Abstract
Purpose
To investigate how effectively density gradient centrifugation (DGC) improves sperm nuclear integrity and to determine whether the sperm chromatin dispersion (SCD) test of sperm nuclear integrity in native or DGC-treated semen can predict the outcome of assisted reproductive technology (ART) in couples undergoing intracytoplasmic sperm injection (ICSI).
Methods
The DNA integrity of spermatozoa from 63 male factor infertility patients undergoing ICSI was analyzed by the SCD test before and after DGC. The predictive value of the sperm DNA fragmentation index (DFI) for ART outcomes was assessed in a cohort of 45 patients who were undergoing fresh embryo transfer. For the analysis, they were divided into pregnant and non-pregnant groups and, independently, into high sperm DFI (DFI > 30 %) and low sperm DFI (DFI ≤ 30 %) groups. Both raw and DGC semen parameters were examined.
Results
In the asthenospermia and oligozoospermia groups, DGC decreased the sperm DFI from 31.5 ± 19.7 and 28.5 ± 10.3 to 19.2 ± 18.3 and 16.0 ± 12.8, respectively (P < 0.01). DGC decreased the sperm DFI in the severe oligozoospermia group from 41.4 ± 19.0 to 36.3 ± 20.6 (P > 0.01). The pregnant and non-pregnant groups did not differ in their fertilization rate and sperm DFI in native or DGC semen (P > 0.05). There was also no significant difference between the high sperm DFI (DFI > 30 %) and low sperm DFI (DFI ≤ 30 %) groups with regard to fertilization rate, implantation rate, and clinical pregnancy rate for both native and DGC semen (P > 0.05). The patients undergoing ICSI with a high sperm DFI had a higher pregnancy loss rate (defined as spontaneous miscarriage or biochemical pregnancy) compared with patients with a low sperm DFI in both the native and DGC semen groups.
Conclusions
DGC highly significantly reduces sperm DNA fragmentation in the semen of ICSI patients, with the exception of those with severe oligozoospermia. The results of the SCD test of sperm DNA fragmentation in native or DGC semen do not correlate with the fertilization rate, implantation rate, or clinical pregnancy rate in patients undergoing ICSI.
Keywords: Sperm chromatin dispersion assay, DNA fragmentation, ICSI, Density gradient centrifugation, Pregnancy outcome
Introduction
The male factor infertility is a common cause of infertility, with sperm defects found in 40–50 % of clinical infertility cases [1]. Intracytoplasmic sperm injection (ICSI) is an efficient treatment modality for male factor infertility subjects with poor sperm quality and those who have failed conventional in vitro fertilization (IVF) cycles. The selection of the spermatozoon to be used for ICSI is based on the judgment of the embryologist, who chooses a motile spermatozoon with the best available morphology. However, the selected spermatozoa may have damaged DNA. Avendano et al. reported that the sperm of infertile men can be morphologically normal according to strict criteria but nevertheless show DNA fragmentation [2]. Sperm DNA fragmentation has been associated with male infertility [3]. ICSI using spermatozoa with damaged DNA may lead to poor embryo development or the transmission of defective genetic material to the offspring [4].
Because sperm DNA integrity is important for both the evaluation and treatment of male infertility, numerous tests have been developed to assess it [5–8]. One of these tests is the sperm chromatin dispersion (SCD) assay. This test is based on the principle that sperm with fragmented DNA fail to produce the characteristic halo of dispersed DNA loops that is observed following acid denaturation and removal of nuclear proteins from sperm with nonfragmented DNA [9]. Originally, fluorescence microscopy was used in the SCD test to determine sperm DNA fragmentation [10]; however, a recent modification allowed for the use of bright-field microscopy [11]. SCD results correlate well with those obtained by other fragmentation assays, such as the sperm chromatin structure assay (SCSA) and terminal transferase-mediated DNA end-labeling (TUNEL) [9]. Not all laboratories have access to a flow cytometer or the technical expertise required to perform the SCSA assay. The TUNEL and SCD assays are simple, less expensive, procedures and can be performed in a short period of time. However, the bright-field SCD microscopy test appears to be more sensitive than the TUNEL assay [12]. Because of its low cost, speed, and reliability, SCD is an appealing method for the assessment of DNA integrity.
Density gradient centrifugation (DGC) is used in most IVF centers to wash raw semen for ICSI. It is important for the sperm processing technique to select functional sperm while causing them minimal damage. The effects of DGC on sperm DNA integrity are controversial, with several studies showing that sperm nuclear integrity is improved, as evaluated by various DNA integrity assays [1, 13–16]. Other studies have not found DGC to be useful for the selection of sperm with high DNA integrity [26, 27]. It is not currently known whether SCD can predict sperm nuclear integrity in the semen of ICSI patients following DGC.
Many studies address the influence of sperm DNA damage on reproductive outcomes after ICSI [2, 4, 13, 15, 17–23], and most of these studies use raw, unprepared semen [2, 13, 17, 19–21, 23]. There are notable exceptions, however; for example, the sperm DNA fragmentation in couples undergoing ICSI has been measured after DGC. Borini et al. reported that sperm DNA fragmentation can affect post-implantation embryo development in patients who undergo ICSI procedures, namely, high sperm DNA fragmentation can compromise ‘embryo viability’, resulting in pregnancy loss [18]. Benchaib et al. also found a statistically significant negative relationship between fertilization rate and percentage of sperm DNA fragmentation [4]. These two studies, which included 50 and 234 ICSI cases, respectively, evaluated the DNA fragmentation of sperm suspensions after DGC by the TUNEL assay. However, Bungum et al. reported that the SCSA performed on semen prepared by DGC could not predict the outcome of assisted reproductive technology (ART). Their study included 510 ART cycles, namely, 197 IUI, 220 IVF, and 93 ICSI cycles [15]. Simon et al. reported that DNA fragmentation could predict the ART outcome associated with IVF but not ICSI. Converting modified bases into further DNA strand breaks increased the test sensitivity, yielding negative correlations between DNA fragmentation and clinical pregnancy in individuals undergoing ICSI or IVF. In their study, DNA fragmentation in 360 couples (230 IVF and 130 ICSI) was measured by the alkaline Comet assay in semen and sperm following DGC [22]. It remains unclear whether measuring sperm chromatin integrity after DGC in ICSI patients has advantages over measurements performed on raw semen samples. Additionally, previous studies did not use the SCD assay to evaluate sperm DNA integrity after DGC.
This study investigated the ability of DGC to improve sperm nuclear integrity and the value of SCD measurements of sperm nuclear integrity, in both native and DGC-processed semen, in predicting pregnancy outcome in couples participating in an ICSI program.
