Abstract
Protein interactions have evolved into highly precise and regulated networks adding an immense layer of complexity to cellular systems. The most accurate atomistic description of protein binding sites can be obtained directly from structures of protein complexes. The availability of structurally characterized protein interfaces significantly improves our understanding of interactomes, and the progress in structural characterization of protein-protein interactions (PPIs) can be measured by calculating the structural coverage of protein domain families. We analyze the coverage of protein domain families (defined according to CDD and Pfam databases) by structures, structural protein-protein complexes and unique protein binding sites. Structural PPI coverage of currently available protein families is about 30% without any signs of saturation in coverage growth dynamics. Given the current growth rates of domain databases and structural PPI deposition, complete domain coverage with PPIs is not expected in the near future. As a result of this study we identify families without any protein-protein interaction evidence (listed on a supporting website http://www.ncbi.nlm.nih.gov/Structure/ibis/coverage/) and propose them as potential targets for structural studies with a focus on protein interactions.
Keywords: Protein-protein interactions, protein binding sites, coverage of protein families, PPI, structural genomics
1. Introduction
To understand the mechanisms of protein function one needs to explore proteins at the molecular level and at the same time analyze their intricate interactions at the interactome level. Although recent advances in experimental high-throughput (HTP) methods have produced unprecedented amounts of protein-protein interaction data, current ‘interactome’ datasets still suffer from a high rate of false positives and low coverage. As a result of these drawbacks, a comprehensive protein interactome mapping for a given organism is still a daunting task with the large majority of protein–protein interactions left to be determined (Venkatesan et al., 2009). Verification of obtained interactions is essential in order to avoid challenges associated with high-throughput studies and further propagation of interaction annotation errors. Structures from Protein Databank (PDB) (Dutta et al., 2009) and related databases (Davis and Sali, 2005; Juettemann and Gerloff, 2011; Kundrotas and Alexov, 2007; Madej et al., 2012; Xu et al., 2006) may provide the most reliable atomic resolution data for protein molecules and their complexes. Ideally, high-throughput data on protein partnerships should be complemented with the details of binding site locations and physicochemical properties of interaction interfaces derived from structures.
Since the first structure of myoglobin was solved in 1958, a large number of protein structures have been resolved and deposited in the PDB (Dutta et al., 2009). Approximately half of these structures represent protein complexes where protein-protein interfaces can be directly observed and protein-binding sites can be extracted. Comparative structural analyses of different protein complexes reveal a recurrence of certain sequence motifs and binding arrangements/modes on protein-protein interfaces (Janin and Rodier, 1995; Jones et al., 2000; Shoemaker et al., 2006). Although binding arrangements evolve quite rapidly as proteins diverge, certain binding modes are conserved among homologs and in some cases even among non-homologous proteins (Aloy et al., 2003; Dayhoff et al., 2010; Keskin et al., 2004; Korkin et al., 2005); (Zhang et al., 2010). Such conserved binding modes reflect the existence of characteristic features on binding interfaces which, in turn, may modulate binding by stabilizing complexes, by providing specific sites for recognition and/or for post-translational modifications (Bhaskara and Srinivasan, 2011; Hashimoto and Panchenko, 2010; Nishi et al., 2011; Reimand et al., 2012). Moreover, some binding sites are found to be promiscuous and involved in interactions with many different proteins (Nobeli et al., 2009), a key functional feature of hubs in interaction networks. Several methods have been developed that use such recurrent motifs to predict protein-protein interactions and to annotate binding sites (Shoemaker et al., 2010; Shoemaker et al., 2012; Tuncbag et al., 2011; Xu and Dunbrack, 2011).
The progress in structural biology and the trends of PDB growth (Berman et al., 2013) are periodically reviewed (Montelione, 2012) and are typically evaluated by analyzing the structural coverage of protein domain families and superfamilies (Finn et al., 2013; Garcia-Serna et al., 2006; Marchler-Bauer et al., 2013; Mistry et al., 2013). While there seems to be an agreement that most structural folds have been sampled and represented by the PDB structures, it is still unclear how fully the protein-protein binding mode arrangements are characterized.
