Abstract
Purpose
To perform Preimplantation Genetic Diagnosis (PGD) on a paternal Brca2 unknown mutation carrier with early-onset breast cancer, whose paternal grandmother and mother had breast cancer at 60s.
Method
Elucidating the linkage via single sperm haplotyping on patient's carrier brother, and identifying the genomic deletion via BLAST followed by PCR screening. PGD was subsequently conducted.
Result
The mutant allele was found by using 4 microsatellite and 2 intragenic SNP markers. Recombination was detected in 8 % of sperms. BLAST was utilized to locate putative hairpin structure(s), followed by PCR screening with seven sets of primers. A novel 2,596 bp deletion containing exon 15 ~ 16 was identified. Due to the severity of phenotype and the integrity of exon 11 encoding RAD51 binding domain, and the fact that the patient's mother also had breast cancer at her 60s, we speculate a possible coexistence of maternal breast cancer risk allele(s). Embryo biopsy was performed on day 3. Unaffected morula and blastocyst were replaced on day 5, resulting in a singleton livebirth. A breast lump appeared in the patient after delivery without the presence of malignant cells.
Conclusion
Concerning the assisted reproductive option for breast cancer patients, the possibility of coexistence of multiple familial risk alleles and the significance of each mutation to the phenotype should be evaluated. To eliminate misdiagnosis resulting from recombination and/or allelic drop-out, both direct mutation detection and linkage analysis approaches may be necessary. BLAST is a very useful and cost-effective tool for identifying large genomic deletion.
Keywords: Breast Cancer, BRCA2 Novel Deletion, PGD, Sperm Haplotyping, BLAST analysis
Introduction
Breast cancer is the most common malignancy and the second leading cause of cancer death among women [1]. Tumor suppressor genes BRCA1 and BRCA2 are among the major susceptibility genes, and their germline mutations confer high risks of breast and ovarian cancer [2, 3]. The lifetime risk of breast cancer and ovarian cancer for BRCA2 mutation carriers by the age of 70 years were 40 ~ 84 %, and 11 ~ 27 %, respectively [4]. The great variation observed in the penetrance of pathogenic mutations of the disease may result from the diverse positions of the mutations, the presence of genetic modifiers, and variations in non-genetic factors, such as environmental factors, reproductive and hormonal factors.
BRCA2 plays a central role in homologous recombination repair eliminating DNA breaks and deleterious lesions by controlling the recombinase RAD51 [5]. The BRCA2 gene has been mapped to chromosome 13q12.3. It contains 27 exons but the first exon is not translated. The BRCA2 polypeptide consists of 3,418 amino acids, with several important domains: a PALB2 binding domain, a RAD51 binding domain harboring eight BRC motifs [6], a highly conserved PhePP motif and a conserved C-terminal region covering a DSS1-DNA binding domain (DBD) and another RAD51-interaction motif. According to the Human Gene Mutation Database, about 1,000 variants have been reported. Great majority (>70 %) of the variants are missense/nonsense, while gross deletions/insertions/duplications only account for 2 ~ 3 %. A relatively small portion of the variants have been characterized or predicted to be potentially deleterious. Most of the variants are of unknown significance.
In addition to facing a life time threat of cancer, BRCA mutation carriers also need to cope with a 50 % chance of transmitting the mutation to their children. Several surveys reveal that passing hereditary cancer predisposition alleles to offspring is one of the major concerns for cancer predisposition mutation carriers, especially for those with strong family histories [7–9], and the concern might prevent the carriers from pursuing parenthood. With advanced technologies, those who do not want to pass the mutation to the next generation have several options, including prenatal diagnosis, preimplantation genetic diagnosis (PGD) or gamete donation.
PGD for BRCA mutations have been reported [10–14]. We report here our PGD approach on a breast cancer patient with unknown BRCA2 mutation, using both linkage analysis and direct mutation detection methods. The genomic breakpoint of the patient has been identified in our clinic, and to the best of our knowledge, this is a novel large genomic deletion.
Materials and methods
Patient
The patient was diagnosed to have cancer in the right breast (pT1cN (0/18) M0, moderately positive to estrogen and progesterone receptors, grade III) in year 2000 (at age 24). Subsequently, modified right mastectomy with axillary dissection and lateral dorsi flag reconstruction, adjuvant chemotherapy and radiotherapy to chest wall were done, followed by tamoxifen treatment for five years. Since both mother and paternal grandmother had CA breast at the age of 60s, genetic screening for Brca1/2 mutations was carried out in patient's family in 2009.
There were no significant findings in her mother. The patient, father and her brother shared a mutant allele with a deletion of exon 15 ~ 16 (c.7436_7805del) in Brca2, detected by Multiplex ligation-dependent probe amplification (MLPA) assay and cDNA sequencing (the exact genomic breakpoints were not identified). Her younger sister was negative for the mutation. Her paternal grandma's genomic DNA was unavailable.