Methods
Subjects
Sixty-three patients undergoing ICSI at the reproductive medical center of International Peace Maternity and Child Health Hospital (IPMCH), affiliated with Shanghai Jiao Tong University, participated in this study between September 2012 and October 2013. Patient ages ranged from 22 to 50 years; the median ages of the women and men were 30.6 ± 4.3 and 32.9 ± 5.4 years, respectively. Male factor infertility patients providing sperm samples for ICSI met at least one of the following World Health Organization (WHO) guideline criteria: sperm concentration <12 × 106/mL, forward motility <31 %, or unexpected complete fertilization failure in previous IVF attempts (WHO, 2010). The 63 patients were divided into the following groups: asthenospermia (sperm concentration ≥12 × 106/mL, forward motility <31 %, n = 32), oligozoospermia (sperm concentration 5 × 106/mL–12 × 106/mL, n = 11), severe oligozoospermia (sperm concentration <5 × 106/mL, n = 17), and unexpected complete fertilization failure in previous IVF attempts (sperm concentration ≥12 × 106/mL, forward motility ≥31 %, n = 3).
Our institutional review board approved this study.
Semen collection and analysis
Semen samples were collected by masturbation after 3–7 days of sexual abstinence, on the day of ovum pick-up. After liquefying the semen at 37 °C in 5 % CO2 in air for 20 min, the samples were examined for concentration and motility according to the WHO guidelines (WHO, 2010) in a Makler® chamber (Sefi Laboratories, Tel Aviv, Israel). Sperm morphology was not assessed,due to it not being routinely performed in our IVF center.
Density gradient centrifugation
Sperm was prepared by standard DGC using 50 and 90 % ISolate (Irvine Scientific; Santa Ana, CA, USA). A maximum of 2 mL of semen was layered on the top of the two layers of ISolate (50 and 90 %), and the sample was centrifuged at 360 g for 20 min. The pellet was then washed in IVF-100TM (Vitrolife, Gothenburg, Sweden) and centrifuged at 360 g. The final dilution of the sample was in IVF-100TM (Vitrolife, Gothenburg, Sweden), and the spermatozoa were incubated in 5 % CO2 in air at 37 °C until insemination.
SCD test
DNA fragmentation was measured by the SCD test, using the SpermFuncTM DNAf kit (BRED Life Science, Shenzhen, China), for both the native and DGC-separated semen. Gelled aliquots of low-melting-point agarose in Eppendorf tubes were provided in the kit for semen sample processing. Eppendorf tubes were placed in a water bath at 80 °C for 20 min to melt the agarose and then transferred to a water bath at 37 °C for 5 min for temperature equilibration. A total of 60 μL of the semen sample was added to the agarose in the Eppendorf tube and mixed. Then, 30 μL of the semen-agarose mix was pipetted onto precoated slides that were provided in the kit and covered with a 22 × 22-mm coverslip. The slides were placed on a cold plate in the refrigerator (4 °C) for 5 min to allow the agarose to produce a microgel in which the sperm cells were embedded. The coverslips were gently removed, and the slides were immediately immersed horizontally in solution A and incubated for 7 min. Next, the slides were horizontally immersed in solution B for 25 min. After washing for 5 min in a tray with abundant distilled water, the slides were dehydrated in increasing concentrations of ethanol (70–90–100 %) for 2 min each, air-dried, and stored at room temperature in opaque closed boxes.
For bright-field microscopy, slides were horizontally covered with a mix of Wright’s staining solution (BRED Life Science, Shenzhen, China) and phosphate buffer solution (BRED Life Science, Shenzhen, China) (1:2) for 15 min with continuous airflow. Then, the slides were washed in running water for 10 s and allowed to dry. Strong staining is preferred to allow the periphery of the dispersed DNA loop halos to be seen more easily. A minimum of 500 spermatozoa per sample were scored under the 100× objective.
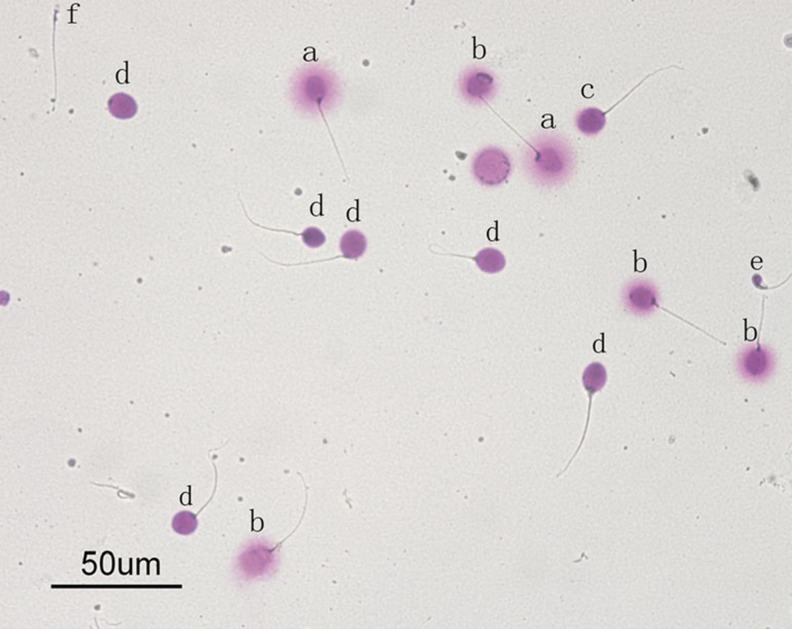
Five SCD patterns are established: (1) sperm cells with large halos, the width of which is similar or larger than the minor diameter of the core; (2) sperm cells with medium-sized halos, the size of which is between that of large and very small halos; (3) sperm cells with very small halos, the width of which is similar to or smaller than 1/3 of the minor diameter of the core; (4) sperm cells without a halo; and (5) sperm cells without a halo and degraded [11, 12]. The nuclei with large-to-medium size halo were considered sperm with nonfragmented DNA, whereas nuclei with small size halo or without halo or without a halo and degraded were considered sperm with fragmented DNA (Fig. 1). Finally, nucleotides that did not correspond to sperm cells were scored separately. Sperm DNA fragmentation was expressed as the percentage of sperm cells with fragmented DNA.
Fig. 1.
The SCD test results in a case of severe oligozoospermia. Sperms (a) show the nuclei with large size halo and (b) show the nuclei with medium size halo that were considered with nonfragmented DNA, whereas (c) show the nuclei with small size halo, (d) show without halo, (e) show without a halo and degraded, and (f) show without halo and pinhead, which were considered sperm with fragmented DNA
Ovarian stimulation and ICSI techniques
For ovarian stimulation, both GnRH agonist and antagonist protocols were used. Briefly, the long GnRH agonist protocol was used as previously described [24], and cetrorelix (Cetrotide; Merk-Serone, Germany) was administered in the antagonist protocol. The ICSI technique has been described previously [25]. Sperm for ICSI were chosen based on criteria for normal morphological sperm (WHO, 2010). At least 3 metaphase II oocytes were retrieved in the ICSI cycles in our study.