Here we consider protein domains as units of protein-protein interactions and use strict criteria to define protein-protein interfaces and binding sites. Based on the binding-site comparisons stored in the IBIS database (Shoemaker et al., 2010; Shoemaker et al., 2012) we identify unique binding sites in protein domains involved in protein-protein interactions. We comprehensively assess the diversity of protein interactions and binding sites in the context of their growth in protein domain families over the last 55 years. Our analysis of unique binding site coverage within protein families from Conserved Domain Database (CDD) and Pfam was able to identify families with no structural evidence of protein-protein interactions. We provide a list of these families which could and should be targeted by structural efforts.
2. Materials and Methods
2.1 Dataset
We downloaded biological assemblies (so called biounits) and structure deposition dates from the MMDB database (Madej et al., 2012). MMDB bioassembly data and deposition dates were in turn taken from the PDB database (Berman et al., 2000). The difference in the number of chains between the asymmetric units (ASU) and biounits is shown in Supplementary Information Figure S1. The deposition dates were used to reconstruct the growth of structural data. We did not consider structures that were obsolete or have been revoked from the PDB. We downloaded Conserved Domains Database (CDD) (Marchler-Bauer et al., 2013) version 3.11 with 9,860 domain models curated at NCBI. We did not consider any CDD models imported from other sources. In this paper the CDD models are called “families”. A CDD superfamily represents a set of similar domain models; we only considered those superfamilies which contained at least one manually curated model. We also downloaded the Pfam-A curated subset of the Pfam database version 27, which contains 14,831 families (Finn et al., 2013). It should be mentioned that not all Pfam families correspond to protein domains; some of them correspond to short repeats and sequence motifs. The CDD domains were mapped on structures using the IBIS database (Shoemaker et al., 2010; Shoemaker et al., 2012). The structural mapping of Pfam families was downloaded from the Pfam FTP server (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/mappings/pdb_pfam_mapping.txt).
2.2 Identifying unique binding sites
We analyzed protein-protein interactions between domains according to the criteria implemented in IBIS database. Namely, CDD domains were mapped onto sequences of protein chains to create "footprint" regions. Protein-protein interactions were defined between two domains (footprint regions) from two different chains if there were at least five contacting residues in each domain within the distance of at most 4 Å between heavy atoms. We did not consider polypeptide chains with less than 20 amino acids as interaction partners. A binding site of a domain was defined as a set of interacting residues on one side of the interaction interface.
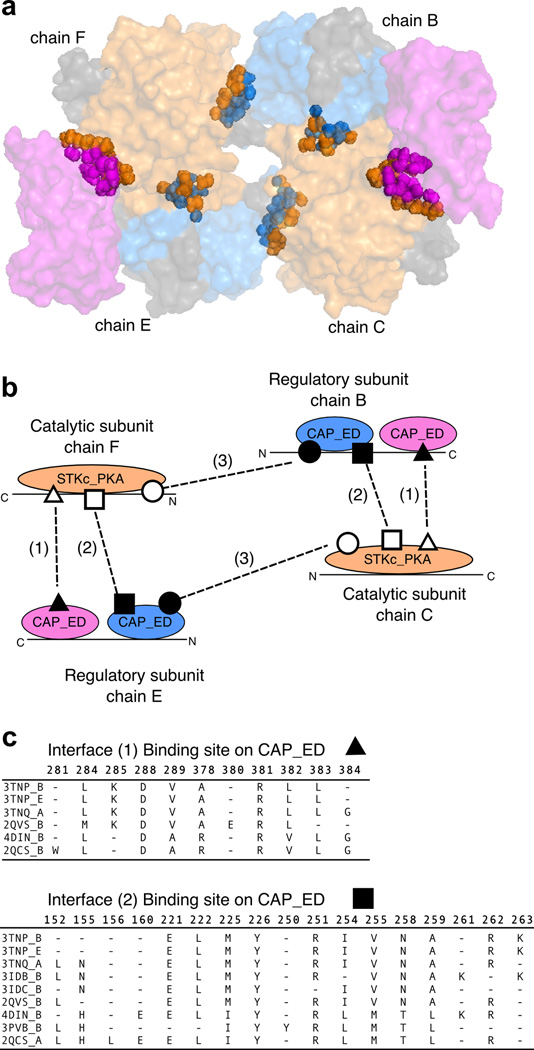
Then we collected domain footprint regions which were mapped to the same CDD domain superfamily, structurally superimposed them and ensured that the sufficient fraction of domain footprints was structurally aligned. Subsequently we clustered their corresponding binding sites based on sequence and structural similarity between the sites and sequence conservation profile of binding site residues (Shoemaker et al., 2010). Binding sites were clustered by a hierarchical complete linkage clustering procedure. To choose the sliding cutoff to define clusters, we used a function which maximizes the mean similarity of members within a cluster and minimizes the complexity of the description provided by cluster membership (number of bits required to describe the data) (Slonim et al., 2005). Binding sites and conserved binding site clusters can be explored online in IBIS database (http://www.ncbi.nlm.nih.gov/Structure/ibis/ibis.cgi). As a result of such clustering, it becomes possible to assess the uniqueness of each binding site. Binding sites from different clusters are called hereafter unique. We consider a binding site as being novel, if no similar binding sites (from the same binding site cluster) were available in PDB prior to its deposition date. For clarity, we illustrate protein-protein interfaces, binding sites and binding site clusters in Figure 1.
