitive sites; S427A;S431A, mTOR motif and Torin1-sensitive sites. Data represent two independent biological replicates from separate dishes and lysate preparations. (f) Cells stably over-expressing the indicated Flag-tagged wild-type (WT) or mutant Foxk1 were grown in nutrient-rich medium. Quantification of at least 88 cells viewed in 5–8 random fields from n = 3 independent experiments is shown to the right. Bar, 50 µm. Data are presented as mean+SEM. * p <0.05, ** p<0.01, *** p<0.001(two-tailed t-test).
Since starvation inhibits the mTOR kinase, rapamycin was used to mimic starvation and induce autophagy. In non-starved cells treated with rapamycin, Foxk1 localized largely to the cytoplasm, with a reduced mobility reminiscent of starvation (Fig. 4a). Furthermore, replenishment of amino acids and serum resulted in the re-accumulation of nuclear Foxk1 to an extent that exceeded pre-starvation levels. In contrast, rapamycin treatment blocked relocation from the cytosol after nutrient addition (Fig. 4a). Treatment with leptomycin B, an inhibitor of CRM1-mediated nuclear export29, provoked the retention of Foxk1 in the nucleus of starved cells (Fig. 4b, d), indicating that its nuclear export is CRM1-dependent. However, treatment with rapamycin followed by leptomycin B did not augment nuclear levels of Foxk1 or diminish its cytosolic levels (Fig. 4c, d), although p62/Sqstm1, another protein exhibiting nucleo-cytoplasmic shuttling30,31, was retained in the nucleus under these conditions. Furthermore, four independent phospho-proteome studies have shown that Foxk1 phosphorylation is sensitive to the mTOR inhibitors, rapamycin and Torin132–35, and Foxk1 is predicted to contain an mTOR phosphorylation motif32. Using a combination of sub-cellular fractionation and immunofluorescence, we showed that serine-to-alanine substitutions at Torin1-sensitive sites diminished nuclear import, and mutations at the predicted mTOR motif nearly abolished nuclear import of ectopically-expressed Foxk1 (Fig. 4e,f). The residues in and around these sites are highly conserved between the Foxk1 and Foxk2 proteins, suggesting that the subcellular localization of Foxk2 may be similarly regulated. Although the effects of mutating the analogous residues in Foxk2 remain to be tested, we did indeed observe relocation of Foxk2 from the nucleus to the cytosol upon starvation, reminiscent of Foxk1 (Supplementary Fig. 3b). Altogether, these data suggest that mTOR activity facilitates nuclear import of Foxk1, whereas mTOR inhibition, either through rapamycin treatment or starvation, abolishes nuclear import of Foxk1.
Starvation removes Foxk-Sin3A complexes from chromatin
The dramatic, starvation-induced relocation of Foxk1 suggested that Foxk1 might be effectively displaced from chromatin. Indeed, within two hours of starvation, Foxk1 was rapidly depleted from genomic targets, and recruitment was further reduced on several loci after four hours of starvation (Figs. 5a). Similarly, Foxk2 was removed from chromatin upon starvation (Supplementary Fig. 3a), suggesting that a common mechanism may underlie transcriptional control by Foxk proteins. Importantly, Sin3A vacated Foxk target genes over the same time frame, although genomic regions devoid of Foxk proteins exhibited no loss of Sin3A (Fig. 5a), confirming that the loss of Sin3A from these targets correlated with the removal of Foxk from chromatin. Consistent with the removal of Foxk1 from the nucleus and Foxk-Sin3A complexes from chromatin during starvation, over-expression of Foxk1 did not have a transcriptional effect on these genes under this condition (Fig. 5b). Sin3A complexes contain histone deacetylases (HDAC1/2), which are able to erase acetylated forms of histone H4 (H4ac)36–39. Therefore, we examined this histone modification on the promoters of autophagy genes as a function of starvation, and given its role in chromatin compaction, we also measured histone density via histone H4 ChIP. Interestingly, starvation-induced loss of Sin3A resulted in reduced histone H4 occupancy (Supplementary Fig. 6a) and a significant increase in H4ac levels (Supplementary Fig.6b), exclusively at Foxk1-Sin3A targets. These results suggest that starvation induced the coordinated loss of HDAC activity and reduced nucleosome occupancy at these loci. More importantly, and consistent with these observations, Foxk1-depletion led to a significant loss of Sin3A (Fig. 5c), reduced occupancy of histone H4 (Fig. 5d), and increased acetylation at the same loci (Fig. 5e). Interestingly, these findings are reminiscent of observations in yeast and mammalian cells, wherein loss of Sin3 was associated with decreased histone occupancy and changes in nucleosome stability40,41. We conclude that Foxk1 represses the expression of autophagy genes through recruitment of Sin3A complexes, leading to localized alterations in histone acetylation and nucleosome structure.
Figure 5. Starvation, via mTOR inhibition, signals the removal of Foxk1 from chromatin.
(a) qChIP indicates that Foxk1 and Sin3A dissociate from chromatin during starvation. Trim63 is a negative control promoter devoid of Foxk1 and Sin3A. (b) Starved cells transfected with Foxk1 did not exhibit transcriptional differences in autophagy gene expression. (c–e) qChIP shows that Foxk1-depletion leads to removal of Foxk1 and Sin3A from chromatin (c), reduction in the levels of histone H4 (d), and enhanced acetylation of H4 (e) at autophagy genes. Data shown in panels c–e are from the same experiments, and IgG controls are presented in Fig. 5c. Trim63 and Ccna1 are negative control promoters devoid of Foxk1 and Sin3A. All qChIP and qRT-PCR data represent n = three independent biological replicates from separate dishes and lysate preparations, and are presented as mean+SEM.* p <0.05; ** p<0.01; *** p<0.001; NS, not significant (one-way ANOVA for panel 5a, two-tailed t-test for panels 5b–e).