As a result, instead of initially planned intensive surveillance, prophylactic left mastectomy was performed combined with sentinel LN biopsy, which was found negative for malignancy later. Meanwhile, ovarian cancer screening was performed. Serial pelvic USG showed a 4 cm anechoic unilocular right ovarian cyst, without solid area or interval change in size. Laparoscopic right ovarian cystectomy was performed in 2010 due to the persistent cyst on ultrasound scanning and raised CA125 (45.5 U/mL). Histology revealed an endometriotic cyst without malignancy.
The patient planned to get pregnant after two years of marriage, but worried about passing on the paternal Brca2 mutation to her offspring. The couple attended our subfertility clinic for counseling. The Mendelian inheritance pattern had been explained and options of natural conception and IVF-PGD discussed. The couple keened for IVF combined with PGD because of unwilling to transmit the paternally derived Brca2 mutant allele to next generation.
Pre-PGD workup
Since the actual genomic breakpoint was not known and the parental DNAs were not available when the patient presented to us, we first tried to establish the linkage with the genomic DNAs of her carrier brother and non-carrier sister. The Qiagen DNA Blood Mini Kit (Qiagen, Upsala, Sweden) was used for genomic DNA extraction. However, the linkage analysis results turned out to be inconclusive because the patient shared no common allele around the BRCA 2 gene with her non-carrier sister, and had exactly the same alleles as her carrier brother. Therefore, the very tedious but effective approach of single sperm (from the carrier bother) haplotyping became our choice for mutant allele identification.
Single-cell PCR
Whole genome amplification (WGA) on single cell was performed according to the published protocol [15]. Four microsatellite markers within 2 Mb flanking the BRCA2 gene (D13S289, D13S1698, D13S1701 and D13S171) and 2 intragenic SNP markers in exon 11 (rs1801406) and exon14 (rs1799955) were used for PGD (Fig. 1). PCR for microsatellite markers and amplification of exon 11 and exon 14 were performed in a 25 μl reaction mixture containing 1 X PCR reaction buffer, 200 μM dNTPs, 0.2 μM forward and reverse primers (except D13S1701, exon 11 and exon 14, where 0.4 μM primers were used), 1 U of Faststart Taq DNA polymerase (Roche) and 1 μl of WGA DNA. PCR products of exon 11 and exon 14 were pooled and purified with the Qiaquick PCR purification system (Qiagen). SNP markers in exon 11 and 14 were determined by minisequencing with SNaPshot Multiplex kit (Applied Biosystems, Foster City, CA); the reaction mixture contained 2.5 μl of SNaPshot multiplex reaction mix, 0.1 μM of minisequencing primers, 0.75 μl of purified pooled PCR products and MilliQ water in a final volume of 5 μl. Minisequencing products were treated with shrimp alkaline phosphatase (USB) before GeneScan analysis. Single cell PCR protocol was validated with 50 lymphocytes (25 from the patient, and 25 from the spouse).
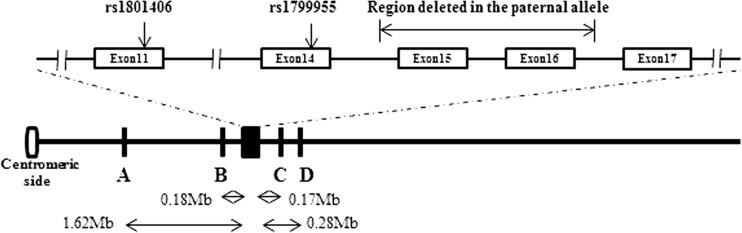
Fig. 1.
Schematic diagram showing the relative positions of the microsatellite markers and the SNP markers. Black box: Brca2 gene; A: D13S289; B: D13S1698; C: D13S1701; D: D13S171. The actual distance (in Mb) of each marker from the Brca2 gene was indicated. The primer sequences for the microsatellite markers were retrieved from NCBI UniSTS. The PCR primers for exon 11 were F: GGAGGTAGCTTCAGAACAGCTTCA; R: AGCATCTCTGCATTCCTCAGAAGTGG and exon 14 F: CCGCACCTGGTCAAGAATTTCTGTCT; R: CAGCTGCTGCTTGATTGGAGTTGT. The minisequencing primers for rs1801406 and rs1799955 were CTCTTCTGCAATATGTAGCTTGG and GATCGGATCGATCGGATCCTGTTCAACTCTGTGAAAATG, respectively
Post-PCR analysis was performed by resolving 0.5 μl of the PCR products in an ABI 3500 DNA Analyzer with the use of GeneScan 500 ROX size standard (Applied Biosystems) for microsatellite markers and with GeneScan 120 LIZ size standard for minisequencing products. Data were analyzed by GeneMapper (v.4.1, Applied Biosystems).