Fertilization rate
At 16–18 h after microinjection, the oocytes were assessed to determine whether fertilization had occurred. Fertilization was considered normal if two pronuclei and two polar bodies were identified. Oocytes without visible pronuclei were considered unfertilized. Oocytes with a single pronucleus or with more than two pronuclei were considered to be abnormally fertilized and were discarded. Fresh embryo transfers were performed at 48 or 72 h after oocyte retrieval.
Pregnancy outcome
Pregnancy was initially detected 2 weeks after embryo transfer by a positive serum β-hCG test. Ultrasound was performed at 6 weeks of gestation to confirm fetal viability. Clinical pregnancy was defined as the presence of a gestational sac detected by ultrasound, and biochemical pregnancy was defined as at least one positive β-hCG test 2 weeks after embryo transfer. Women with clinical pregnancies who miscarried before the 12th week were defined as having had a spontaneous abortion. The pregnancy loss rate was the number of biochemical pregnancies and spontaneous miscarriages divided by the number of β-HCG-positive patients [18].
Statistical analysis
Results are expressed as the mean ± SD. The paired-samples t-test was used to analyze the semen parameters before and after DGC. One-way analysis of variance (ANOVA) was used to compare the male age and sperm DNA fragmentation between the asthenospermia, oligozoospermia, and severe oligozoospermia groups. Inter-group (positive and negative pregnancy groups) differences in clinical and semen parameters were assessed by the independent-samples t-test. The difference in male age, female age, number of oocytes retrieved, oocyte maturation, metaphase II oocytes, fertilization rate, and sperm count between couples with a low sperm DFI (DFI ≤ 30 %) and high sperm DFI (DFI > 30 %) in both native and DGC semen were compared using an independent-samples t-test. Pearson’s chi-square test or Fisher’s exact test was used to compare the rates of clinical pregnancy, implantation, and pregnancy loss between groups. All hypothesis testing was two-sided. Differences were considered significant at P < 0.05 and highly significant at P < 0.01. Statistical analysis was performed using Sigma Stat software (SPSS, Chicago, IL, USA).
Results
Effect of DGC on sperm DNA fragmentation and sperm characteristics
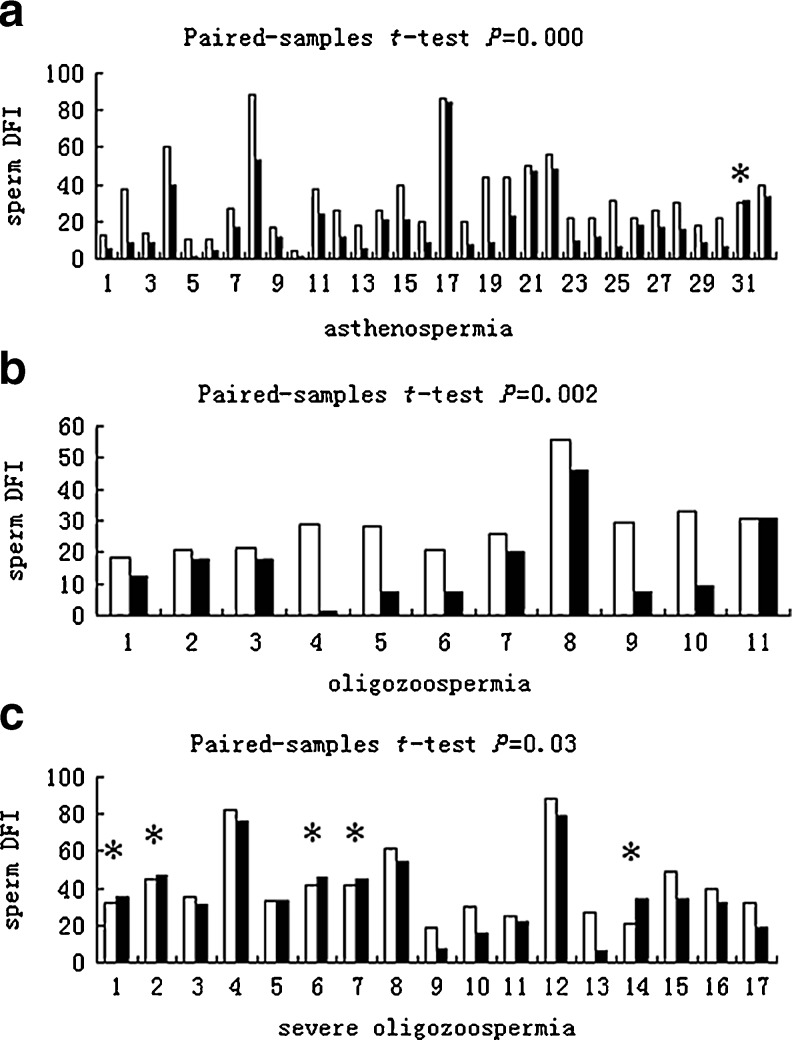
To analyze the effect of DGC on sperm DNA fragmentation and sperm characteristics, the 63 patients were divided into the following groups: asthenospermia (n = 32), oligozoospermia (n = 11), severe oligozoospermia (n = 17), and unexpected complete fertilization failure in previous IVF attempts (n = 3). Semen parameters in these patients, except the 3 patients with unexpected complete fertilization failure in previous IVF attempts, are presented in Table 1. The progressive motility of severe oligozoospermia was not examined. There were no significant differences between the three groups with regard to male age (P = 0.084; one-way ANOVA). There was a significant decrease in the sperm concentration of the native versus DGC-processed samples from the asthenospermia (P < 0.01; paired-samples t-test) and oligozoospermia (P < 0.05; paired-samples t-test) groups, but not the severe oligozoospermia group (P > 0.05; paired-samples t-test). There was also a highly significant increase in progressive motility in the asthenospermia (P < 0.01; paired-samples t-test) and oligozoospermia (P < 0.01; paired-samples t-test) groups. The majority of the samples showed an improvement in sperm DFI following DGC (Fig. 2), whereas sperm DFI increased after preparation in only 6 (5 from the severe oligozoospermia group and 1 from the asthenospermia group) of the 60 samples. In the asthenospermia group, sperm DFI following DGC decreased from 31.5 ± 19.7 to 19.2 ± 18.3 (P < 0.01; paired-samples t-test). In the oligozoospermia group, sperm DFI following DGC decreased from 28.5 ± 10.3 to 16.0 ± 12.8 (P < 0.01; paired-samples t-test). However, in the severe oligozoospermia group, sperm DFI following DGC decreased from 41.4 ± 19.0 to 36.3 ± 20.6 (P = 0.03; paired-samples t-test). The DFI of raw sperm did not differ between the three groups (P = 0.131; one-way ANOVA). Following DGC, the sperm DFI was significantly higher in the severe oligozoospermia group than in any other group (P = 0.004; one-way ANOVA).