Figure 1. Definition of a binding site.
(a) Example of protein interfaces and binding sites in Camp-dependent protein kinase type I (PDB 3tnp). The structure is a heterotetramer where chains C and F represent catalytic subunit while chains B and E represent regulatory subunits. The former subunit has one domain STKc_PKA (orange), the latter has two domains belonging to the same CAP_ED domain family (blue and magenta with the linkers between domains shown in gray). (b) Schematic representation of binding sites (encoded by shapes), each side of the interface is shown in different shade patterns. There are six interfaces; only three of them are unique (dashed lines labeled with numbers in brackets). Out of total 12 binding sites, six sites are unique within the complex (distinguished by shade). (c) Examples of two binding sites clusters show that there exist binding sites in other complexes similar to the sites in our example (two sites in CAP_ED domains in chain C). The clusters are shown as alignments of binding residues (the residue numbers in PDB 3tnp are shown as column names). The first row in the alignment corresponds to the 3tnp structure.
2.3 Analysis of domain database and structure growth
An average yearly growth rate of CDD database (1200 families per year) was estimated by counting the number of new families and superfamilies that were added during the last five years. However, we did not count families that were eventually removed from the database, and only considered NCBI-curated families and superfamilies containing at least one CDD-curated family. We estimated the growth of Pfam database (900 new families per year) starting from Pfam release 23 in 2008 to release 27 in 2013. More details on database growth and coverage growth rates are provided in Supplementary Information Table S1.
The structural coverage was analyzed for families in the most recent release of CDD 3.11 and Pfam 27 using the PDB deposition dates as a reference point allowing us to look back in time. Each family was assigned two dates: (i) year when the first structure matching the family was deposited and (ii) year when the first structure with PPI complex was deposited. Then we aggregated the number of covered families by year. The analysis of CDD superfamilies was done following the same logic, considering a superfamily to be structurally covered when at least one of the families had a representative structure. We assigned CDD family annotations to structures using the most specific best matching family models. Due to the hierarchical nature of family classifications in CDD, the models representing the intermediate nodes in the hierarchy might appear as lacking structural coverage and therefore the number of CDD families without any structural representatives may be slightly overestimated.
3. Results
3.1 Unique binding sites and interfaces
We define protein-protein interfaces based on the contacts between domains located on different chains in macromolecular assemblies. While the interface is a characteristic of a pair of interacting proteins/domains, a protein-binding site describes each interaction partner. Therefore, we express the diversity of protein interactions via the diversity of binding sites. Figure 1 illustrates binding sites and their conservation among homologous complexes. Camp-dependent protein kinase type I (PDB 3tnp) represents a heterotetramer and consists of two pairs of identical subunits: catalytic (chains C and F, shown in orange) and regulatory (chains B and E shown in blue and magenta). Each regulatory subunit consists of two domains from the same CAP_ED family (blue and magenta). We illustrate protein-protein interfaces by showing them in spheres, whereas the rest of the protein is shown as semitransparent surface (Figure 1a). Each interface consists of two binding sites and therefore is depicted in two colors.