Foxk1 and Foxo3 coordinately regulate a common set of atrophy and autophagy genes
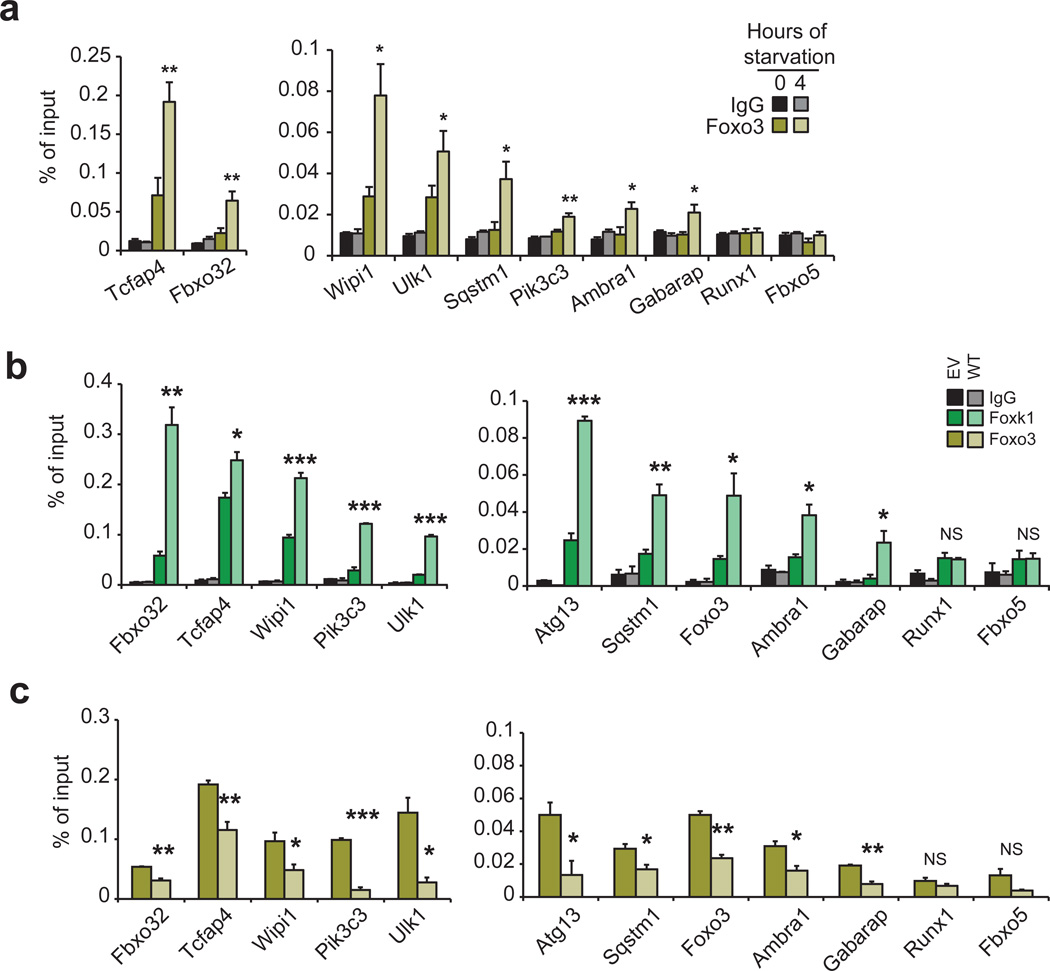
It has been shown that Foxo3, another forkhead/winged-helix transcription factor, coordinately initiates atrophy and autophagy programs by directly binding the promoters of Fbxo32 and autophagy genes (such as Gabarap1l, Map1lc3b, Atg12l), inducing their expression and resulting in proteolysis of skeletal muscle proteins6,42. In starved cells, Foxo3 enters the nucleus, binds the promoters of several autophagy genes, and up-regulates their expression6,28,42. Since Foxo3 recognizes a forkhead consensus motif43 highly related to the Foxk1 binding sequence deduced by our ChIP-seq experiments, it is possible that Foxo3 and Foxk1 bind to a common set of targets. Interestingly, upon starvation, Foxo3 was enriched at each of the promoters bound, and then vacated, by Foxk1 (Fig. 6a), including autophagy genes, indicating that starvation promotes Foxo3 binding to an overlapping set of genes that are repressed by Foxk1 in nutrient-rich conditions. In addition, over-expression of Foxk1 in starved cells displaced Foxo3 at these same loci (Fig. 6b, c), suggesting that Foxk1 and Foxo3 compete for the same binding sites. These data strongly suggest that transcriptional de-repression due to nutrient deprivation correlates with the concerted departure of Foxk1 from the nucleus and the simultaneous entry of Foxo3 into that compartment.
Figure 6. Foxk1 and Foxo3 are recruited to the same atrophy and autophagy genes.
(a) qChIP shows increased Foxo3 recruitment at Foxk1 sites during starvation. (b) Starved cells stably overexpressing wild-type Foxk1 (WT) exhibit higher levels of Foxk1 on chromatin than control cells expressing an empty vector (EV). (c) Overexpression of Foxk1 displaces Foxo3 from chromatin in starved cells. Data represent n = three independent biological replicates from separate dishes and lysate preparations, and are presented as mean+SEM. * p <0.05; ** p<0.01; *** p<0.001; NS, not significant (two-tailed t-test).