Table 1.
Semen characteristics and DNA damage assessed by the SCD assay
| asthenospermia (n = 32) | oligozoospermia (n = 11) | severe oligozoospermia (i = 17) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean male age ± SD | 34.0 ± 5.7 a | 33.4 ± 6.8 a | 30.4 ± 3.5 a | ||||||
| Sample type analyzed | Native | DGC | P d | Native | DGC | P d | Native | DGC | P d |
| Sperm concentration mean ± SD | 37.7 ± 25.7 | 7.5 ± 2.8 | 0.000 | 8.2 ± 2.2 | 5.7 ± 2.7 | 0.048 | 1.6 ± 1.4 | 1.2 ± 1.2 | 0.236 |
| Progressive motility ± SD | 21.9 ± 9.1 | 60.4 ± 24.5 | 0.000 | 33.3 ± 18.0 | 68.5 ± 24.8 | 0.000 | _ | _ | _ |
| Sperm DFI ± SD | 31.5 ± 19.7 b | 19.2 ± 18.3 c | 0.000 | 28.5 ± 10.3 b | 16.0 ± 12.8 c | 0.002 | 41.4 ± 19.0 b | 36.3 ± 20.6 c | 0.03 |
a,b,cOne-way ANOVA test; d paired-samples t-test
Fig. 2.
The sperm DFI of native semen (white columns) and DGC-processed samples (black columns). The patients are divided into asthenospermia (a), oligozoospermia (b), and severe oligozoospermia (c) groups. Statistical comparisons using the paired-samples t-test are included in the figure. * Sperm DFI increased after DGC preparation
In the 3 samples from individuals with unexpected complete fertilization failure in previous IVF attempts, the mean male age was 33.3 ± 2.1. Sperm concentration following DGC decreased from 69.3 ± 51.0 to 23.7 ± 15.5 (P = 0.187; paired-samples t-test). The progressive motility following DGC increased from 43.3 ± 7.6 to 62.8 ± 25.6 (P = 0.345; paired-samples t-test). There was also a decrease in sperm DFI following DGC from 35.5 ± 1.8 to 20.2 ± 16.6 (P = 0.217; paired-samples t-test).
Sperm DNA fragmentation and pregnancy outcome
The predictive value of sperm DFI for ART outcomes was assessed in 45 cycles with fresh embryo transfer (comprising 24 asthenospermia cases, 9 oligozoospermia cases, 11 severe oligozoospermia cases, and 1 case of unexpected complete fertilization failure in previous IVF attempts). The other 18 cycles were freeze-all IVF cycles.
We did not identify any significant differences between the couples who did or did not achieve pregnancy with regard to female and male age, mean sperm count, number of oocytes retrieved, oocyte maturation, metaphase II oocytes, fertilization rate, or sperm DFI in native or DGC semen (Table 2, P > 0.05; independent-samples t-test).
Table 2.
Clinical and semen parameters in couples who did or did not achieve clinical pregnancy
| Pregnant (n = 24) | Not pregnant (n = 21) | P a | |
|---|---|---|---|
| Mean male age ± SD | 32.8 ± 5.7 | 33.1 ± 5.1 | 0.853 |
| Sperm concentration mean ± SD | 27.2 ± 29.6 | 20.4 ± 20.3 | 0.377 |
| Mean female age ± SD | 30.4 ± 4.1 | 31.6 ± 4.2 | 0.323 |
| Oocytes retrieved | 13.2 ± 6.2 | 10.4 ± 5.3 | 0.116 |
| Metaphase II oocytes | 10.4 ± 3.9 | 8.5 ± 4.3 | 0.130 |
| Oocyte maturation (%) | 82.9 ± 13.3 | 82.8 ± 12.9 | 0.969 |
| Embryo transferred | 2 | 1.8 | _ |
| Fertilization rate (%) | 77.3 ± 17.3 | 71.6 ± 19.8 | 0.309 |
| SDFI ± SD in native semen | 34.3 ± 17.3 | 32.3 ± 21.1 | 0.732 |
| SDFI ± SD in DGC semen | 22.6 ± 16.8 | 23.8 ± 24.1 | 0.847 |
aIndependent-samples t-test
A threshold of 30 % was used for sperm DFI to discriminate between samples with normal and elevated levels of DNA-damaged spermatozoa [9]. The relationships between the fertilization rate, pregnancy outcome, and sperm DNA fragmentation were separately examined for raw and DGC semen (Table 3). There were no significant differences between the high (>30 %) and low (≤30 %) sperm DFI groups with regard to female and male age, mean sperm count, number of oocytes retrieved, oocyte maturation, or metaphase II oocytes with respect to whether native or DGC-processed sperm were used (P > 0.05; independent-samples t-test). There was no significant difference between the high and low sperm DFI groups with regard to fertilization rate (P > 0.05; independent-samples t-test), implantation rate (P > 0.05; Pearson’s chi-square test), or clinical pregnancy rate (P > 0.05; Fisher’s exact test), regardless of whether native or DGC-processed sperm were used. Regarding native semen, 5 couples with a sperm DFI > 30 % had a pregnancy loss (3 spontaneous abortions and 2 biochemical pregnancies) and two couples with a sperm DFI ≤ 30 % had a pregnancy loss (2 spontaneous abortions), corresponding to pregnancy loss rates of 35.7 and 16.7 %, respectively. However, there was no statistically significant difference between the two groups with regard to the pregnancy loss rate (P > 0.05; Fisher’s exact test). Regarding DGC semen, 4 couples with a sperm DFI > 30 % had a pregnancy loss (3 spontaneous abortions and 1 biochemical pregnancy) and 3 couples with a sperm DFI ≤ 30 % had a pregnancy loss (2 spontaneous abortions and 1 biochemical pregnancy), corresponding to pregnancy loss rates of 44.4 and 17.6 %, respectively. There was also no statistically significant difference between the two groups with regard to pregnancy loss rate (P > 0.05; Fisher’s exact test).
Table 3.