Although there are six interfaces in the complex, only three of them are distinct or unique (shown as dashed lines with numbers 1–3 on Figure 1b). Consecutively, on each chain there are six unique binding sites (shown as different shapes with different shades). There can be binding sites in other protein complexes that are similar to the ones observed in the example structure. Figure 1c shows two clusters of protein binding sites from the regulatory subunit for interfaces number 1 and 2. Domains from other protein complexes are structurally superimprosed on domains from regulatory subunits (those that cannot be structurally superimposed are disregarded) and the gaps (dashes) in the sequence alignment of binding site regions indicate those residues that are not structurally aligned. Considering binding sites per domain is very important because it provides spatially localized binding sites on each domain (separate binding site patches corresponding to different domains in Figure 1a,b) and distinct protein interfaces. If we consider a whole protein chain as a unit of interaction, there would be only one interface between STKc_PKA in chain C and two CAP_ED domains in chain B, which does not fully represent the nature of the interaction between these two chains and makes it rather hard to compare binding sites between different protein families where domain recombination is a common evolutionary event.
3.2 Growth dynamics of coverage of CDD superfamilies by structural complexes
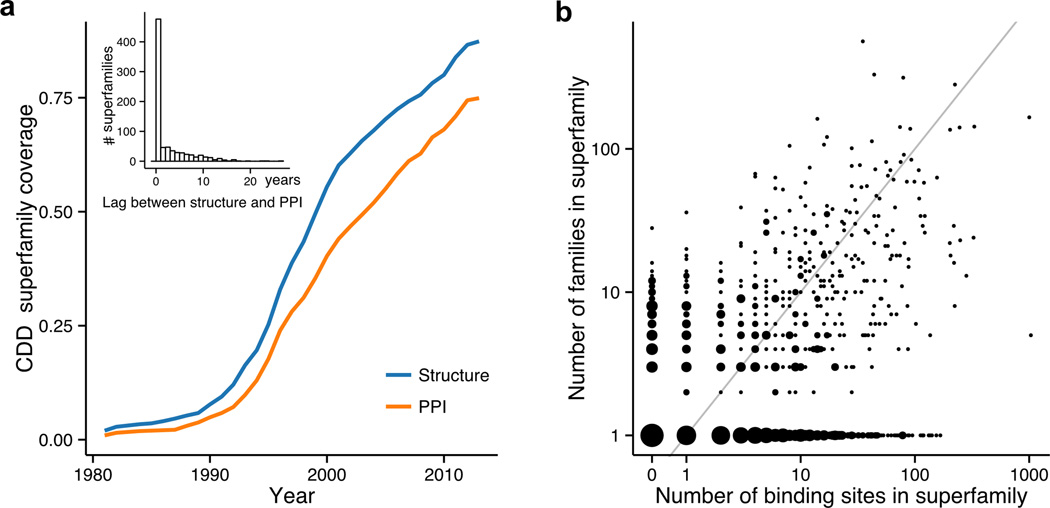
Here we analyze the coverage of CDD superfamilies by structural complexes and unique binding sites since this characteristic can be used as an indicator of success of structural genomics efforts. Domain coverage is defined as a fraction of all domain families (or superfamilies) with at least one structure or structural evidence of PPI with a well-defined binding site (see binding site definitions in Methods). As shown in Figure 2a, starting from the early 1990s the structural coverage has dramatically increased with two thirds of all superfamilies exemplified by at least one structure by the year 2000. The rate of improvement in structural coverage remains impressive due to the guiding efforts of structural genomics initiatives (Burley et al., 2008), reaching 90% of coverage for currently available superfamilies. Figure 2a also shows the growth of CDD superfamily coverage with protein-protein interactions. As shown in the inset, in some cases the structures are first solved as monomers and only later are deposited as PPIs, thereby creating some significant delays between monomer and assembly deposition dates (time lag in years is shown in Figure 2a inset). It may seem that the current superfamily coverage with PPIs of more than 75% should provide a comprehensive description of the diversity of binding sites. However, the protein binding sites diverge rather fast in evolution and some of them might be characteristic for only specific protein families. Certain superfamilies have very heterogeneous binding sites while for others binding sites and binding partners are much more conserved. In general the diversity of PPI binding sites in a superfamily should depend on the role of protein-protein interactions in its function. Therefore in the next section we explore the association between the diversity of superfamilies and the number of binding sites.