Foxk1 deficiency increases autophagic flux
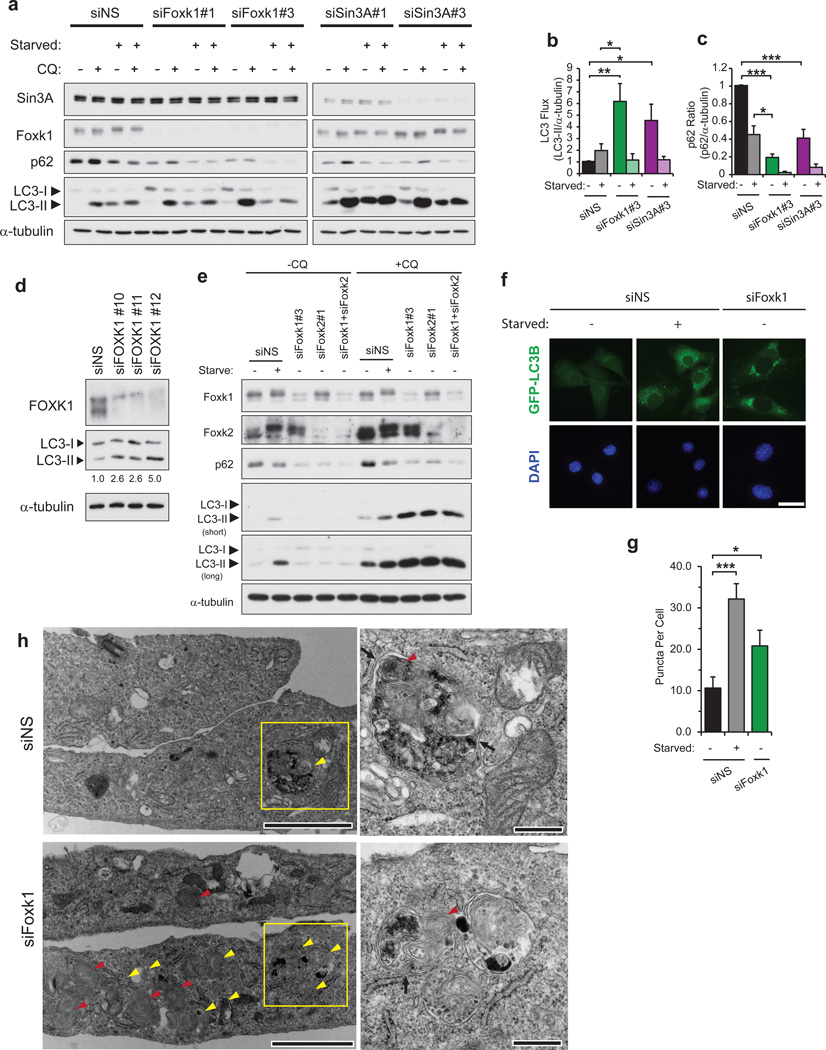
Since autophagy and lysosome-mediated protein degradation make significant contributions to skeletal muscle atrophy6, we speculated that Foxk proteins could play an important role in regulating this autophagic arm. The loss of Foxk1 or Foxk2 results in the up-regulation of several genes controlling the initiation and nucleation of autophagosome formation (e.g., Ulk1, Pik3c3) and a slight reduction in expression of genes that control subsequent autophagic events (e.g., Sqstm1, Nbr1; Fig. 3b,e–g). To clarify the role of Foxk in regulating autophagic processes, myoblasts were either grown in nutrient-rich medium or starved of amino acids and serum. We followed the conversion of LC3-I to LC3-II in the presence of chloroquine, which neutralizes the pH of lysosomes, thereby preventing the degradation of autophagosomal contents, such as LC3-II and p6244,45. This treatment allowed us to measure autophagic flux in cells depleted of Foxk1 or Sin3A. Remarkably, in nutrient-rich conditions, the loss of Foxk1 or Sin3A resulted in the dramatic enhancement of basal autophagy, as shown by increased LC3-II levels (Fig. 7a, b). We tested the generality of these findings by performing analogous knock-down experiments in human IMR90 fibroblasts. Importantly, our observations in fibroblasts recapitulated those seen in myoblasts (Fig. 7d). Furthermore, in nutrient-rich conditions, p62 levels were reduced to below starvation levels in Foxk1- and Sin3A-depleted cells (Fig. 7a, c). Although Foxk1-depletion slightly down-regulates p62/Sqstm1 mRNA expression (80% of control; Fig. 3e), the extreme reduction of p62 protein levels to 20% of the control suggests that autophagic degradation is likely the more important factor contributing to p62 reduction, supporting a persistent enhancement in autophagic flux in cells depleted of Foxk1 or Sin3A. In striking contrast, in starved cells, suppression of Foxk1 or Sin3A failed to produce a significant difference in LC3 flux as compared to controls (Fig. 7b). This observation confirms that Foxk1-Sin3A complexes actively repress autophagy genes only when Foxk1 is nuclear, that is, in nutrient-rich conditions. Similarly, depletion of Foxk2 or co-depletion of Foxk1 and Foxk2 increased autophagic flux to levels comparable to those observed in Foxk1-depleted cells (Fig. 7e, Supplementary Fig. 3c), further indicating that the Foxk proteins redundantly – yet independently – repress autophagic processes. Hence, the Foxk-Sin3A complex represses autophagy to basal levels found in nutrient-rich conditions, but their repressive effects are relieved when autophagy is induced by amino acid and serum starvation.
Figure 7. Foxk/Sin3A complexes suppress autophagic flux in nutrient-rich conditions.
(a,b) Control (siNS) cells or cells depleted of either Foxk1 or Sin3A were grown in nutrient-rich or starvation medium in the presence or absence of chloroquine (CQ) for 90 min. LC3 flux was calculated as the ratio of LC3-II to α-tubulin levels in cells treated with chloroquine (CQ). LC3 and α-tubulin immunoblots correspond to the same gels in each group, and LC3-II/α -tubulin ratios were calculated from a single film containing both LC3 and the corresponding α–tubulin blots. Quantitation is from n = 3 independent experiments. (c) Foxk1 or Sin3A loss leads to reduced p62 protein levels. The ratio of p62 to α-tubulin levels was calculated using cells that were not treated with CQ. Quantitation is from n = 3 independent experiments. (d) Foxk1-depletion by siRNAs against three distinct target sequences increased autophagic flux in non-starved human IMR90 fibroblasts incubated in the presence of 30 µM CQ for 90 min. Numbers indicate LC3 flux, calculated as the ratio of LC3-II to α-tubulin levels. (e) Control (siNS) cells, cells singly depleted of either Foxk1 or Foxk2, or cells co-depleted of Foxk1 and Foxk2 were grown in nutrient-rich or starvation medium in the presence or absence of CQ for 90 min. Experiments were performed independently three times. (f) Representative images of direct fluorescence of stably-expressed GFP-LC3B in control (siNS), starved, or Foxk1-depleted myoblasts. Bar, 20 µm. (g) Quantification of the number of puncta in cells stably expressing GFP-LC3B. Puncta were counted in at least 36 cells viewed in 3–4 random fields over n = three independent experiments for each condition. (h) Foxk1 loss produced an abundance of autophagic vacuoles (yellow arrowheads) and multi-lamellar bodies (red arrowheads). Black arrows indicate double membranes of autophagosomes. Right panels are magnified views of boxed regions indicated to the left. Bars, 2 and 0.5 µm for the left and right panels, respectively. Data are presented as mean+SEM. *p<0.05, ** p<0.01, *** p<0.001 (two-tailed t-test).