Clinical and semen parameters of the native and DGC-processed semen samples divided according to DFI (DFI ≤ 30 % versus DFI > 30 %)
| Native | DGC | |||||
|---|---|---|---|---|---|---|
| DFI ≤ 30 % (n = 23) | DFI > 30 % (n = 22) | P | DFI ≤ 30 % (n = 29) | DFI > 30 % (n = 16) | P | |
| Mean male age ± SD | 32.0 ± 4.8 | 34.0 ± 5.9 | 0.217 a | 31.8 ± 4.3 | 34.8 ± 6.7 | 0.082 a |
| Sperm concentration mean ± SD | 22.1 ± 21.5 | 26.2 ± 29.6 | 0.596 a | 6.3 ± 3.4 | 6.2 ± 8.6 | 0.965 a |
| Mean female age ± SD | 30.7 ± 3.8 | 31.2 ± 4.6 | 0.673 a | 30.7 ± 3.6 | 31.3 ± 5.1 | 0.674 a |
| Oocytes retrieved | 11.3 ± 5.5 | 12.5 ± 6.4 | 0.519 a | 12.2 ± 5.9 | 11.4 ± 6.0 | 0.656 a |
| Metaphase II oocytes | 9.3 ± 4.0 | 9.8 ± 4.4 | 0.711 a | 10.0 ± 4.1 | 9.0 ± 4.3 | 0.395 a |
| Oocyte maturation (%) | 83.7 ± 12.4 | 81.9 ± 13.8 | 0.642 a | 83.9 ± 12.3 | 80.9 ± 14.3 | 0.461 a |
| Embryos transferred | 1.9 | 1.9 | _ | 1.9 | 1.9 | _ |
| Fertilization rate (%) | 74.2 ± 17.3 | 74.8 ± 20.0 | 0.606 a | 76.9 ± 17.1 | 70 ± 20.4 | 0.235 a |
| Implantation rate (%) | 38.6 (17/44) | 31.0 (13/42) | 0.455 b | 38.2 (21/55) | 29.0 (9/31) | 0.122 b |
| Clinical pregnancy rate | 52.2 (12/23) | 54.5 (12/22) | 1.0 c | 55.2 (16/29) | 50.0 (8/16) | 0.765 c |
| Pregnancy loss rate | 16.7 (2/12) | 35.7 (5/14) | 0.391 c | 17.6 (3/17) | 44.4 (4/9) | 0.188 c |
aIndependent-samples t-test; bPerson’s chi-square; cFisher’s exact test
Discussion
We used ISolate DGC to wash semen, and sperm DNA integrity was evaluated by the SCD assay. With the exception of the severe oligozoospermia samples, the sperm preparation improved sperm motility and decreased sperm concentration; these results are in agreement with the findings of other studies [15].
Most studies also showed that sperm DNA integrity was improved by DGC processing [1, 13–16], although Zini et al. found no differences [26, 27]. The present study showed that DGC improved the sperm DNA fragmentation index (DFI) in most couples undergoing ICSI (Fig. 2). In the asthenospermia and oligozoospermia groups, the reduction in sperm DFI following DGC was highly significant (P < 0.01). In the 3 samples from couples with unexpected complete fertilization failure in previous IVF attempts, DGC decreased the sperm DFI; however, the sample number was too low to enable the statistical significance of this result. However, in the severe oligozoospermia group, DGC decreased the sperm DFI from 41.4 ± 19.0 to 36.3 ± 20.6 (P > 0.01). Of the 60 samples tested, only 6 showed an increase in sperm DFI after DGC; 5 of these samples were from the severe oligozoospermia group (Fig. 2). The effect of DGC on sperm nuclear integrity may relate to the initial sperm concentration. When the initial sperm concentration is too low, DGC cannot effectively improve sperm nuclear integrity. This result was also noted by Zini, who suggested that the potential adverse effects of DGC on sperm DNA integrity may be related to initial semen quality [28], which is generally poor in cases of severe oligozoospermia. Our data indicate that DGC can reduce the DNA fragmentation rate, significantly reducing the probability that sperm with low DNA integrity will be selected for ICSI. However, DGC may not benefit severe oligozoospermia patients. After the DGC washing procedure, the sperm DFI in the severe oligozoospermia group was also significantly higher than that in the other groups (P < 0.01). There is some concern that ICSI bypasses the process of natural sperm selection. A number of studies have suggested a link between sperm DNA damage and genetic or epigenetic disorders [29–31]. Severe oligozoospermia patients undergoing ICSI may, therefore, have a higher probability of transmitting defective genetic material to their offspring.
The predictive value of sperm DFI in native and DGC sperm for ICSI outcomes was assessed in a cohort of 45 patients who were undergoing fresh embryo transfer. For analysis, they were first divided into pregnant and non-pregnant groups. The biological impact of an abnormal sperm chromatin structure depends on the combined effects of the extent of DNA damage in the spermatozoa and the capacity of the oocyte to repair the damage [32]. We therefore compared not only male age, mean sperm count, and sperm DFI in native or DGC semen between the two groups but also female age, number of oocytes retrieved, oocyte maturation, metaphase II oocytes, and fertilization rate. There were no significant differences in these parameters between the couples who achieved or did not achieve pregnancy (P > 0.05). Our study supports a previous study by Avendano et al., who assessed sperm DFI using the TUNEL assay in 36 patients undergoing ICSI [2], as well as a study by Simon et al., who used the Comet assay to assess sperm DFI in 130 patients undergoing ICSI [22].