Figure 2. Structural coverage and binding sites of CDD superfamilies.
(a) Cumulative CDD superfamily coverage. Inset: lag in years between deposition of the first structure representing the superfamily and the structure with at least one observed proteinprotein interaction. (b) Number of families in superfamily versus the number of binding sites. The superfamilies are shown as circles. The size of the circle is proportional to the number of superfamilies (the largest circle contains 216 superfamilies, the smallest – one). The gray diagonal shows one-to-one correspondence between the number of sites and the number of families.
3.3 Relationship between the diversity of CDD superfamilies and the number of unique binding sites
Next we compared the number of CDD families in a given superfamily with the number of unique binding sites within the superfamily. Figure 2b shows that there are many superfamilies (shown below the main diagonal), where the number of binding sites is up to an order of magnitude higher than the number of families within a given superfamily. Moreover, there is a significant number of superfamilies with only one family but a large number of binding sites. All of these examples constitute families with multiple binding sites. It is consistent with the previous observation that paralogous proteins from the same family have a tendency to bind different partners using different binding sites (Dessailly et al., 2013; Hamp and Rost, 2012; Lewis et al., 2012; Reid et al., 2010). This binding specialization could prevent the undesirable cross-talk between similar pathways involving paralogs with different specificities. Such families might benefit from their classification into several functional domain subfamilies. Superfamilies containing at least several different domain families but only one binding site presumably possess a very conserved interface. However, it could also be the result of the limited structural coverage of protein interfaces in different families. About 25% of the superfamilies have no structural evidence of protein-protein interactions and could be potential targets for structural genomics. Based on two parameters described so far, we have arranged all superfamilies into 12 groups (Table 1). The lists of superfamilies from each group are available on the supporting website, where we provide links to corresponding examples from the CDD and IBIS databases.
Table 1.
The number of CDD superfamilies grouped by the number of families (rows) and protein-protein interaction binding sites (columns).
| Number of families per superfamily |
Total number of superfamilies |
Number of binding sites per superfamily |
|||
|---|---|---|---|---|---|
| No known sites |
1–10 sites |
11–100 sites |
101+ sites |
||
| 1–10 | 874 | 262 | 421 | 178 | 13 |
| 11–100 | 170 | 9 | 53 | 93 | 15 |
| 101+ | 16 | 0 | 1 | 10 | 5 |
| Total | 1060 | 271 | 457 | 281 | 33 |
3.4 Growth dynamics of coverage of CDD and Pfam families with structural complexes
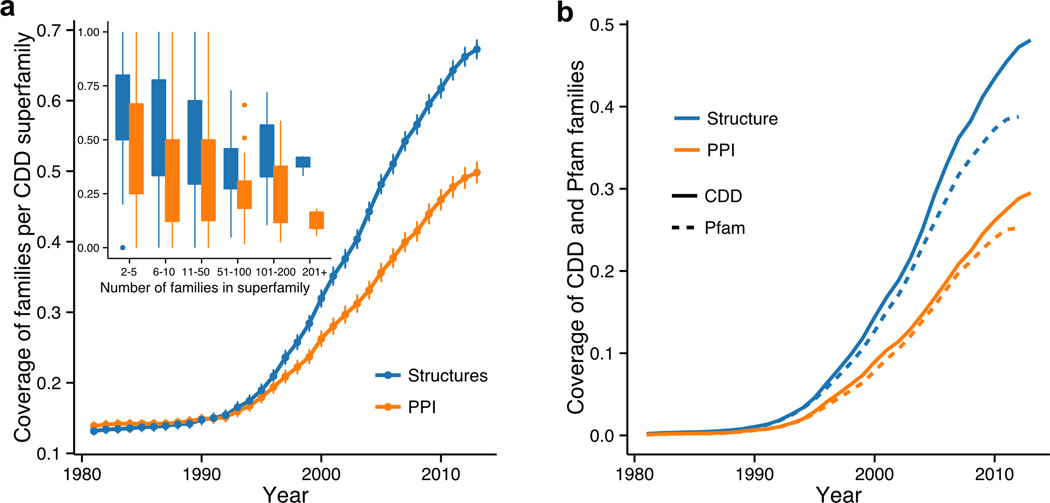
Since the number of families within one superfamily varies greatly, we calculated the structural coverage on the domain family level (Figure 3a). While about 75% of the superfamilies are covered by structures of complexes (Figure 2), about 45% of all families within one superfamily are covered by at least one structural complex. As evident from an inset of Figure 3a superfamilies with large number of families are not well covered neither by structures nor by structural complexes. This is especially pronounced for PPI data, where the average coverage barely reaches 20% in large superfamilies.