Targeting putative genomic breakpoints
Randomly searching for mutation sites within introns of BRCA2 could be labor-intensive, because of the relatively large size of the introns with lots of common repetitive sequences. As hairpin structure is commonly associated with genome rearrangements, we used BLAST (Basic Local Alignment Search Tool, NIH) to analyze intron 14 and 16, and found that a 135 bp fragment of intron 14 (IVS14-947 ~ IVS14-1081) were nearly 100 % complementary to a sequence of intron 16 (IVS16-581 ~ IVS16-715) (Fig. 4), suggesting a possibility of forming a hairpin structure susceptible to deletion. Therefore, a panel of PCR primers within the flanking regions was designed for mutation screening.
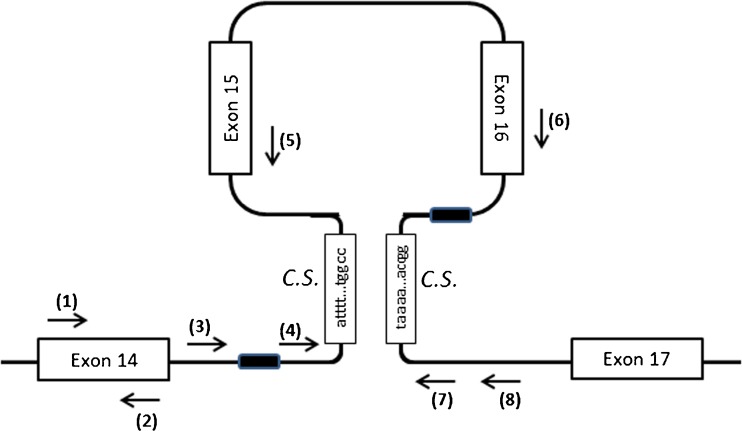
Fig. 4.
Complementary sequences (C.S., 135 bp in length) in intron 14 & 16 and the locations of the primers designed to target the mutation sites. The locations of the primers are indicated with arrows: (1) Brca2e14-42 F (5′-CCGCACCTGGTCAAGAATTTC); (2) Brca2e14-242R (5′-CTAACACACTGTTCAACTCTGTG); (3) Brca2i14-216 F (5′-CAATCTAGGACTGCTGTTACTGGA); (4) Brca2i14-699 F (5′-AGGAGAGCATGTAAACTTCGAG); (5) Brca2e15-10R (5′-CTCTGGCATTCTGAAGACTTG); (6) Brca2e16-143 F (5′-TCATACCCTCCAATGATGGAAAG); (7) Brca2i16-864R (5′-TACGGGCATGCATCACCATAC); (8) Brca2i16-1161R (5′-TAAGTGGGATTGCAGGCGCGTG). Primer pairs of (1)&(2), (3)&(7), (3)&(8), (4)&(7), (4)&(8), (4)&(5), (6)&(7) had been used for PCR reaction. The primer pair of (3)&(7) gave rise to an abnormal PCR product in patient's samples. Sequencing the abnormal PCR product indicated that the elements of IVS14-491 ~ IVS14-512 and IVS16-441 ~ IVS16-462 (Black boxes) were identical (i.e., ggaggctgaggcaggagaatcg), and the mutant allele lost 2,596 nucleotides in total, including one of the elements and the sequence in between
PCR for mutation detection
Genomic DNA was extracted from peripheral blood samples (patient, sister, and brother) using the Qiagen mini DNA extraction kit. After dilution, 1 μl (10 ng/μl) of the extracted DNA was used for PCR reaction. Several sets of primers surrounding the mentioned complementary sequences were designed for the screening purpose (Fig. 4). Without knowing the exact breakpoints, the size of PCR product for each pair of primers was not predictable. However, the different PCR products derived from the patient or the brother (with deletion), comparing to those from her sister (normal), would be informative, and the corresponding PCR products would further be analyzed for the deletion. A PCR reaction of 25 μl was conducted in the following condition: 1 X PCR reaction buffer, 2 μM dNTPs, 0.2 μM each of the forward and reverse primer, 5 % DMSO, and 1 U of Faststart Taq DNA polymerase (Roche), using touch-down PCR method (95 °C for 5 min; 10 cycles of 96 °C for 45 s, 65 °C for 45 s (decreased 0.5 °C per cycle), 72 °C for 45 s; 30 cycles of 94 °C for 45 s, 60 °C for 45 s, 72 °C for 45 s; followed by 72 °C for 7 min). When the mutant allele was detected by the primer set of BRCA2i14-216 F & BRCA2i14-864R, the following PCR program was used to reduce non-specific amplification: 95 °C for 5 min; 32 cycles of 94 °C for 45 s, 64 °C for 45 s, 72 °C for 45 s; followed by 72 °C for 7 min. The PCR products were then purified by the Qiaquick PCR purification system (Qiagen), followed by sequencing.