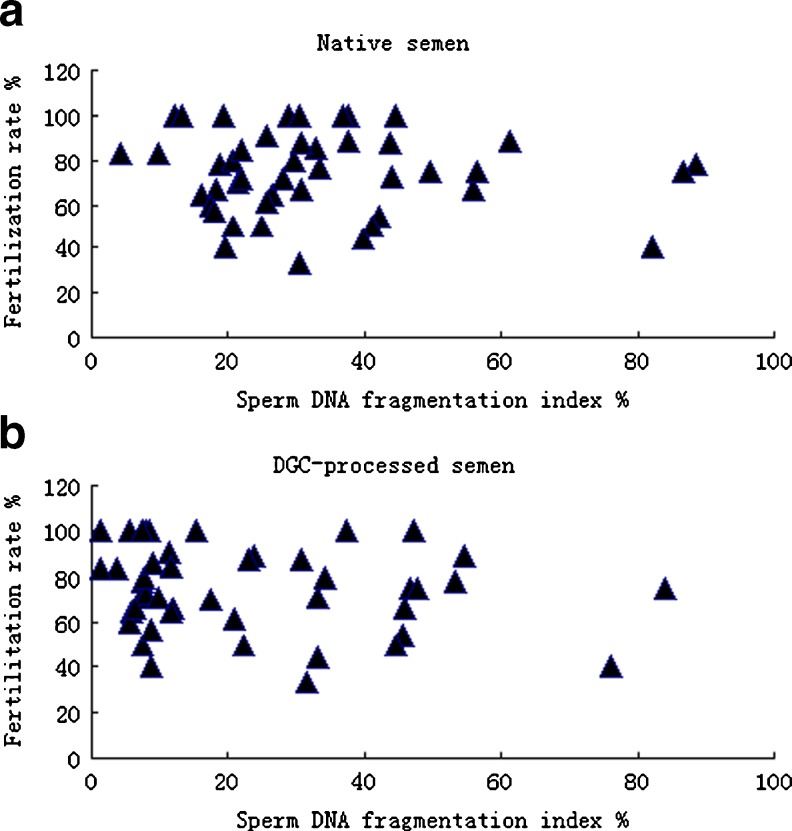
The 45 patients undergoing fresh embryo transfer were separated into high (>30 %) and low (≤30 %) sperm DFI groups. Both the raw and DGC semen parameters were examined. The 30 % threshold used to separate ‘low DFI’ from ‘high DFI’ was based on previous studies in which a SCSA was performed [9, 15, 19]. Chohan et al. reported that the TUNEL and SCD assays strongly correlated with the SCSA for sperm DNA fragmentation (r > 0.866; P < 0.001), both for infertile men and donors with known fertility. The breakdown of the DFI into 3 categories (≤15 %, >15 % – < 30 %, and ≥30 %) showed that the SCSA, TUNEL assay, and SCD test predict the same levels of DNA fragmentation [9]. In our study, there were also no significant differences between the high (>30 %) and low (≤30) sperm DFI groups, regardless of whether native or DGC-processed semen was used, with respect to the female or male age, mean sperm count, number of oocytes retrieved, oocyte maturation, and metaphase II oocytes (P > 0.05). In agreement with previous studies [20, 33, 34], we found no differences between the high and low sperm DFI groups with respect to the fertilization rate for both native and DGC semen (P > 0.05). With raw scattered plots, the fertilization rate distributions of native and DGC semen were not different between the high (>30 %) and low (≤30 %) sperm DFI groups (Fig. 3). Lin et al. suggested that normal fertilization does not ensure high-quality DNA in the paternal genome because no relationship exists between the fertilization rate and DNA fragmentation [20]. In contrast, other studies report that sperm DNA fragmentation negatively correlates with fertilization rate [6, 21, 35, 36].
Fig. 3.
Raw scattered plots showing the fertilization rates of native semen (a) and DGC-processed semen (b) in correlation with sperm DNA fragmentation rates
We observed no statistically significant differences between native and DGC semen with regard to implantation rate (P > 0.05), regardless of whether the sperm DFI was high or low. Speyer et al. also found no differences between the high and low sperm DFI groups with regard to implantation rate, with a threshold of 30 %, in patients undergoing ICSI [23].
The existing data regarding the relationship between sperm DNA integrity and pregnancy rate are conflicting. Previous investigations suggested that pregnancy is unlikely to occur when sperm nuclear DFI values are high [15, 34, 36, 37]. However, we found that a DFI level >30 % was still compatible with pregnancy. Of the 22 patients with a DFI value >30 % in the native semen group, 54.5 % (12 of 22) achieved a clinical pregnancy (Table 3). Of the 16 patients with a DFI value >30 % in the DGC semen group, 50 % (8 of 16) achieved a clinical pregnancy (Table 3). There was no difference between the high and low sperm DFI samples with regard to pregnancy rate in patients undergoing ICSI, with a threshold of 30 %, in both the native and DGC semen groups. Several published studies reached similar conclusions [17, 20, 38].
Some studies indicated that both fertilization and early embryo development did not appear to depend on sperm DNA integrity, as the embryonic genome is expressed subsequent to the second cleavage division [20, 39]. Furthermore, the selection of morphologically normal sperm for ICSI and good-quality embryos for transfer may reduce the potentially adverse effects of sperm DNA damage on the outcome of ART. Our data also support the hypothesis that ICSI is able to compensate for existing DNA strand breaks as well as sperm that are deficient according to conventional parameters [15, 40].
Recently, a meta-analysis showed a significant relationship between a high frequency of sperm with elevated DNA damage and miscarriage [41]. Robinson et al. analyzed the data with regard to the use of prepared or raw semen and found that both groups showed a significant increase in the miscarriage rate in men with high rates of DNA damage in their sperm. We also found that patients undergoing ICSI with a high DFI had a higher pregnancy loss rate (defined as spontaneous miscarriage or biochemical pregnancy) compared with patients with a low DFI in both the native and DGC semen groups (Table 3). The difference did not reach statistical significance, possibly because the number of subjects was too low. Further studies with larger sample sizes will be needed to correlate sperm DNA integrity with pregnancy loss rate.
Borini et al. suggested that a biochemical pregnancy or early miscarriage may result from blocked embryo development due to paternal genomic anomalies [18]. The oocyte is able to trigger repair mechanisms when it recognizes that the sperm DNA is damaged. However, if the number of double-strand DNA breaks in sperm is high, the oocyte cannot fully repair them, generating genetic mutations that later block or alter embryo development [42].
Tests for sperm DNA damage and interventions to decrease DNA damage should be considered part of the diagnostic and treatment resources offered to those suffering from pregnancy loss during ART procedures. A major cause of sperm DNA damage is oxidative stress (OS), caused by the generation of reactive oxygen species (ROS) resulting from contaminating leucocytes, defective sperm, and antioxidant depletion [43, 44]. A recent review showed a statistically significant increase in the pregnancy and live birth rates with the use of antioxidants. Further research is needed on the mechanisms and prevention of DNA damage, including antioxidant therapy [45].
In conclusion, we showed, for the first time, that DGC can highly significantly reduce the sperm DNA fragmentation in the semen of ICSI patients, as assessed by SCD, with the exception of patients with severe oligozoospermia. Sperm DNA fragmentation in both native and DGC semen is not related to fertilization rate, implantation rate, or clinical pregnancy rate in ICSI.
Acknowledgment
The authors thank International Peace Maternity and Child Health Hospital Affiliated for funding the study.
Footnotes
Min Wang and Jian Sun contributed equally to this work.