To further validate that the loss of Foxk1 promotes basal autophagy, we monitored autophagosome formation in control, starved, and Foxk1-depleted cells using direct fluorescence of GFP-tagged LC3B. Both starvation and Foxk1 depletion generated considerably more punctate GFP-LC3B dots than control cells, signifying increased autophagosome formation (Fig. 7f, g). This indicates that the loss of Foxk1, like starvation, increased autophagosome production and autophagic flux.
We also examined control and Foxk1-depleted cells using electron microscopy. Foxk1-depletion in myoblasts vastly increased the number of autophagic vacuoles, many of which contained electron-dense ribosomes undergoing degradation, indicative of autolysosomal assembly and activity (Fig. 7h; Supplementary Fig. 7). Since starvation-induced autophagosomal membranes are at least partially supplied by the outer membranes of mitochondria to yield multi-lamellar bodies (MLB)46, we found it interesting that Foxk1-depleted cells exhibited substantially increased numbers of MLBs. In addition, mitochondria of Foxk1-depleted cells were smaller and more electron-dense (Supplementary Fig. 7), possibly due to the loss of bulk outer mitochondrial membranes to form autophagosomes. The increased number of autophagic vacuoles and MLBs in Foxk1-depleted cells further supports the notion that Foxk1 loss up-regulates Ulk1 and Pik3c3 complexes to enhance the nucleation and formation of autophagosomal membranes, which sustains the prolonged elevation of basal autophagy observed in cells depleted of Foxk1.
Discussion
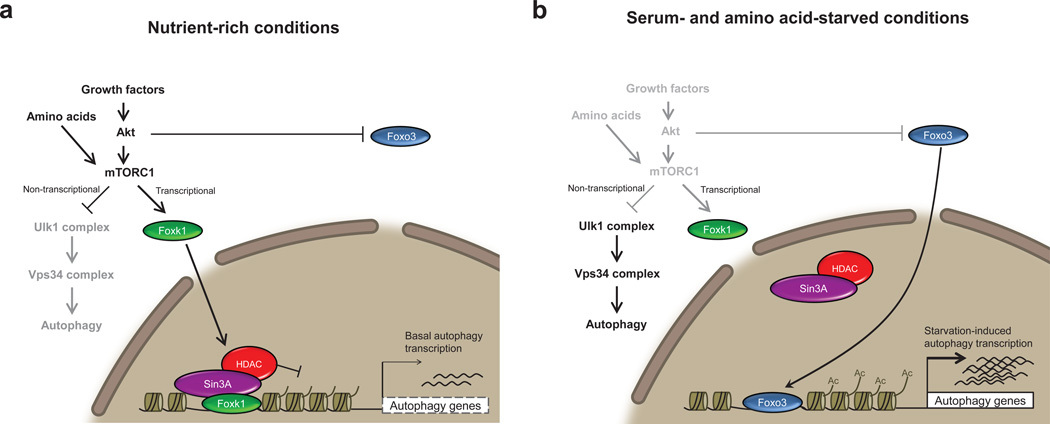
Our studies demonstrate that recruitment of Foxk and Sin3A co-repressor complexes act as central players to transcriptionally dampen autophagy to basal levels (Fig. 8a). Interestingly, the yeast Ume6-Sin3-Rpd3 complex, homologous to mammalian Sin3-HDAC, has been shown to negatively regulate autophagy50. Deletion of the sequence-specific factor Ume6 increases autophagic activity and produces larger autophagic bodies even in the absence of starvation, strongly resembling our observations in Foxk-depleted cells. Since Ume6 is not conserved in mammals, we suggest that the Foxk-Sin3-HDAC complexes may have supplanted this ancient autophagy-regulating pathway, and in this setting, the Foxk proteins may have assumed the role of yeast Ume6.
Figure 8. Model illustrating a role for Foxk1 repression of autophagy and atrophy induction.
(a) Autophagy genes are transcriptionally repressed through the direct binding of Foxk proteins and recruitment of Sin3A-HDAC complexes, resulting in the reduction of activating H4ac marks. Gene repression exerted through Foxk proteins is controlled downstream of mTOR complex 1 (mTORC1) activity, as mTORC1 inhibition, either through rapamycin or starvation treatment, prevents nuclear import of Foxk1. These observations suggest that mTORC1 negatively regulates autophagy by (1) inhibiting the activation of autophagy-initiating Ulk1 complexes47,48 and (2) promoting the nuclear import and transcriptional repression of Foxk targets, including components of the Ulk1 and Vps34 complexes. mTORC1 thus effectively down-regulates autophagy through non-transcriptional and transcriptional means. (b) Starvation leads to the inhibition of the Akt-mTORC1-Foxk1 axis. Inhibition of mTORC1 prevents nuclear import of the Foxk proteins and, hence, leads to de-repression of autophagy and atrophy genes, mirroring depletion of Foxk. In addition, Akt inhibition permits nuclear import and subsequent binding of Foxo3 at the promoters of these same genes, leading to the upregulation of their expression.
Autophagic induction via inhibition of activity of Foxk-Sin3A-HDAC complexes has important implications for muscle development, repair, and homeostasis. Autophagic induction through mTORC1 inhibition prevents myogenesis51,52, suggesting that Foxk1 repression of autophagy, brought about by active mTORC1, may be important for the progression of the myogenic differentiation program. Additionally, viable Foxk1-null mice exhibit growth and muscle regeneration defects, presumably due to defective satellite cell proliferation15,16. Foxk1-deficiency could contribute to these phenotypes by elevating or prolonging autophagy, resulting in autophagy-associated death of satellite cells necessary for muscle repair. Lastly, we suggest that Foxk-Sin3A complexes are important for the repression of atrophic and catabolic processes, such as autophagy, since the disruption of this repressive mechanism may explain the occurrence of severely disrupted sarcomeres and lethality in Sin3A-depleted muscle10, even in the absence of starvation. Altogether, these data suggest that Foxk-Sin3A complexes are vital for the repression of autophagy to basal levels, and its deregulation may lead to pathological conditions wrought by over-stimulation or abnormal persistence of autophagy.