Results
Haplotype analysis and elucidation of first-degree family members' genotypes
Twenty-five sperms were isolated by micromanipulator for haplotype analysis, using the six markers as indicated (Fig. 1). The procedures were described in detail in the Materials and Methods section. The results were shown in Fig. 2. Recombination was detected between D13S1698 and rs1801406 in 8 % of the sperms studied (2/25). No recombination was detected on the telomeric side of the BRCA2 gene. The six markers had also been tested in the patient and her sister, and the allelic patterns for the family members were determined (Fig. 2).
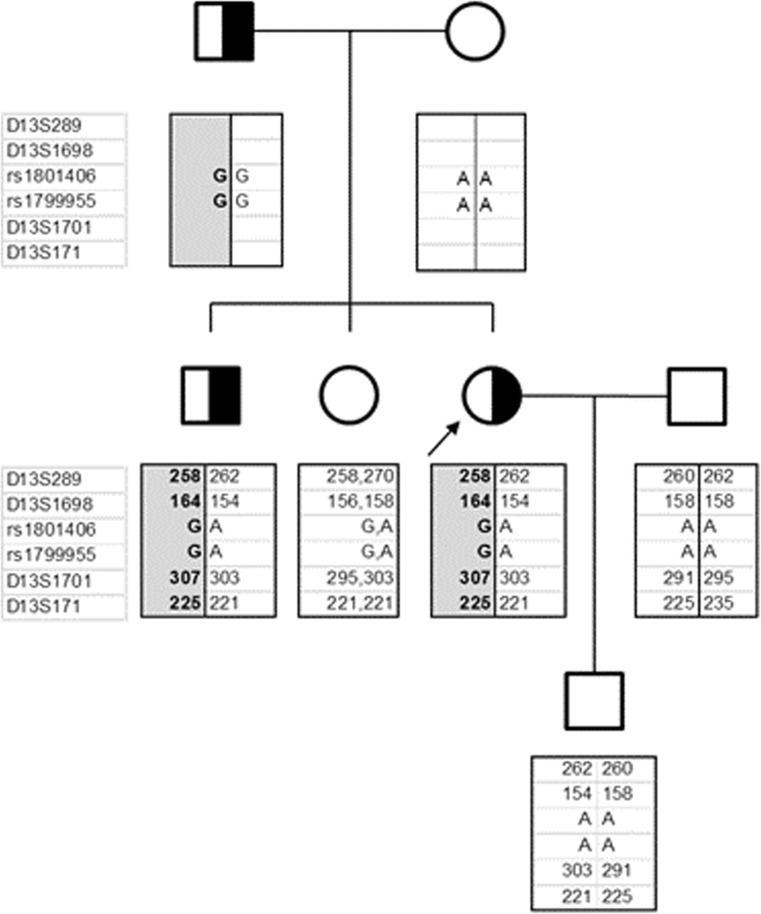
Fig. 2.
Family Pedigree. Patient is indicated by the arrow sign. Linkage of the mutant allele to marker loci (D13S289; D13S1698; rs1801406; rs1799955; D13S1701; D13S171) was deduced by haplotype analysis of the sperms derived from the carrier brother. The mutant allele was shown in shaded box
Treatment cycle
The patient underwent her first PGD cycle using an antagonist protocol. Letrozole 5 mg daily was started from day 2 till the day of human chorionic gonadotrophin to suppress the serum estradiol concentration arising from ovarian stimulation to reduce the chance of recurrence of breast cancer. Human menopausal gonadotrophin was given for nine days. Fifteen oocytes were retrieved and all were mature for intracytoplasmic sperm injection. Twelve oocytes were fertilized and eight good quality embryos were available for biopsy on day 3 after the oocyte pickup. The procedures for blastomere biopsy and WGA of blastomeres were reported previously [15]. Three embryos were found to carry mutant allele while five inherited normal allele. Among the five embryos without mutant allele, one morula and one blastocyst were replaced on day 5, resulting in a singleton live birth; one blastocyst was vitrified on day 6, and the remaining 2 embryos with normal allele were of poor quality and discarded. The patient declined invasive prenatal diagnosis because of the risk of miscarriage following the prenatal diagnostic procedure. A baby boy was delivered by lower segment caesarean section at term. Cord blood was collected and the genetic test conducted by an overseas accredited laboratory confirmed that the mutant allele could not be detected (data not shown).
Identification of the genomic breakpoint for the deleted allele
In order to improve the accuracy of the diagnosis and the cancer risk estimate in future cases, an effort was made to identify the genomic breakpoint of the mutant allele. Several sets of primers were designed for simultaneous screening of the speculated hairpin region identified via BLAST analysis. The screening results are shown in Fig. 3. Abnormal PCR products of >700 bp appeared in the patient's samples, with the Brca2i14-216 F & Brca2i16-864R primer pair. The PCR program was then fine-tuned to eliminate background noise. Sequencing of the informative PCR products revealed a common fragment (ggaggctgaggcaggagaatcg) in intron14 (IVS14-486 ~ IVS14-507) and intron16 (IVS16-441 ~ IVS16-462) (Fig. 4). A genomic sequence of 2,596 nucleotides in between the two fragments, including exon 15 and 16, plus one of the two fragments was deleted.
