Abstract
Biosurfactant screening was made among the eight halophilic bacterial genera isolated from Kovalam solar salt works in Kanyakumari of India. After initial screening, Kocuria sp. (Km), Kurthia sp. (Ku) and Halococcus sp. (Hc) were found to have positive biosurfactant activity. Biosurfactant derived from Kocuria sp. emulsified more than 50% of the crude oil, coconut oil, sunflower oil, olive oil and kerosene when compared to the other strains. Further, Kocuria marina BS-15 derived biosurfactant was purified and characterized by TLC, FTIR and GC–MS analysis. The TLC analysis revealed that, the purified biosurfactants belong to the lipopeptide group. The IR spectrum results revealed that functional groups are R2C N N, alkenes and N–H. The GC–MS analysis confirmed the compound as Nonanoic acid and Cyclopropane with the retention time of 12.78 and 24.65, respectively.
Keywords: Kocuria marina, Biosurfactant, Emulsification, Nonanoic acid
1. Introduction
Biosurfactants are amphiphilic compounds produced on living surfaces, mostly on microbial cell surfaces or excreted extracellularly and contain hydrophobic and hydrophilic moieties. They reduce surface tension and interfacial tension between individual molecules at the surface and interface respectively (Karanth et al., 1999). The microbial surfactants (MS) are complex molecules covering a wide range of chemical types including peptides, fatty acids, phospholipids, glycolipids, antibiotics, lipopeptides, etc (Cooper and Zajic, 1980). These properties resulted in detergency, emulsifying, foaming and disparity traits as well as increase in solubility and mobility of hydrophobic organic compounds (singh et al., 2007).
Research in the area of biosurfactants has expanded quite a lot in recent years due to its potential use in different areas. In recent years increasing global environmental awareness has led to much more interest in microbial surfactants compared to their chemical counter parts. It is due to the unique properties of biosurfactants including biodegradability, low toxicity, mild production conditions, and environmental acceptability, lower critical micelle concentration, higher selectivity as well as better activity at extreme temperature, pH and salinity (Das and Mukherjee, 2007).
Biosurfactant producing organisms are very diverse and have been isolated from a wide variety of environments, including soil, sea water, marine sediments, oil fields (Bodour et al., 2003) and even extreme environments (Cameotra and Makkar, 1998). The synthesis of these surface active molecules takes place by de novo pathway and/or assembly from substrates (Syldatk and Wagner, 1987). This wide range of structural diversity results in a broad spectrum of potential industrial applications including production of food, cosmetics, and pharmaceuticals, agriculture, mining, enhanced oil recovery, transportation of crude oil, cleaning oil storage tanks and pipelines and soil remediation (Pastewski et al., 2006).
The search for novel biosurfactants in extremophiles seems to be particularly promising since they have particular adaptations to increased stability in adverse environments and the microbial products are highly stable and important in medical biotechnology. In the solar salt works of southern India, there is a wide diversity of moderate and extreme halo bacterial sp. having pharmacologically important compounds including biosurfactants (Donio et al., 2013). There are very few reports on biosurfactant producers in hypersaline environments and in the recent years, there has been a greater increase in interest and importance in halophilic bacteria for biomolecules. Halotolerant or halophilic microorganisms, able to live in saline environments, offer a multitude of actual or potential applications in various fields of biotechnology (Margesin and Schinner, 2001). Halophiles, which have a unique lipid composition, may have an important role to play as surface-active agents. The archae bacterial ether-linked phytanyl membrane lipid of the extremely halophilic bacteria has been shown to have surfactant properties (Post and Collins, 1982). Yakimov et al. (1995) reported the production of biosurfactant by a halotolerant Bacillus species and its potential in enhanced oil recovery; Bacillus licheniformis strain BAS 50 was able to grow and produce a lipopeptide surfactant when cultured on a variety of substrates at salinities up to 13% NaCl.
Individual wastes alone have resulted in high yields of biosurfactants by new efficient microbial isolates viz. Kocuria turfanesis strain BS-J and Pseudomonas aeruginosa strain BS-P (Dubey et al., 2012). Microbial communities like Acinetobacter, Arthrobacter, Pseudomonas, Halomonas, Bacillus, Rhodococcus, Enterobacter and yeast have been reported to produce biosurfactants (BS)/bioemulsifiers (BE) (Das et al., 2008a). About 56 reports including 35 for bioemulsifier, 12 for glycolipid and 9 for other types are available on different types of BS/BE produced by marine microorganisms. The present study intends screening of the biosurfactants from the eight halobacterial species which were isolated from the solar salt works in Kanyakumari district. Among the eight bacterial isolates that were screened for potential biosurfactant producer(s), K. marina BS-15 was found to be able to produce biosurfactant by various screening methods.