Prolonged autophagy decreases genome-wide levels of H4K16ac and subsequently leads to reduced expression of autophagy genes3. Interference with H4K16ac modulation promotes cell death, implying that this reduction is a part of the normal starvation-induced autophagy program that prevents prolonged autophagy and subsequent lethality. Our studies suggest that starvation initiates a rapid response that increases, rather than decreases, both H4ac levels and corresponding gene expression, and our Foxk1 and Sin3A-depletion studies support the role of H4ac in determining the fate of cells that have initiated autophagy. Cells deficient in Foxk or Sin3A exhibit unchecked, prolonged autophagy in nutrient-rich conditions (Fig. 7). This ultimately results in cell death, which is significantly exacerbated by starvation (data not shown). Likewise, prolonged inhibition of class I HDACs similarly induces autophagy in yeast and mammalian cells, where it also promotes cell death53–55. Taken together, these studies imply that autophagy induction through abrogation of HDAC activity may override mechanisms that are ultimately necessary to down-regulate autophagy and thereby prevent cell death associated with prolonged autophagy56. Our work highlights the importance of Foxk-Sin3A-HDAC complexes in restraining autophagy and emphasizes the lethality of disrupting this repression in the absence of starvation.
Our studies provide a better understanding of the mechanisms underlying life and death decisions in autophagy-associated cell death, supporting the use of HDAC inhibitors to treat various cancers. Currently, the National Cancer Institute reports the use of SAHA (vorinostat), a class I and II HDAC inhibitor57, in clinical trials either alone or in combination with other drugs to combat various hematological and solid tumors, including T-cell lymphoma, multiple myeloma, glioblastoma multiforme, and colorectal cancers (http://www.cancer.gov/clinicaltrials/search/results?protocolsearchid=12463342). Our study unveils a chromatin-dependent mechanism controlled by mTORC1 that may disconnect autophagy induction from normal compensatory pathways. Our work suggests avenues for implementation of autophagy- and HDAC-modulating drugs to improve current cancer therapies by taking advantage of mechanisms that push a cell toward autophagy-associated cell death.
Materials & Methods
Cell Culture
C2C12 myoblasts (Sigma) and IMR90 normal fibroblasts (NIA Aging Cell Repository) were grown in DMEM supplemented with 10% fetal bovine serum, 20 mM L-glutamine, 100 IU penicillin, and 100 μg/mL streptomycin. Cells were starved by first rinsing twice with pre-warmed Hank’s Balanced Salt Solution (HBSS) and incubating in pre-warmed HBSS supplemented with 10 mM HEPES pH 7.5, 100 IU penicillin, and 100 µg/mL streptomycin for 4 h, unless otherwise indicated. Cells were treated with 100 nM rapamycin for 16 h. Cells were treated with 30 μM chloroquine for 90 min. All cells were routinely confirmed to be free of mycoplasma.
Antibodies
Antibodies used were FOXK1 (Santa Cruz sc-134550, 1:2000), FOXK2 (Bethyl A301-730A-1, 1:1000), Sin3A58 (1:5000), SIN3B (Santa Cruz sc-768, 1:1000), HDAC2 (Abcam ab7029, 1:10000), SAP3059 (1:1000), Sds3/SAP4560 (1:1000), Pf1/Phf12 (Bethyl A301-647A, 1:1000), RBP2/Kdm5a61 (1:1000), MRG15 (1:1000), LC3B (Cell Signaling #2775, 1:2000), p62/Sqstm1 (MBL PM045, 1:5000), α-tubulin (Sigma #T5168, 1:5000), H2B (Abcam ab1790, 1:2000), H4 (Millipore 05-858), pan-acetyl H4 (Millipore 06-866), FOXO343, and Flag (Sigma #F7425, 1:500). The specificities of the Foxk1 and Foxk2 antibodies were validated by their specific recognition of their intended antigens in co-immunoprecipitation experiments (Fig. 1c, Supplementary Figure 1), as well as in lysates from cells depleted of either Foxk1 or Foxk2 (Fig. 1d).
Preparation of Cytosolic, Nuclear, and Solubilized Chromatin Fractions for Immunoprecipitation
Harvested cells were rinsed with ice-cold PBS, resuspended in 5 cell pellet volumes of homogenization buffer (20 mM HEPES pH 7.9, 10 mM potassium chloride, 1.5 mM magnesium chloride, 1 mM DTT, and protease and phosphatase inhibitors (aprotinin, leupeptin, pepstatin A, PMSF, sodium fluoride, and sodium orthovanadate)), and allowed to swell on ice for 15 minutes. The cells were lysed with 45 strokes in a Dounce homogenizer using a B-type pestle to release intact nuclei. The nuclei were then spun down at 500×g for 15 minutes at 4 °C, and the resulting supernatant (cytoplasmic fraction) was collected. The nuclear pellet was washed once with homogenization buffer, extracted in buffer (20 mM HEPES pH 7.9, 0.4 M potassium chloride, 1.5 mM magnesium chloride, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, and protease and phosphatase inhibitors) for 30 minutes at 4 °C. The nuclear extract was spun at 20,000×g for 30 minutes at 4 °C, and the resulting supernatant (nuclear fraction) was collected. The pellet was quickly washed once with MNase buffer (20 mM Tris pH 7.5, 100 mM potassium chloride, 2 mM magnesium chloride, 1 mM calcium chloride, 0.3 M sucrose, 0.1 % Triton X-100). The pellet was resuspended in 3 pellet volumes of MNase buffer plus 10 U MNase per 100 μL of resuspension and agitated at room temperature for 30 minutes. The MNase digested pellet was quenched by the addition of EDTA and EGTA to a final concentration of 5 mM, centrifuged at 4 °C for 15 minutes, and the supernatant was saved as the solubilized chromatin fraction. All samples were dialyzed against dialysis buffer (50 mM HEPES pH 7.9, 150 mM sodium chloride, 0.2 mM EDTA, 10 % glycerol, 0.1% NP40, 0.5 mM DTT, 0.2 mM PMSF) before use. Protein concentrations were quantified by the Bradford method. For immunoprecipitations, 1 mg of protein after centrifugation was agitated with the appropriate antibody (previously coupled to protein A sepharose with dimethyl pimelimidate (DMP)) overnight at 4 °C. The beads were then washed four times with dialysis buffer, and the bound proteins were analyzed by SDS-PAGE and immunoblotting.