References
- 1.Jackson RE, Bormann CL, Hassun PA, Rocha AM, Motta EL, Serafini PC, et al. Effects of semen storage and separation techniques on sperm DNA fragmentation. Fertil Steril. 2010;94(7):2626–30. doi: 10.1016/j.fertnstert.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 2.Avendano C, Franchi A, Duran H, Oehninger S. DNA fragmentation of normal spermatozoa negatively impacts embryo quality and intracytoplasmic sperm injection outcome. Fertil Steril. 2010;94(2):549–57. doi: 10.1016/j.fertnstert.2009.02.050. [DOI] [PubMed] [Google Scholar]
- 3.Erenpreiss J, Elzanaty S, Giwercman A. Sperm DNA damage in men from infertile couples. Asian j of androl. 2008;10(5):786–90. doi: 10.1111/j.1745-7262.2008.00417.x. [DOI] [PubMed] [Google Scholar]
- 4.Benchaib M, Lornage J, Mazoyer C, Lejeune H, Salle B, Francois GJ. Sperm deoxyribonucleic acid fragmentation as a prognostic indicator of assisted reproductive technology outcome. Fertil Steril. 2007;87(1):93–100. doi: 10.1016/j.fertnstert.2006.05.057. [DOI] [PubMed] [Google Scholar]
- 5.Hughes CM, Lewis SE, McKelvey-Martin VJ, Thompson W. A comparison of baseline and induced DNA damage in human spermatozoa from fertile and infertile men, using a modified comet assay. Mol Hum Reprod. 1996;2(8):613–9. doi: 10.1093/molehr/2.8.613. [DOI] [PubMed] [Google Scholar]
- 6.Sun JG, Jurisicova A, Casper RF. Detection of deoxyribonucleic acid fragmentation in human sperm: correlation with fertilization in vitro. Biol Reprod. 1997;56(3):602–7. doi: 10.1095/biolreprod56.3.602. [DOI] [PubMed] [Google Scholar]
- 7.Manicardi GC, Bianchi PG, Pantano S, Azzoni P, Bizzaro D, Bianchi U, et al. Presence of endogenous nicks in DNA of ejaculated human spermatozoa and its relationship to chromomycin A3 accessibility. Biol Reprod. 1995;52(4):864–7. doi: 10.1095/biolreprod52.4.864. [DOI] [PubMed] [Google Scholar]
- 8.Evenson DP, Jost LK, Marshall D, Zinaman MJ, Clegg E, Purvis K, et al. Utility of the sperm chromatin structure assay as a diagnostic and prognostic tool in the human fertility clinic. Hum reprod (Oxf, Engl) 1999;14(4):1039–49. doi: 10.1093/humrep/14.4.1039. [DOI] [PubMed] [Google Scholar]
- 9.Chohan KR, Griffin JT, Lafromboise M, De Jonge CJ, Carrell DT. Comparison of chromatin assays for DNA fragmentation evaluation in human sperm. J Androl. 2006;27(1):53–9. doi: 10.2164/jandrol.05068. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez JL, Muriel L, Rivero MT, Goyanes V, Vazquez R, Alvarez JG. The sperm chromatin dispersion test: a simple method for the determination of sperm DNA fragmentation. J Androl. 2003;24(1):59–66. [PubMed] [Google Scholar]
- 11.Fernandez JL, Muriel L, Goyanes V, Segrelles E, Gosalvez J, Enciso M, et al. Simple determination of human sperm DNA fragmentation with an improved sperm chromatin dispersion test. Fertil Steril. 2005;84(4):833–42. doi: 10.1016/j.fertnstert.2004.11.089. [DOI] [PubMed] [Google Scholar]
- 12.Zhang LH, Qiu Y, Wang KH, Wang Q, Tao G, Wang LG. Measurement of sperm DNA fragmentation using bright-field microscopy: comparison between sperm chromatin dispersion test and terminal uridine nick-end labeling assay. Fertil Steril. 2010;94(3):1027–32. doi: 10.1016/j.fertnstert.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Larson KL, DeJonge CJ, Barnes AM, Jost LK, Evenson DP. Sperm chromatin structure assay parameters as predictors of failed pregnancy following assisted reproductive techniques. Hum Reprod. 2000;15(8):1717–22. doi: 10.1093/humrep/15.8.1717. [DOI] [PubMed] [Google Scholar]
- 14.Tomlinson MJ, Moffatt O, Manicardi GC, Bizzaro D, Afnan M, Sakkas D. Interrelationships between seminal parameters and sperm nuclear DNA damage before and after density gradient centrifugation: implications for assisted conception. Hum Reprod. 2001;16(10):2160–5. doi: 10.1093/humrep/16.10.2160. [DOI] [PubMed] [Google Scholar]
- 15.Bungum M, Spano M, Humaidan P, Eleuteri P, Rescia M, Giwercman A. Sperm chromatin structure assay parameters measured after density gradient centrifugation are not predictive for the outcome of ART. Hum reprod (Oxf, Engl) 2008;23(1):4–10. doi: 10.1093/humrep/dem353. [DOI] [PubMed] [Google Scholar]
- 16.Jayaraman V, Upadhya D, Narayan PK, Adiga SK. Sperm processing by swim-up and density gradient is effective in elimination of sperm with DNA damage. J Assist Reprod Genet. 2012;29(6):557–63. doi: 10.1007/s10815-012-9742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zini A, Meriano J, Kader K, Jarvi K, Laskin CA, Cadesky K. Potential adverse effect of sperm DNA damage on embryo quality after ICSI. Hum Reprod. 2005;20(12):3476–80. doi: 10.1093/humrep/dei266. [DOI] [PubMed] [Google Scholar]
- 18.Borini A, Tarozzi N, Bizzaro D, Bonu MA, Fava L, Flamigni C, et al. Sperm DNA fragmentation: paternal effect on early post-implantation embryo development in ART. Hum Reprod. 2006;21(11):2876–81. doi: 10.1093/humrep/del251. [DOI] [PubMed] [Google Scholar]
- 19.Bungum M, Humaidan P, Axmon A, Spano M, Bungum L, Erenpreiss J, et al. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum Reprod. 2007;22(1):174–9. doi: 10.1093/humrep/del326. [DOI] [PubMed] [Google Scholar]
- 20.Lin MH, Kuo-Kuang Lee R, Li SH, Lu CH, Sun FJ, Hwu YM. Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil Steril. 2008;90(2):352–9. doi: 10.1016/j.fertnstert.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 21.de la Calle JF V, Muller A, Walschaerts M, Clavere JL, Jimenez C, Wittemer C, et al. Sperm deoxyribonucleic acid fragmentation as assessed by the sperm chromatin dispersion test in assisted reproductive technology programs: results of a large prospective multicenter study. Fertil Steril. 2008;90(5):1792–9. doi: 10.1016/j.fertnstert.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Simon L, Brunborg G, Stevenson M, Lutton D, McManus J, Lewis SE. Clinical significance of sperm DNA damage in assisted reproduction outcome. Hum Reprod. 2010;25(7):1594–608. doi: 10.1093/humrep/deq103. [DOI] [PubMed] [Google Scholar]