2. Materials and methods
2.1. Sampling and isolation of biosurfactant producing bacteria
Brine water had the salinity of 230‰ and 9 pH was collected from the condenser pond of Kovalam (8°05′04.35″ N 77°31′17.07″ E) solar salt works in India. The samples were serially diluted and the decimal dilutions spread on nutrient agar medium containing 5%, 10%, 15% of NaCl with a thin film of crude oil (Himedia, Mumbai, India). Bacterial colonies surrounded by an emulsified halo were identified as a biosurfactant producer, after 37 °C incubation (Morikawa et al., 1992). The biosurfactant producing bacterial colonies were picked up and inoculated into the nutrient broth containing 10% of NaCl. After 24 h, the bacterial cells were harvested by centrifugation at 5000 rpm and were subjected to different biosurfactant screening assays.
2.2. Screening assays for biosurfactant production
The oil displacement test is a method used to determine the surface activity by measuring the diameter of the clear zone, which occurs after dropping a surfactant-containing solution on a thin layer of oil on water. The binomial diameter allows an evaluation of the surface tension reduction efficiency of a given biosurfactant (Rodrigues et al., 2006). Drop-collapse test by adding mineral oil in 96-well microtitre plates (Jain et al., 1991); Emulsification activity by adding kerosene and equal volume of cell free supernatant (Cooper and Goldenberg, 1987) and hemolytic activity in 5% blood agar plate.
2.3. Biosurfactant detection by methylene blue method
Assay was carried out using the method of Jones and Esposito (2000) with some modifications. One milliliter of mineral salt medium culture was vigorously shaken for 30 s with 0.003% methylene blue, and then an equal amount of chloroform was added to the sample. The mixture was left for 20 min to extract the methylene blue anionic surfactant ion pair into chloroform layer. At this point, it is necessary to note that all the blue dye has migrated into the chloroform layer. The tube was centrifuged at 3000 rpm for 5 min. After the extraction with chloroform, the absorbance of each sample was measured at 625 nm against a reference of pure grade chloroform.
2.4. Emulsification activity
The emulsification activity index was measured using the method described by Cooper and Goldenberg (1987) in which 2 ml of the kerosene was added to equal volume of cell free supernatant and homogenized in a vortex at high speed for 2 min. The emulsification stability was measured after 24 h and the emulsification index was calculated by dividing the measured height of the emulsion layer by the total height of the liquid layer and multiplying by 100. The emulsification activity of the strain was compared with the standards including SDS and Tween 80.
2.5. Phenotypic and genomic identification of BS-15
Based on the higher biosurfactant activity, the BYS2 bacterial strain was identified based on Gram staining, motility, and other biochemical tests according to Bergey’s Manual of Systematic Bacteriology (Holt et al., 1994).
One hundred nano gram of genomic DNA was extracted from the BS-15 bacterial strain and the 16S rRNA gene was amplified using the universal primer with standard PCR protocol. The PCR products were purified by Gel extraction kit (Medox Biotech India Pvt. Ltd) and sequenced (Chromos Biotech Pvt. Ltd, Bangalore). The nucleotides of the 16S rRNA sequence were matched with the other microbes in the NCBI database using BLAST program. The construction of phylogenetic tree carried out by Geneious Basic software and evolutionary history were inferred using the neighbor-joining method (Sneath and Sokal, 1973).
2.6. Extraction and partial purification of biosurfactants produced by K. marina BS-15
After the removal of the bacterial cells from the culture media by centrifugation, the obtained supernatant was treated by acidification to pH 2.0 using 6 M HCl solution, and the acidified supernatant was left overnight at 4 °C for the complete precipitation of the biosurfactant. After centrifugation at 8500 rpm for 20 min, the precipitate was dissolved in distilled water at pH 7.0, followed by the biosurfactant extraction step with a solvent having a 65:15 chloroform-to-methanol ratio at room temperature. Then the organic phase was transferred to a round-bottom flask connected to a rotary evaporator to remove the solvent, yielding biosurfactant product. About 1.97 mg of the biosurfactant was extracted per liter of culture medium. The components in the extracted biosurfactant were separated on silica gel 60 plates (Merck) using a solvent system having a 65:25:4 chloroform-to methanol-to-water ratios. The separated components were detected by iodine vapor and UV light exposure (Nitschke and Pastore, 2006).