Figure 3. Structural coverage of CDD and Pfam families.
(a) Cumulative average coverage of CDD families within the CDD superfamilies with structural data and with structural evidence of protein interactions; the error bars show standard errors. An inset shows the coverage (for the year of 2013) for different groups of superfamilies depending on the number of families in them. Superfamilies with more families tend to have worse coverage, even though almost half of the families in large superfamilies have structures, PPI data is available for only 15–20% of them. (b) Growth of coverage of CDD (solid line) and Pfam families (dashed line) in terms of structures (blue) and protein interactions (orange).
Considering the large coverage variance within superfamilies for the CDD database, we decided to compare CDD coverage per domain level with another widely used high-quality domain annotation set from the Pfam database. In terms of absolute number, the coverage trends for CDD and Pfam agree surprisingly well with each other. Namely, the average rates of structural coverage for CDD and Pfam families are 218 CDD and 210 Pfam families per year respectively. Despite different philosophies and construction methods employed in these databases, similar trend persists for the coverage by structural protein complexes (157 CDD and 161 Pfam families are covered by structural protein complexes per year). This similarity is suggestive of the robustness of our rate estimates. However, if we consider the fractional structural coverage (a fraction of all CDD and Pfam families with structural evidence), there are certain differences between CDD and Pfam (Figure 3b) which can be explained by different sizes of these databases (Table 2). As one can see from comparing Figures 3a and 3b, the growth dynamics of CDD domain structural coverage is slower for domains compared to superfamilies, reaching 50% of structural coverage and 30% of PPI coverage by families.
Table 2.
Coverage of CDD superfamilies and CDD/Pfam families with structures and structural protein-protein interactions
| Total | With structure and PPI |
With structure, no PPI |
Without structure |
|
|---|---|---|---|---|
| CDD superfamilies | 1060 | 794 | 133 | 133 |
| CDD families | 9860 | 2912 | 1838 | 5110 |
| Pfam families | 14831 | 3740 | 2009 | 9082 |
We identified families and superfamilies that are largely underrepresented by solved structural complexes and unique binding interfaces. Such proteins could serve as potential targets for future structural discoveries. We provide the complete lists of families on the supplementary website (http://www.ncbi.nlm.nih.gov/Structure/ibis/coverage/). The families without any structures are probably first on the priority list, followed by the families with structures and without the evidence of protein interactions. Families in large superfamilies are of particular interest, because they are likely to have diverse binding sites.
4. Discussion
The ultimate objective of improving the structural coverage of protein-protein interfaces and binding sites is an understanding of molecular mechanisms of protein function and protein recognition. Characterization and classification of structures of protein complexes and protein-protein binding sites is at least as important as the classification of folds. Now, when most of the folds are already exemplified by experimentally determined structures and more than 70% of sequences can be structurally modeled at least in part (Levitt, 2009), the focus in structural biology gradually shifts towards the characterization of protein assemblies, especially assemblies with novel protein interfaces. Novel complexes and interfaces are especially important for drug design of protein interaction inhibitors and for the rational protein design to create protein complexes with novel specificities (Huang et al., 2007; Khare and Fleishman, 2013).
Interactomes represent the networks characterizing all potential protein-protein interactions of a given species. The unit of interaction can correspond to a protein or a domain and the availability of high-quality structural information on protein-protein and domaindomain interactions is critical for building reliable interactomes. Here we analyze how the structural and PPI coverage of domain families change over time. We use currently available sets of domain families and trace back in time their structural coverage employing the structure deposition dates.