Purification and Mass Spectrometry Analysis of Sin3A Complexes
Normal rabbit serum, anti-hemagglutinin (12CA5), and anti-Sin3A antibodies were coupled to protein A sepharose using DMP. Five milligrams of solubilized chromatin was pre-cleared by agitating with normal rabbit serum for 1 h, anti-hemagglutinin antibody for 1 h, and protein A sepharose for 30 min, all at 4 °C. Sin3A complexes were then captured by incubating the pre-cleared supernatant with 5 µg of anti-Sin3A antibody for 2 h at 4 °C. The resin was washed six times in high-salt wash buffer (50 mM Tris pH 7.9, 0.6 M potassium chloride, 5 mM EDTA, 0.1% NP40, and protease and phosphatase inhibitors) and then agitated in a chaotropic solution (50 mM Tris pH 7.9, 2 M guanidine hydrochloride, 150 mM potassium chloride, 5 mM EDTA, 0.1% NP40) for 15 min at room temperature to elute Sin3A complexes. The resulting eluate was TCA precipitated and fractionated by SDS-PAGE until the dye front entered 1 cm into the resolving gel. After Coomassie Blue staining, six gel slices were subjected to mass spectrometric analyses at the Harvard Mass Spectrometry and Proteomics Resource Laboratory by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a Thermo LTQ-Orbitrap mass spectrometer.
Chromatin Immunoprecipitation
Sub-confluent cells were cross-linked at room temperature with 1% formaldehyde for 10 minutes, then quenched by incubating in 0.125 M glycine. The cells were washed twice with ice-cold PBS and resuspended in 3 mL ChIP Lysis Buffer (10 mM Tris pH 8, 1 mM EDTA, 0.5 mM EGTA, 0.5% N-lauroyl sarcosine, and protease and phosphatase inhibitors) per ten 15-cm dishes. Each 1 mL cell resuspension was individually sonicated on ice for 1–2 rounds using a Branson Sonifier 450 at Output 2 and a duty cycle of 90% for 6 pulses. An average fragment size of 350 bp was obtained by further sonicating the chromatin in a Diagenode Bioruptor for 30 minutes on high power with a 30 s ON/30 s OFF cycle. Debris was then spun down at 20,000×g for 15 min at 4C, and the chromatin in the supernatant was quantified and used for subsequent ChIP experiments.
Chromatin was diluted to a final volume of 500 µL in ChIP Lysis Buffer plus 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, and protease inhibitors before pre-clearing with 10 μL protein A sepharose (previously blocked in 1 mg/mL BSA) for 4 h at 4 °C. Pre-cleared chromatin was incubated with antibody overnight at 4 °C. For Foxk1, Foxk2, Sin3A, and Foxo3 ChIP, 20 μg of chromatin and 2 μg of antibody were used. For pan-acetyl H4 ChIP, 10 μg of chromatin and 2 μL of serum were used. For histone H4 ChIP, 2 μg of chromatin and 4 μg of antibody were used. Immuno-complexes were captured by incubating with 10 μL protein A sepharose for 4 h at 4 °C. Immunoprecipitates were washed eight times with RIPA buffer (50 mM HEPES pH7.6, 10 mM EDTA, 0.7 % sodium deoxycholate, 1% NP40, 0.5 M lithium chloride, and protease inhibitors), once with a low-salt wash (50 mM Tris pH 8, 10 mM EDTA, 50 mM sodium chloride), and eluted in elution buffer (50 mM Tris pH 8, 10 mM EDTA, 1% SDS) at 65 °C for 15 min. The supernatant was incubated overnight at 65 °C to reverse cross-links, diluted two-fold in 50 mM Tris pH 8 plus 10 mM EDTA, and then sequentially digested with 80 μg RNase A for 2 h at 37 °C and 80 μg proteinase K for 30 min at 55 °C. DNA was extracted with phenol:chloroform:isoamyl alcohol and ethanol precipitated. DNA pellets were resuspended in 300 μL 10 mM Tris pH 8 and assayed by quantitative PCR using the SYBR Green method.
ChIP-seq and RNA-seq Library Preparation and Data Analyses
ChIP-seq libraries were prepared as previously described62 and sequenced (single-read, 51 bp) using an Illumina HiSeq 2000 machine. Raw reads were aligned to the mm9 genome using BWA 0.7.5a with default options. All ChIP-seq data have been submitted to the Gene Expression Omnibus (GEO). Non-duplicate reads mapping to a single genomic locus were analyzed using the Qeseq algorithm63 with the following parameters: “-s 100 -c 15 -p 0.00005” where s, c, and p denote one-third of the sonicated DNA fragment size, the minimum number of reads required to constitute an enriched region (peak), and the p-value used to consider a peak as significantly enriched, respectively. MEME-ChIP64 was used to identify enriched DNA motifs within 100 bp centered around the summit of each Foxk1 peak. Peaks were subjected to GREAT analysis to identify Gene Ontologies (GO). Each gene was assigned a basal “regulatory domain” defined as a region 3 kb upstream and 1 kb downstream of the transcription start site (TSS), which was extended up to 100 kb or to the boundary of the nearest gene’s basal domain. Each peak was annotated to genes in whose regulatory domains it overlapped. All C2C12 histone modification (H3K4me1, H3K4me3, H3K27ac, H3K27me3), c-Jun, and RNA PolII ChIP-seq data were previously published17,62. C2C12 MyoD1 ChIP-seq data are available from ENCODE. Autophagy gene databases used were the Human Autophagy Database (http://autophagy.lu) and the mouse Autophagy Database (http://www.tanpaku.org/autophagy). The list of atrophy genes was obtained from http://agoldberg.med.harvard.edu/muscledatabase.