2.7. Partial characterization
2.7.1. Fourier transmission infra red spectroscopy (FT-IR)
The basic functional groups of the purified Biosurfactants from halophilic K. marina BS-15 were analyzed qualitatively by the Fourier transform infra red (FTIR) method described by Kemp (1991).
2.7.2. Gas chromatography–mass spectrometry (GC–MS)
GC–MS analysis of partially purified biosurfactants was analyzed individually using Agilent GC–MS 5975 Inert XL MSD (United States) gas chromatography equipped with J and W 122–5532G DB-5 ms 30 × 0.25 mm × 0.25 μm and mass detector (EM with replaceable horn) that operated in EMV mode. Helium was used as carrier gas with the flow rate of 1.0 ml min−1. The injection port temperature was operated at 250 °C. The column oven temperature was held at 80 °C for 2 min then programmed at 10 °C min−1 to 250 °C, which was held for 0 min, and then at 5 °C min−1 to 280 °C which was held for 9 min. Electron impact spectra in positive ionization mode were acquired between m/z 40 and 450.
3. Results and discussion
3.1. Biosurfactant screening from halophilic bacteria
Seven halophilic bacterial sps isolated from the solar salt works were screened for biosurfactant production. Among the strains, K. marina BS-15 was highly positive for drop collapse test, oil displacement test and hemolytic activity. The other strain, Kurthia sp. (Ku) was highly positive for drop collapse test and Halococcus sp. (Hc) was highly positive for oil displacement test. The other strains such as Proteus sp. (Pr), Photobacterium sp. (Pb), Aerococcus sp. (Ac), Clavibactor sp. (Cb) failed to produce biosurfactants (Table 1). Satpute et al. (2008) suggested that more screening methods are essential to identify different types of biosurfactants from potential biosurfactant producers. The test conducted in our earlier works (Donio et al., 2013) like drop collapse test, Oil spreading technique and hemolytic activity helped to screen the lipopeptides and polymeric type of biosurfactants from the Halomonas sp. BS4. It is also essential to perform different biosurfactant screening methods for the different biosurfactant producing microbes, because biosurfactants are a heterogeneous nature of secondary metabolites with surface active properties. The drop collapse and oil displacement methods are the most effective tools to prove the biosurfactant production in many of the bacterial strains.
Table 1.
Biosurfactant screening of halophilic bacterial sp. isolated from solar salt works in India.
| Halophilic bacterial sp. | Screening methods |
||
|---|---|---|---|
| Drops collapse | Oil displacement | Hemolytic activity | |
| Proteus sp. (Pr) | _ | _ | _ |
| K. marina BS-15 | +++ | ++ | +++ |
| Photobacterium sp. (Pb) | _ | _ | _ |
| Aerococcus sp. (Ac) | _ | _ | _ |
| Kurthia sp. (Ku) | +++ | ++ | + |
| Coprococcus sp. (Cp) | ++ | ++ | _ |
| Clavibacter sp. (Cb) | _ | + | _ |
| Halococcus sp. (Hc) | ++ | +++ | ++ |
+++, higher activity; ++: medium activity; +, less activity; –, no activity.
3.2. Biosurfactant detection by methylene blue method
The reactivity between biosurfactants and methylene blue is given in Fig. 1. The OD at 625 nm revealed that, the strains of K. marina BS-15, Kurthia sp. (Ku), Coprococcus sp. (Cc) and Halococcus sp. (Hc) were able to produce biosurfactants. The strains of Proteus sp. (Pr), Photobacterium sp. (Pb) and Clavibacter sp. (Cb) were unable to produce biosurfactants. Among the bacterial strains, K. marina BS-15 and Halococcus sp. had a highly significant (P <= 0.001) biosurfactant production when compared to the others. Methylene blue detection is one of the efficient methods to detect anionic surfactants and the biosurfactants produced from microbes react with methylene blue and form anionic surfactant ion pair (Siegmund and Wagner, 1991). This was migrated into the chloroform layer and confirmed the production of biosurfactants in the production medium. Aparna et al. (2012) detected the irhamnolipid type of biosurfactants from Pseudomonas sp. 2B using the CTAB–Methylene blue agar medium based method.