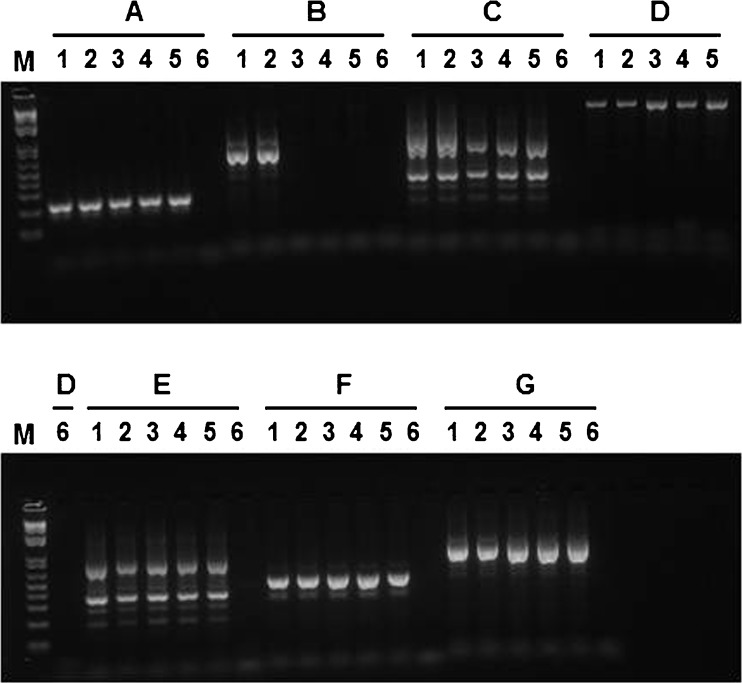
Fig. 3.
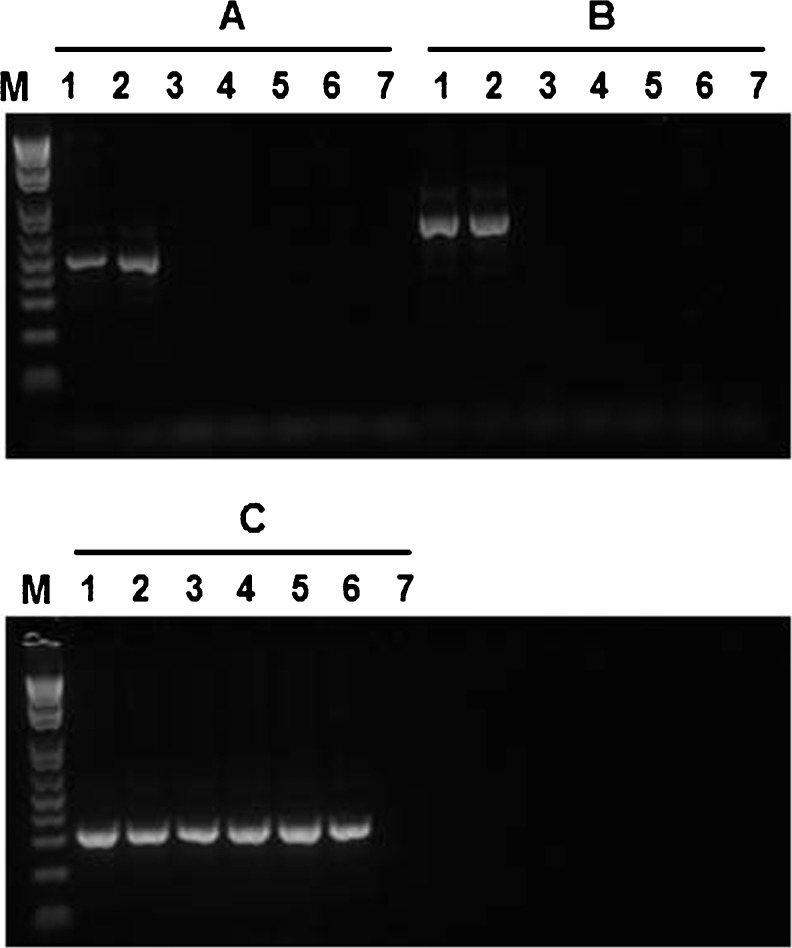
PCR screening of the genomic breakpoint. Seven pairs (A ~ G) of primers were used to amplify the putative mutation regions. Primer set A: Brca2e14-42 F & Brca2e14-242R; B: Brca2i14-216 F& Brca2i16-864R; C: Brca2i14-216 F & Brca2i16-1161R; D: Brca2i14-699 F & Brca2i16-864R; E: Brca2i14-699 F & Brca2i16-1161R; F: Brca2i14-699 F & Brca2e15-10R; and G: Brca2e16-143 F & Brca2i16-864R. Please refer to Fig. 4 for the location and sequence of each primer. The six samples from left to right were patients (1,2); sister (3,4); unrelated individual (5); H2O (6). M: 1Kb Plus DNA ladder (Invitrogen). The primer set B produced informative PCR results; an abnormal PCR product (>700 bp visualized from the agarose gel) was clearly observed in the patient's samples, but not in the samples of sister and unrelated individual
Confirmation of the genotypes of the eight embryos and the baby born
PCR was performed on the WGA products of the eight biopsied embryos mentioned as above, with the primer pair of BRCA2i14-216 F & BRCA2i14-864R. The mutant allele resulting in an abnormal PCR product of 714 bp was clearly detected in 3 embryos (data not shown), which was consistent with the results obtained from the haplotype analysis. Genetic analysis on the cord blood confirmed that the baby did not carry the mutant allele (Fig. 5).