For RNA-seq, cells were rinsed in PBS, and RNA was isolated using the RNeasy Mini Kit (QIAgen) with QIAshredder columns and in-column DNaseI digestion as per the manufacturer’s instructions. After polyA selection, the TruSeq method was used to generate RNA libraries. RNA-seq reads for each library were aligned to the mouse genome build mm9 using Tophat 2.0.9 and Bowtie 2.1.0 with the “-G genes.gtf” and “–no-novel-junctions” options, where genes.gtf contains the UCSC coding transcripts in GTF format. The transcriptome for each library was assembled using Cufflinks 2.1.1 with the default options. The transcriptomes were merged using Cuffmerge, and differential expression analyses was performed using Cuffdiff with the “-u” and “-M mask.gtf” options, where mask.gtf contains mitochondrial RNA, tRNA, or rRNA transcripts that were masked from these analyses. All subsequent ChIP- and RNA-seq analyses and heatmap generation were completed using customized Mathematica and Python scripts.
Whole Cell Lysate (WCL) Preparation and Subcellular Fractionation
All cells were briefly rinsed twice with ice-cold PBS before harvesting. For WCLs, the cells were resuspended in Triton Lysis Buffer (TLB; 50 mM Tris pH 7.9, 0.5% Triton X-100, 137.5 mM sodium chloride, 10% glycerol, 5 mM EDTA, 1 mM DTT, and protease and phosphatase inhibitors) and sonicated using a Branson Sonifier 450 at Output 2 and a duty cycle of 90% for 6 pulses. For cytosolic and nuclear fractions, the cells were lysed in 100 μL TLB by pipetting 15 times and spun at 500×g for 5 min at 4 °C. The supernatant (cytosolic fraction) was removed to a separate tube. The nuclear pellet was rinsed with 300 μL TLB and spun down at 500×g for 5 min at 4 °C. The supernatant was discarded and the pellet resuspended in 100 μL TLB. The cytosolic and nuclear fractions were sonicated as for the WCLs. After centrifugation, protein concentrations were quantified by the Bradford method and analyzed by SDS-PAGE and immunoblotting. Immunoblots displayed in Figures 4a, 4e, and Supplementary Figure 5b were obtained after electrophoresing samples through larger polyacrylamide gels to achieve higher resolution. This allowed us to better differentiate between phosphorylated forms of Foxk1. All other immunoblots were generated after electrophoresis of samples through mini-gels.
Stable cell lines and Transient siRNA and Plasmid Transfections
Cells were seeded at 20×103 or 10×103 cells/cm2 for 24 or 48 h transfections, respectively. For transfection of siRNA, cells were transfected with 3 μL RNAiMax per 1.3 mL medium and 15 nM siRNA using the fast-forward method. siRNA target sequences are provided in Supplementary Table 5. For transfection of plasmids, cells were plated at 10×103 cells/cm2 and transfected 24 h later with 4 μL Lipofectamine 2000 and 1 μg DNA each per 1.3 mL medium. The medium was always changed to fresh growth medium the next day.
To generate stable cell lines, retroviral particles were first obtained by transfecting pBABE-puro plasmids into Phoenix ecotropic packaging cells. Viral supernatants were collected at 48 and 72 h post-transfection. Cultured cells were transduced with virus and 8 μg/mL polybrene for 4–6 h. After transduction, cells were allowed to grow in virus-free medium for 24 h before selecting with 1 μg/mL puromycin for one week. Cells were grown in puromycin-free medium for at least two days prior to use.
mRNA Expression Analyses
Cells grown in 6-well dishes were quickly rinsed twice with pre-warmed PBS and harvested in 1 mL TRIzol (Ambion) per well. Total RNA was isolated as per the manufacturer’s instructions. The Verso cDNA Synthesis Kit (Thermo Scientific) was used to synthesize cDNA from 250 ng total RNA as per the manufacturer’s instructions. cDNA was diluted to 400 μL in 10 mM Tris pH 8 and assayed by quantitative PCR using the SYBR Green method. Eci2 was used as a loading control for qRT-PCR expression analyses as it was found to be a relatively highly expressed gene that was unchanged after starvation or Foxk1-depletion.
Phosphatase Assays
Foxk1 immunoprecipitates were washed three times with dialysis buffer and incubated in a total volume of 50 µL containing 400 U λ protein phosphatase (NEB), 1X PMP buffer, 1 mM manganese chloride, and phosphatase inhibitors (sodium orthovanadate, sodium fluoride, β-glycerophosphate) for 1 h at 30 °C. Immunoprecipitates were then washed three times with dialysis buffer to remove non-interacting proteins.
Immunofluorescence
Cells grown on glass coverslips were fixed with 4% formaldehyde (Sigma) for 15 min at room temperature. To visualize over-expressed Flag-tagged Foxk1, cells were permeabilized in PBS plus 1% Triton for 5 min and blocked in PBS, 0.1% Triton, and 5% BSA for 30 min at room temperature. Cells were then successively incubated with anti-Flag antibody (Sigma #F7425) and secondary antibody coupled to AlexaFluor 488 (Life Technologies). Cells were stained with DAPI, mounted on glass slides using ProLong Gold antifade reagent (Molecular Probes), and visualized on a Nikon Eclipse E800 microscope equipped with a Photometrics Coolsnap HQ CCD camera. Images were acquired with MetaMorph v7.8 (Molecular Devices).
Transmission Electron Microscopy
Cells were fixed in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde and 2% paraformaldehyde for 2 hours and post-fixed with 1% osmium tetroxide for 1.5 hours at room temperature, then processed in a standard manner and embedded in EMbed 812 (Electron Microscopy Sciences, Hatfield, PA). Semi-thin sections were cut at 1 μm and stained with 1% Toluidine Blue to evaluate the quality of preservation. Ultrathin sections (60 nm) were cut, mounted on copper grids and stained with uranyl acetate and lead citrate by standard methods. Stained grids were examined under a Philips CM-12 electron microscope (FEI; Eindhoven, The Netherlands) and photographed with a Gatan (4k x2.7k) digital camera (Gatan, Inc., Pleasanton, CA).