Given that roughly 30% of currently available domain families have at least one structural PPI representative, we investigated whether the current growth (well approximated by a linear function over the last ten years) can be extrapolated to find the point in time with near-complete structural coverage of all domain families. Certainly, we do not expect that all domain families will be covered by structures and/or structural complexes. It is a rather unrealistic assumption given that there are families which do not function while interacting with other protein partners and there are families comprising intrinsically disordered proteins which are not present in PDB. If we take into account the growth rate of domain family databases (about 1200 new CDD and 900 new Pfam domain families added per year on average), we can estimate that about 3900 CDD families will have at least one structural PPIs by the year 2020 (2912 CDD families are currently covered by PPIs). If the growth rate persists, the CDD and Pfam databases may grow to approximately 17000 and 21000 families by 2020, respectively. In addition, many structures of proteins with unknown functions are currently being solved, which may provide an evidence for new CDD or Pfam families (Marchler-Bauer et al., 2013; Mistry et al., 2013). Therefore, given a current 30% structural PPI coverage and a 23% projected PPI coverage by the year 2020, we do not foresee in the near future any considerable increase in the percentage of families with structural PPIs. This conclusion only holds true if the current domain and structure deposition rates do not change. However, we might expect that the domain family databases will soon reach a saturation point and will not grow anymore, while, with the help of structural initiatives, the sampling of different PPIs will continue to increase. Right now, as protein domain family databases continue to grow, complete structural PPI coverage of domain families remains a moving target.
5. Conclusions
The availability of structurally characterized protein interfaces is critical and significantly improves the reliability of interactomes. We analyze the coverage of protein domain families and superfamilies with structures and structural PPIs and assess their growth dynamics. We show that protein interaction coverage of domain families is lagging behind structural coverage. While the overall number of possible protein interactions is unknown, the PPI coverage of protein domain families with structural data serves as a good measure of progress. Currently, only about 30% of protein families have structural PPI coverage, and this number is not likely to improve in the nearest future due to high growth rate in the number of protein domain families. Finally, we identify families and superfamilies without protein-protein interaction evidence and families without any structural data (listed on a supporting website http://www.ncbi.nlm.nih.gov/Structure/ibis/coverage/). These families could be used as potential targets for structural initiatives with the focus on protein interactions.
Supplementary Material
Acknowledgements
We thank Thomas Madej and Christopher Lanczycki for help with the MMDB database. This work was supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- Berman HM, Coimbatore Narayanan B, Di Costanzo L, Dutta S, Ghosh S, Hudson BP, Lawson CL, Peisach E, Prlic A, Rose PW, Shao C, Yang H, Young J, Zardecki C. Trendspotting in the Protein Data Bank. FEBS Lett. 2013;587:1036–1045. doi: 10.1016/j.febslet.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara RM, Srinivasan N. Stability of domain structures in multi-domain proteins. Sci Rep. 2011;1:40. doi: 10.1038/srep00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Joachimiak A, Montelione GT, Wilson IA. Contributions to the NIH-NIGMS Protein Structure Initiative from the PSI Production Centers. Structure. 2008;16:5–11. doi: 10.1016/j.str.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- Dayhoff JE, Shoemaker BA, Bryant SH, Panchenko AR. Evolution of protein binding modes in homooligomers. J Mol Biol. 2010;395:860–870. doi: 10.1016/j.jmb.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessailly BH, Dawson NL, Mizuguchi K, Orengo CA. Functional site plasticity in domain superfamilies. Biochim Biophys Acta. 2013;1834:874–889. doi: 10.1016/j.bbapap.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Serna R, Opatowski L, Mestres J. FCP: functional coverage of the proteome by structures. Bioinformatics. 2006;22:1792–1793. doi: 10.1093/bioinformatics/btl188. [DOI] [PubMed] [Google Scholar]