Fig. 5.
PCR on cord blood DNA. PCR was performed with BRCA2i14-216 F & BRCA2i16-636R (A, 5′-TCCTGTGCTCAAGAGATCTGC), BRCA2i14-216 F & BRCA2i16-864R B, or HBA-F & HBA-R (C, 5′- GCGATCTGGGCTCTGTGTTCT, and 5′- GTTCCCTGAGCCCCGACACG). If the DNA templates contain the mutant allele, 486 bp and 714 bp PCR products should be obtained, respectively. The additional primer of BRCA2i16-636R was designed and applied in order to reduce a possible misdiagnosis due to non-specific amplification. The primer C was complimentary to hemoglobin alpha gene and the PCR product was 312 bp, which was used to indicate the presence of genomic DNA templates in each reaction. Lane 1 ~ 2, patient (mutant allele carrier); lane 3, patient' sister (normal); lanes 4 ~ 5, baby; lanes 6, unrelated normal; lane 7, H2O. As ADO with genomic DNA has never been reported, negative findings should demonstrate the baby's normal condition
Discussion
The PGD was carried out upon a request from an early-onset breast cancer patient carrying a paternally derived Brca2 mutant allele. Direct mutation detection and linkage analysis are common strategies for PGD. However, during the pre-PGD workup, we encountered problems in either way with routine protocols, due to unknown mutations, unavailability of parental DNAs, and uninformative genotype data obtained from the lymphocytes of her carrier and non-carrier siblings. Fortunately, the patient's brother is a carrier, making the sperm haplotyping an effective solution for linkage analysis.
It is noteworthy that genomic recombination is not a rare event, which could potentially cause misdiagnosis with linkage analysis approach. In this case, recombination between the BRCA2 gene and the selected microsatellite markers occurred at a rate of 8 % on the centromeric side. Although such a misdiagnosis could be detected by prenatal diagnosis, the invasive nature of the procedure would likely be rejected by PGD patients according to a recent report [14], so did our patient.
Knowing the precise mutation for direct mutation detection could avoid such a possible misdiagnosis. Meanwhile, for genetic counseling purpose, identification of the mutation is also important for the estimation of cancer risk among the family members, as the position of mutations is often associated with cancer risk and severity of cancer [6, 16]. However, identification of genomic breakpoints for a large deletion could be complex and time-consuming. Exome sequencing is not cost-effective, and may not work here, as the targeted breakpoints locate within introns. In this study, we identified the mutation very rapidly, via employing BLAST to search for potential hairpin structures, which had been frequently associated with genomic deletion. The rapid localization of the speculated regions enabled us to design PCR primers unique for these regions, which greatly shortened the time for the discovery. Our success in this case indicated that using BLAST searching for putative hairpin structure would be very simple and useful for the identification of genomic deletion, and apart from basic molecular laboratory techniques, understanding the mechanisms underlying mutations and knowing related bioinformatics tools would be essential to achieve a rapid and accurate diagnosis.
It is worth to mention, allelic dropout (i.e., failed to amplify one allele among the two) is a common systematic error with whole genome amplification of single cell during PGD. Direct mutation detection for large genomic deletion, using PCR primers aligned to both flanking regions of the breakpoints, can only amplify the mutant allele but not the long normal allele. The negative PCR result on normal allele cannot be distinguished from the situation of allelic dropout of mutant allele. Therefore, under such circumstances, a combination of linkage analysis and direct mutation detection method should be the method of choice for the best diagnostic accuracy.
Based on the Human Genetic Mutation Database and extensive literature search, the genomic deletion we identified here had not been reported previously. Interestingly, we found one report describing a missing of exon 15 and 16 of the BRCA2 mRNA in one of the 335 Spanish moderate to high-risk breast/ovarian cancer families previously screened negative for point mutations of the BRCA genes by conventional methods [17]. The mutant allele was present in three individuals of the same family, two of which (one male, one female) had breast cancer at the age of 60 years. However, further investigation at the genome level was not conducted in the study. It is unclear whether that family had the same genomic deletion.