Statistical Analyses
All qChIP and qRT-PCR data are from three biological replicates from separate dishes and lysate preparations. Quantitative PCR samples exhibiting multiple PCR products were discarded since the corresponding cycle thresholds are not reliable. All experiments were performed independently a minimum of three times with the exception of those depicted in Figures 4e, 7d, 7h, and Supplementary Figure 7, which were performed in duplicate. Investigators were not blinded to group allocations during data analyses. P-values were calculated using a two-tailed Student’s t-test or one-way analysis of variance (ANOVA) for multiple comparisons (Fig. 5a). Differences are significant for p< 0.05, unless otherwise indicated.
To determine the significance of Foxk1 and Sin3A peak overlap (Fig. 2b), we determined the likelihood that randomly placed Foxk1 peaks overlap with Sin3A peaks. First, we determined the mappable genomic regions by pooling all 51-bp single-end sequence reads from several input and ChIP libraries. Then we randomly assigned each Foxk1 peak to a new mappable region within the same chromosome and determined the number of Foxk1 peaks that overlap with Sin3A peaks. This process was repeated 10,000 times to determine the probability that Foxk1 randomly overlaps with Sin3A peaks. The converse computational experiment was also performed. In these experiments, the number of randomly assigned Foxk1 peaks overlapping Sin3A peaks was 150 +/− 12.1 (mean +/− s.d.), and the number of randomly assigned Sin3A peaks overlapping Foxk1 peaks was 162 +/− 12.3. For each of these simulations, the random likelihoods of having an overlap of 2503 peaks are p = 10−8215 (z = 195, one-tailed z-test) and p = 10−7869 (z = 190, one-tailed z-test), respectively. Thus, obtaining an overlap of at least 2503 peaks has a corresponding p-value that is essentially 0.
Supplementary Material
Acknowledgements
We thank W. Lane for mass spectrometric analysis; A. Heguy, E. Venturini, and O. Aminova of the New York University Langone Medical Center (NYULMC) Genome Technology Center for ChIP-seq and RNA-seq library sequencing; F. Liang, C. Petzold, and K. Dancel of the NYULMC OCS Microscopy Core for the preparation of samples and imaging for TEM; D. Garry for Foxk1 cDNA; Y. Zhang, D. Reinberg, E. Benevolenskaya, and A. Brunet for their generous gifts of antibodies. This work utilized computing resources at the High Performance Computing Facility of the NYULMC Center for Health Informatics and Bioinformatics. This work was supported by NIH grants 2R01CA077245-16 and 2R01GM067132-09A1 to B.D.D. and F30AG040894 to C.J.B.
Footnotes
Author Contributions
C.J.B. performed the experiments and bioinformatics analyses. D.E.A. provided Sin3A and Sds3 antibodies. C.J.B and B.D.D. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. All authors reviewed and approved the manuscript.
Competing Financial Interest
The authors declare no competing financial interests.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 3.Füllgrabe J, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietrocola F, et al. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan S, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannenberg J-H, et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonel P, Demmers J, Tan DWM, Watt F, Hendrich BD. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 2012;363:62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Oevelen C, et al. The Mammalian Sin3 Proteins Are Required for Muscle Development and Sarcomere Specification. Mol. Cell. Biol. 2010;30:5686–5697. doi: 10.1128/MCB.00975-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, et al. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell. 2014;53:979–992. doi: 10.1016/j.molcel.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Q, et al. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem. J. 2000;345:335–343. [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Seldin DC, Garry DJ. Foxk1 recruits the Sds3 complex and represses gene expression in myogenic progenitors. Biochem. J. 2012;446:349–357. doi: 10.1042/BJ20120563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X, Garry DJ. Sin3 interacts with Foxk1 and regulates myogenic progenitors. Mol. Cell. Biochem. 2012;366:251–258. doi: 10.1007/s11010-012-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garry DJ, et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meeson AP, et al. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- 17.Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 21.Sacheck JM, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 22.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 23.Young ARJ, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 24.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan EYW, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 29.Kudo N, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pankiv S, et al. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakar K, Karaca S, Port SA, Urlaub H, Kehlenbach RH. Identification of CRM1-dependent Nuclear Export Cargos Using Quantitative Mass Spectrometry. Mol. Cell. Proteomics. 2013;12:664–678. doi: 10.1074/mcp.M112.024877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigbolt KT, et al. Characterization of early autophagy signaling by quantitative phosphoproteomics. Autophagy. 2014;10:1–16. doi: 10.4161/auto.26864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harder LM, Bunkenborg J, Andersen JS. Inducing autophagy: A comparative phosphoproteomic study of the cellular response to ammonia and rapamycin. Autophagy. 2014;10:17–33. doi: 10.4161/auto.26863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan J-S, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi T, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Oevelen C, et al. A role for mammalian Sin3 in permanent gene silencing. Mol. Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen XF, et al. The Rpd3 core complex is a chromatin stabilization module. Curr. Biol. 2012;22:56–63. doi: 10.1016/j.cub.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mammucari C, et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Webb AE, et al. FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hailey DW, et al. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 49.Georges AB, Benayoun BA, Caburet S, Veitia RA. Generic binding sites, generic DNA-binding domains: where does specific promoter recognition come from? FASEB J. 2010;24:346–356. doi: 10.1096/fj.09-142117. [DOI] [PubMed] [Google Scholar]
- 50.Bartholomew CR, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erbay E, Chen J. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 2001;276:36079–36082. doi: 10.1074/jbc.C100406200. [DOI] [PubMed] [Google Scholar]
- 52.Yoon M-S, Chen J. Distinct amino acid-sensing mTOR pathways regulate skeletal myogenesis. Mol. Biol. Cell. 2013;24:3754–3763. doi: 10.1091/mbc.E13-06-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gammoh N, et al. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robert T, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scherz-Shouval R, et al. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 58.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone Deacetylase Activity Is Required for Full Transcriptional Repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, et al. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 60.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 2003;23:3456–3467. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Bigas N, et al. Genome-wide Analysis of the H3K4 Histone Demethylase RBP2 Reveals a Transcriptional Program Controlling Differentiation. Mol. Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Micsinai M, et al. Picking ChIP-seq peak detectors for analyzing chromatin modification experiments. Nucleic Acids Res. 2012;40:e70. doi: 10.1093/nar/gks048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.