- 23.Speyer BE, Pizzey AR, Ranieri M, Joshi R, Delhanty JD, Serhal P. Fall in implantation rates following ICSI with sperm with high DNA fragmentation. Hum Reprod. 2010;25(7):1609–18. doi: 10.1093/humrep/deq116. [DOI] [PubMed] [Google Scholar]
- 24.Lu X, Wu Y, Gao XH, Wang YW, Wang L, Sun XX. Effect of letrozole on estradiol production and P450 aromatase messenger RNA expression of cultured luteinized granulosa cells from women with and without endometriosis. Fertil Steril. 2012;98(1):131–5. doi: 10.1016/j.fertnstert.2012.03.055. [DOI] [PubMed] [Google Scholar]
- 25.Greenblatt EM, Meriano JS, Casper RF. Type of stimulation protocol affects oocyte maturity, fertilization rate, and cleavage rate after intracytoplasmic sperm injection. Fertil Steril. 1995;64(3):557–63. doi: 10.1016/s0015-0282(16)57792-9. [DOI] [PubMed] [Google Scholar]
- 26.Zini A, Mak V, Phang D, Jarvi K. Potential adverse effect of semen processing on human sperm deoxyribonucleic acid integrity. Fertil Steril. 1999;72(3):496–9. doi: 10.1016/S0015-0282(99)00295-2. [DOI] [PubMed] [Google Scholar]
- 27.Zini A, Finelli A, Phang D, Jarvi K. Influence of semen processing technique on human sperm DNA integrity. Urology. 2000;56(6):1081–4. doi: 10.1016/S0090-4295(00)00770-6. [DOI] [PubMed] [Google Scholar]
- 28.Zini A, Nam RK, Mak V, Phang D, Jarvi K. Influence of initial semen quality on the integrity of human sperm DNA following semen processing. Fertil Steril. 2000;74(4):824–7. doi: 10.1016/S0015-0282(00)01495-3. [DOI] [PubMed] [Google Scholar]
- 29.Aitken RJ, Krausz C. Oxidative stress, DNA damage and the Y chromosome. Reprod (Camb, Engl) 2001;122(4):497–506. doi: 10.1530/rep.0.1220497. [DOI] [PubMed] [Google Scholar]
- 30.Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71(1):162–4. doi: 10.1086/341096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enciso M, Alfarawati S, Wells D. Increased numbers of DNA-damaged spermatozoa in samples presenting an elevated rate of numerical chromosome abnormalities. Hum reprod (Oxf, Engl) 2013;28(6):1707–15. doi: 10.1093/humrep/det077. [DOI] [PubMed] [Google Scholar]
- 32.Meseguer M, Santiso R, Garrido N, Garcia-Herrero S, Remohi J, Fernandez JL. Effect of sperm DNA fragmentation on pregnancy outcome depends on oocyte quality. Fertil Steril. 2011;95(1):124–8. doi: 10.1016/j.fertnstert.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 33.Morris ID, Ilott S, Dixon L, Brison DR. The spectrum of DNA damage in human sperm assessed by single cell gel electrophoresis (Comet assay) and its relationship to fertilization and embryo development. Hum reprod (Oxf, Engl) 2002;17(4):990–8. doi: 10.1093/humrep/17.4.990. [DOI] [PubMed] [Google Scholar]
- 34.Larson-Cook KL, Brannian JD, Hansen KA, Kasperson KM, Aamold ET, Evenson DP. Relationship between the outcomes of assisted reproductive techniques and sperm DNA fragmentation as measured by the sperm chromatin structure assay. Fertil Steril. 2003;80(4):895–902. doi: 10.1016/S0015-0282(03)01116-6. [DOI] [PubMed] [Google Scholar]
- 35.Huang CC, Lin DP, Tsao HM, Cheng TC, Liu CH, Lee MS. Sperm DNA fragmentation negatively correlates with velocity and fertilization rates but might not affect pregnancy rates. Fertil Steril. 2005;84(1):130–40. doi: 10.1016/j.fertnstert.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 36.Simon L, Lutton D, McManus J, Lewis SE. Sperm DNA damage measured by the alkaline Comet assay as an independent predictor of male infertility and in vitro fertilization success. Fertil Steril. 2011;95(2):652–7. doi: 10.1016/j.fertnstert.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Virro MR, Larson-Cook KL, Evenson DP. Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil Steril. 2004;81(5):1289–95. doi: 10.1016/j.fertnstert.2003.09.063. [DOI] [PubMed] [Google Scholar]
- 38.Payne JF, Raburn DJ, Couchman GM, Price TM, Jamison MG, Walmer DK. Redefining the relationship between sperm deoxyribonucleic acid fragmentation as measured by the sperm chromatin structure assay and outcomes of assisted reproductive techniques. Fertil Steril. 2005;84(2):356–64. doi: 10.1016/j.fertnstert.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 39.Tesarik J, Mendoza C, Greco E. Paternal effects acting during the first cell cycle of human preimplantation development after ICSI. Hum Reprod. 2002;17(1):184–9. doi: 10.1093/humrep/17.1.184. [DOI] [PubMed] [Google Scholar]
- 40.Ozmen B, Koutlaki N, Youssry M, Diedrich K, Al-Hasani S. DNA damage of human spermatozoa in assisted reproduction: origins, diagnosis, impacts and safety. Reprod biomed online. 2007;14(3):384–95. doi: 10.1016/S1472-6483(10)60883-8. [DOI] [PubMed] [Google Scholar]
- 41.Robinson L, Gallos ID, Conner SJ, Rajkhowa M, Miller D, Lewis S, et al. The effect of sperm DNA fragmentation on miscarriage rates: a systematic review and meta-analysis. Hum Reprod. 2012;27(10):2908–17. doi: 10.1093/humrep/des261. [DOI] [PubMed] [Google Scholar]
- 42.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–61. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 43.Lewis SE, Boyle PM, McKinney KA, Young IS, Thompson W. Total antioxidant capacity of seminal plasma is different in fertile and infertile men. Fertil Steril. 1995;64(4):868–70. doi: 10.1016/s0015-0282(16)57870-4. [DOI] [PubMed] [Google Scholar]
- 44.Tremellen K. Oxidative stress and male infertility–a clinical perspective. Hum Reprod Update. 2008;14(3):243–58. doi: 10.1093/humupd/dmn004. [DOI] [PubMed] [Google Scholar]
- 45.Showell MG, Brown J, Yazdani A, Stankiewicz MT, Hart RJ. Antioxidants for male subfertility. The Cochrane database of systematic reviews. 2011(1):CD007411. doi:10.1002/14651858.CD007411.pub2 [DOI] [PubMed]