Figure 1.
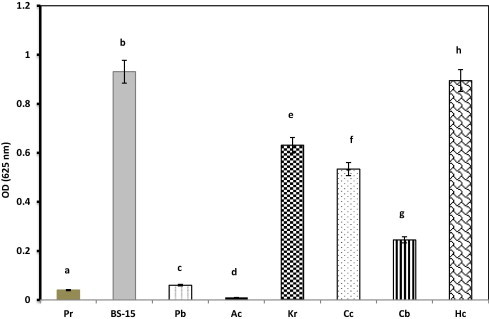
Methylene blue method detection of biosurfactants from halobacterial sp. Means with the same superscript do not significantly (P <= 0.001) each other’s – One Way ANOVA.
3.3. Emulsification activity of biosurfactant
The emulsification activities of the halophilic bacterial strains are given in Table 2. The better emulsification activities (E24) like 70.90% by Halococcus sp. (Hc) against kerosene, 68.02% by K. marina BS-15 against sunflower oil, 65.12% by Halococcus sp. (Hc) against olive oil, 63.27% by K. marina BS-15 against coconut oil, 56.06% by K. marina BS-15 against crude oil and 56.05% by Halococcus sp. (Hc) S8 against sunflower oil were observed. Two way ANOVA revealed that, the values significantly differed from each other’s (F = 18.37; P <= 0.001). The results indicated that the biosurfactant was capable of effectively emulsifying both aromatic and aliphatic hydrocarbons. The present results also revealed that, the hydrocarbon enriched oils and plant oils were the suitable source for oil degradation and biosurfactant production respectively. Emulsification potential of Rhodococcus sp. TA6 was studied successfully by Shavandi et al. (2011) using several hydrocarbon substrates as a sole carbon source including pentane to light motor oil. Prieto et al. (2008) have reported that soybean oil, diesel oil, gasoline and cyclohexane are good substrates for emulsification by rhamnolipid-type biosurfactant of P. aeruginosa while n-hexane and fish oil result in poor emulsification.
Table 2.
Emulsification activity (%) of crude biosurfactant from halobacterial strains against oils.
| Oils used | Control |
Emulsification activity (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SDS | Tween 80 | Pr | BS-15 | Pb | Ac | Kr | Cc | Cb | Hc | |
| Crude oil | 50.06 ± 0.50 | 56.04 ± 0.05 | 27.19 ± 0.26 | 52.06 ± 0.05 | 23.15 ± 0.21 | 25.21 ± 0.33 | 50.08 ± 0.10 | 19.15 ± 0.21 | 21.23 ± 0.20 | 37.28 ± 0.44 |
| Coconut oil | 65.1 ± 0.1 | 59.27 ± 0.45 | 32.05 ± 0.04 | 63.27 ± 0.45 | 18.12 ± 0.15 | 17.22 ± 0.33 | 46.1 ± ‘ 0.1 | 22.21 ± 0.33 | 15.05 ± 0.05 | 43.05 ± 0.05 |
| Sunflower oil | 63.17 ± 0.28 | 61.24 ± 0.39 | 29.16 ± 0.20 | 68.02 ± 0.02 | 26.29 ± 0.49 | 28.24 ± 0.39 | 34.1 ± 0.26 | 18.30 ± 0.51 | 27.15 ± 0.22 | 56.05 ± 0.05 |
| Olive oil | 71.25 ± 0.38 | 68.21 ± 0.33 | 25.28 ± 0.44 | 50.43 ± 0.37 | 15.25 ± 0.38 | 30.08 ± 0.10 | 29.30 ± 0.51 | 27.1 ± 0.16 | 12.15 ± 0.21 | 65.12 ± 0.15 |
| Kerosene | 52.46 ± 0.41 | 53.2 ± 0.26 | 31.15 ± 0.21 | 37.09 ± 0.10 | 32.15 ± 0.21 | 14.17 ± 0.28 | 43.19 ± 0.26 | 29.22 ± 0.32 | 31.17 ± 0.28 | 70.90 ± 0.27 |
The values are significantly differed each other’s (F = 18.37; P <= 0.001) – Two Way ANOVA.