- Hamp T, Rost B. Alternative protein-protein interfaces are frequent exceptions. PLoS Comput Biol. 2012;8:e1002623. doi: 10.1371/journal.pcbi.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Panchenko AR. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc Natl Acad Sci U S A. 2010;107:20352–20357. doi: 10.1073/pnas.1012999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PS, Love JJ, Mayo SL. A de novo designed protein protein interface. Protein Sci. 2007;16:2770–2774. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J, Rodier F. Protein-protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Jones S, Marin A, Thornton JM. Protein domain interfaces: characterization and comparison with oligomeric protein interfaces. Protein Eng. 2000;13:77–82. doi: 10.1093/protein/13.2.77. [DOI] [PubMed] [Google Scholar]
- Juettemann T, Gerloff DL. BISC: binary subcomplexes in proteins database. Nucleic Acids Res. 2011;39:D705–D711. doi: 10.1093/nar/gkq859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, Tsai CJ, Wolfson H, Nussinov R. A new, structurally nonredundant, diverse data set of protein-protein interfaces and its implications. Protein Sci. 2004;13:1043–1055. doi: 10.1110/ps.03484604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare SD, Fleishman SJ. Emerging themes in the computational design of novel enzymes and protein-protein interfaces. FEBS Lett. 2013;587:1147–1154. doi: 10.1016/j.febslet.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Korkin D, Davis FP, Sali A. Localization of protein-binding sites within families of proteins. Protein Sci. 2005;14:2350–2360. doi: 10.1110/ps.051571905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrotas PJ, Alexov E. PROTCOM: searchable database of protein complexes enhanced with domain-domain structures. Nucleic Acids Res. 2007;35:D575–D579. doi: 10.1093/nar/gkl768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AC, Jones NS, Porter MA, Deane CM. What evidence is there for the homology of protein-protein interactions? . PLoS Comput Biol. 2012;8:e1002645. doi: 10.1371/journal.pcbi.1002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madej T, Addess KJ, Fong JH, Geer LY, Geer RC, Lanczycki CJ, Liu C, Lu S, Marchler-Bauer A, Panchenko AR, Chen J, Thiessen PA, Wang Y, Zhang D, Bryant SH. MMDB: 3D structures and macromolecular interactions. Nucleic Acids Res. 2012;40:D461–D464. doi: 10.1093/nar/gkr1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Kloppmann E, Rost B, Punta M. An estimated 5% of new protein structures solved today represent a new Pfam family. Acta Crystallogr D Biol Crystallogr. 2013;69:2186–2193. doi: 10.1107/S0907444913027157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT. The Protein Structure Initiative: achievements and visions for the future. F1000 Biol Rep. 2012;4:7. doi: 10.3410/B4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- Reid AJ, Ranea JA, Orengo CA. Comparative evolutionary analysis of protein complexes in E. coli and yeast. BMC Genomics. 2010;11:79. doi: 10.1186/1471-2164-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Hui S, Jain S, Law B, Bader GD. Domain-mediated protein interaction prediction: From genome to network. FEBS Lett. 2012;586:2751–2763. doi: 10.1016/j.febslet.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci. 2006;15:352–361. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred Biomolecular Interaction Server--a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker BA, Zhang D, Tyagi M, Thangudu RR, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. IBIS (Inferred Biomolecular Interaction Server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic Acids Res. 2012;40:D834–D840. doi: 10.1093/nar/gkr997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonim N, Atwal GS, Tkacik G, Bialek W. Information-based clustering. Proc Natl Acad Sci U S A. 2005;102:18297–18302. doi: 10.1073/pnas.0507432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncbag N, Gursoy A, Nussinov R, Keskin O. Predicting protein-protein interactions on a proteome scale by matching evolutionary and structural similarities at interfaces using PRISM. Nat Protoc. 2011;6:1341–1354. doi: 10.1038/nprot.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, Yildirim MA, Simonis N, Heinzmann K, Gebreab F, Sahalie JM, Cevik S, Simon C, de Smet AS, Dann E, Smolyar A, Vinayagam A, Yu H, Szeto D, Borick H, Dricot A, Klitgord N, Murray RR, Lin C, Lalowski M, Timm J, Rau K, Boone C, Braun P, Cusick ME, Roth FP, Hill DE, Tavernier J, Wanker EE, Barabasi AL, Vidal M. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Canutescu A, Obradovic Z, Dunbrack RL., Jr ProtBuD: a database of biological unit structures of protein families and superfamilies. Bioinformatics. 2006;22:2876–2882. doi: 10.1093/bioinformatics/btl490. [DOI] [PubMed] [Google Scholar]
- Xu Q, Dunbrack RL., Jr The protein common interface database (ProtCID)--a comprehensive database of interactions of homologous proteins in multiple crystal forms. Nucleic Acids Res. 2011;39:D761–D770. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci U S A. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.