Loss of exon 15 and 16 in the BRCA2 mRNA (nucleotides 7,663 ~ 8,032 in mRNA) would produce a truncated protein consisting of a normal N-terminal of 2,478 amino acids followed by an altered 45 amino acids (due to frameshift) and a putative premature stop codon TAA. Partial BRCA2 activity might be preserved in the truncated polypeptide as the important RAD51 binding domain encoded by exon 11 remains intact. This possibility is supported by the fact that the two Spanish carriers did not develop breast cancers until reaching their 60s, though further functional studies are required for confirmation.
Our patient has the deletion of exon 15 and exon 16 in the BRCA2 gene. In view of the phenotypes of the Spanish carriers and the lack of phenotype in the carrier brother and father, it is reasonable to speculate that the paternally derived mutant allele itself might not be fully responsible for the early onset breast cancer incurred in our patient. One fact should not be ignored is that the patient's mother had breast cancer at her 60s. The previous investigation on the mother's BRCA1/2 revealed some variants with unknown significance but no obvious pathogenic mutations.
Although the role of BRCA1/2 mutations in breast cancer has been well defined, the deleterious mutations in these two genes were identified by DNA full-sequence analysis in only 12.5 % of 46,276 women of high risk group from 1996 ~ 2006 [18], and the percentage is similar across diverse ethnicities. Many other high or moderate risk genes such as PTEN, TP53, CHEK2, ATM, BRIP, PALB2, etc. have also been identified [19] to be associated with breast cancer. In addition, more than 40 novel breast cancer susceptibility loci have been found in a most recent meta-analysis of nine GWAS studies [20, 21]. As breast cancer involves multiple cancer susceptibility alleles, the patient's mother might carry another breast cancer gene mutation that had not yet be identified. Therefore, the possibility of the patient carrying another maternally derived cancer-related allele cannot be excluded. As a result, although our PGD service had helped the new born boy to be free from the paternally derived mutant allele, his cancer risk arising on a possible mutant allele from the grandmother still exists.
Since 2006 when the UK government approved the first PGD for BRCA1 pathogenic mutation carrier, PGD for BRCA1/2 genes is increasingly performed worldwide because it is demanded by the patients. However, it seems that not much attention had been paid to the possibility of co-existing familial risk alleles during PGD work-up and genetic counseling.
In fact, as more and more susceptibility loci have been identified in breast cancer, a multifactorial disorder, the chance for coexistence of several risk alleles should not be ignored. In this case, if maternal deleterious mutation(s) of other associated genes did exist yet not been identified, actually we do not know which mutation (paternal or maternal) contributed more to the severe early onset breast cancer incurred in our patient. Certainly, the PGD performed to eliminate the cancer risk resulting from the paternal mutant allele is not questionable, as such a mutation of large deletion had been clearly associated with breast cancer (e.g., patient's paternal grandma, two Spanish carriers). Nevertheless, when such multi-factorial disorders as breast cancer involved in IVF clinic for PGD, it is important to consider all possible familial risk alleles, and to evaluate the significance of each mutation contributing to particular phenotypes, if possible, before therapeutic decisions could be made.
Another concern is whether or not the recurrence of breast cancer could result from the hormonal treatment during IVF. In a recent report on the largest cohort of PGD on BRCA mutation, 2 out of 70 women were diagnosed to have breast cancer within 3 months post-PGD treatment while breast screening had been negative before treatment [10]. But Kotsopoulos et al. [22] reported no elevated risk of recurrence of the cancer after fertility treatment. A breast lump was found in our patient after delivery of the baby. During the follow-up, the breast lump became smaller, and the biopsy results did not show malignant cells but did presence of some atypical cells. More studies are needed to determine the cancer recurrence rate following the hormonal treatment during PGD.
In conclusion, this work-up provides a clear example of using integrated approaches to establish a reliable and accurate protocol for a complex PGD case. BLAST analysis is a very useful and cost-effective tool to achieve a rapid identification of large genomic deletion. Both direct mutation detection and linkage analysis approaches might be necessary on some occasions, in order to eliminate misdiagnosis resulting from recombination and/or allelic drop-out. Concerning the assisted reproductive option for breast cancer patients, the possibility of coexistence of multiple familial cancer risk alleles should be considered and the cancer recurrence rate following hormone treatment during IVF should be evaluated.
Acknowledgments
The study was supported by the Department Fund.
Footnotes
Qingxue Wang and Judy FC Chow are co-first authors.
Capsule The possibility of coexistence of multiple cancer risk alleles should be considered during IVF-PGD workup for cancer patients. Both direct mutation detection and linkage analysis approaches may be necessary to eliminate misdiagnosis resulting from recombination and/or allelic drop-out in PGD.
Contributor Information
Qingxue Wang, Phone: +852 2255 3398, Email: qxwang@hku.hk.
William SB Yeung, Phone: +852 2255 3405, Email: wsbyeung@hkucc.hku.hk.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–30. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 4.Barnes DR, Antoniou AC. Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. J Intern Med. 2012;271(4):331–43. doi: 10.1111/j.1365-2796.2011.02502.x. [DOI] [PubMed] [Google Scholar]
- 5.Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18(7):748–54. doi: 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baer R, Lee WH. Functional domains of the BRCA1 and BRCA2 proteins. J Mammary Gland Biol Neoplasia. 1998;3(4):403–12. doi: 10.1023/A:1018736115722. [DOI] [PubMed] [Google Scholar]
- 7.Partridge AH, Gelber S, Peppercorn J, Sampson E, Knudsen K, Laufer M, et al. Web-based survey of fertility issues in young women with breast cancer. J Clin Oncol. 2004;22(20):4174–83. doi: 10.1200/JCO.2004.01.159. [DOI] [PubMed] [Google Scholar]
- 8.Partridge AH, Gelber S, Peppercorn J, Ginsburg E, Sampson E, Rosenberg R, et al. Fertility and menopausal outcomes in young breast cancer survivors. Clin Breast Cancer. 2008;8(1):65–9. doi: 10.3816/CBC.2008.n.004. [DOI] [PubMed] [Google Scholar]
- 9.Smith KR, Ellington L, Chan AY, Croyle RT, Botkin JR. Fertility intentions following testing for a BRCA1 gene mutation. Cancer Epidemiol Biomarkers Prev. 2004;13(5):733–40. [PubMed] [Google Scholar]
- 10.Wilkinson E. Preimplantation genetic diagnosis for mutated BRCA genes. Lancet Oncol. 2012;13(8):e331. doi: 10.1016/S1470-2045(12)70321-2. [DOI] [PubMed] [Google Scholar]
- 11.Jasper MJ, Liebelt J, Hussey ND. Preimplantation genetic diagnosis for BRCA1 exon 13 duplication mutation using linked polymorphic markers resulting in a live birth. Prenat Diagn. 2008;28(4):292–8. doi: 10.1002/pd.1925. [DOI] [PubMed] [Google Scholar]
- 12.Offit K, Sagi M, Hurley K. Preimplantation genetic diagnosis for cancer syndromes: a new challenge for preventive medicine. JAMA. 2006;296(22):2727–30. doi: 10.1001/jama.296.22.2727. [DOI] [PubMed] [Google Scholar]
- 13.Spits C, De Rycke M, Van Ranst N, Verpoest W, Lissens W, Van Steirteghem A, et al. Preimplantation genetic diagnosis for cancer predisposition syndromes. Prenat Diagn. 2007;27(5):447–56. doi: 10.1002/pd.1708. [DOI] [PubMed] [Google Scholar]
- 14.Sagi M, Weinberg N, Eilat A, Aizenman E, Werner M, Girsh E, et al. Preimplantation genetic diagnosis for BRCA1/2–a novel clinical experience. Prenat Diagn. 2009;29(5):508. doi: 10.1002/pd.2232. [DOI] [PubMed] [Google Scholar]
- 15.Chow JF, Yeung WS, Lau EY, Lam ST, Tong T, Ng EH, et al. Singleton birth after preimplantation genetic diagnosis for Huntington disease using whole genome amplification. Fertil Steril 2009;92(2):828 e7-10. [DOI] [PubMed]
- 16.Lubinski J, Phelan CM, Ghadirian P, Lynch HT, Garber J, Weber B, et al. Cancer variation associated with the position of the mutation in the BRCA2 gene. Fam Cancer. 2004;3(1):1–10. doi: 10.1023/B:FAME.0000026816.32400.45. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez-Enriquez S, de la Hoya M, Martinez-Bouzas C. Sanchez de Abajo A, Ramon Y, Cajal T, et al. Screening for large rearrangements of the BRCA2 gene in Spanish families with breast/ovarian cancer. Breast Cancer Res Treat. 2007;103(1):103–7. doi: 10.1007/s10549-006-9376-8. [DOI] [PubMed] [Google Scholar]
- 18.Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115(10):2222–33. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanale D, Amodeo V, Corsini LR, Rizzo S, Bazan V, Russo A. Breast cancer genome-wide association studies: there is strength in numbers. Oncogene. 2012;31(17):2121–8. doi: 10.1038/onc.2011.408. [DOI] [PubMed] [Google Scholar]
- 20.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies more than 40 novel breast cancer susceptibility loci. American Society of Human Genetics, 2012 Annual Meeting, 2012
- 21.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21(24):5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotsopoulos J, Librach CL, Lubinski J, Gronwald J, Kim-Sing C, Ghadirian P, et al. Infertility, treatment of infertility, and the risk of breast cancer among women with BRCA1 and BRCA2 mutations: a case–control study. Cancer Causes Control. 2008;19(10):1111–9. doi: 10.1007/s10552-008-9175-0. [DOI] [PubMed] [Google Scholar]