Pr, Proteus sp.; Km, K. marina BS-15; Pr, Photobacterium sp.; Ac, Aerococcus sp.; Kr–Kurthia sp.; Cp, Coprococcus sp.; Cb, Clavibacter sp.; and Hc, Halococcus sp.
3.4. Phenotypic identification of biosurfactant producing halophilic bacteria
The morphological, biochemical and physiological tests revealed that, the eight strains isolated from solar salt works were Proteus sp. (Pr), Kokuria sp. (Km), Photobacterium sp. (Pb), Aerococcus sp. (Ac), Kurthia sp. (Ku), Coprococcus sp. (Cp), Clavibacter sp. (Cb) and Halococcus sp. (Hc) etc (Table 3). Among these bacterial strains, Kokuria sp. (Km) was highly positive for biosurfactant production whereas Kurthia sp. (Ku) and Halococcus sp. (Hc) were moderately positive for biosurfactant production. Kokuria sp. (Km) was a Gram positive cocci, non motile, positive for indole, methyl red and VP tests. Also they ferment glucose, sucrose and fructose etc. Kocuria spp. are the members of the Micrococcaceae family that are frequently found in the environment (Lee et al., 2009). Kim et al. (2004) isolated and identified the K. marina KMM 3905T from Troitsa Bay marine sediment. This strain grows in the temperature range of 4–43 °C, tolerates maximum of 15% NaCl, positive for urease and nitrate reduction, negative for oxidase and alkaline phosphatase, and negative for acid production from glucose, lactose, or sucrose. At present the genus Kocuria comprises of 18 species with validly published names (http://www.bacterio.cict.fr/k/kocuria.html) and all these species have been isolated from different environmental sources including K. rosea, K. varians and K. kristinae (Stackebrandt et al., 1995), K. himachalensis (Mayilraj et al., 2006), K. halotolerans (Tang et al., 2009) and K. assamensis (Kaur et al., 2011).
Table 3.
Phenotypic identification of biosurfactant producing Halobacterial sp. from Kovalam solar salt works.
| Test | Isolates | |||||||
|---|---|---|---|---|---|---|---|---|
| Pr | BS-15 | Pb | Ac | Kr | Cc | Cb | Hc | |
| Gram staining | _ | + | _ | _ | + | _ | + | + |
| Cell shape | Long rod | Cocci | Shot rod | Cocci | Rod | Cocci | Cocci | Cocci |
| Motility | Motile | Non motile | Non motile | Non motile | Motile | Non motile | Non motile | Non motile |
| Indole | + | + | _ | _ | _ | _ | _ | + |
| Methyl red | + | + | _ | + | + | + | + | + |
| VP | + | + | _ | _ | _ | + | _ | + |
| Citrate | + | + | _ | + | + | _ | _ | + |
| Oxidase | + | _ | _ | _ | + | _ | _ | _ |
| Catalase | _ | + | _ | _ | + | _ | _ | + |
| Nitrate | _ | + | _ | _ | _ | + | _ | + |
| Urease | + | + | _ | _ | + | + | _ | _ |
| TSI | + | + | + | + | + | _ | _ | _ |
| Gelatin | _ | _ | _ | _ | _ | _ | _ | _ |
| Starch | _ | _ | _ | _ | _ | _ | _ | _ |
| CHO fermentation | ||||||||
| Glucose | _ | + | _ | _ | _ | + | + | + |
| Sucrose | _ | + | _ | _ | _ | + | + | _ |
| Galactose | _ | _ | _ | _ | + | + | _ | + |
| Maltose | _ | _ | _ | _ | + | _ | _ | _ |
| Fructose | _ | + | _ | _ | _ | _ | _ | + |
3.5. Genomic identification of K. marina BS-15
Phylogenetic and evolutionary analysis of the 16S rRNA sequence revealed that, K. marina BS-15 shared high similarity to other K. marina strains. K. marina BS-15 also had high similarity to the strains of K. marina RB-210, Kocuria sp. QW12 and K. marina S48 (Fig. 2). The strain was deposited in NCBI database. The strain name and GenBank accession number are K. marina BS-15, KC594576.1 respectively. K. marina is also most closely related to K. rhizophila DSM 11926, K. varians DSM 20033, and K. carniphila CCM 132 based on the report by Kim et al. (2004) and constructing phylogenetic analysis using 16S rRNA gene sequences. However, K. marina forms an independent phylogenetic lineage within Kocuria (Tvrzova et al., 2005).
Figure 2.
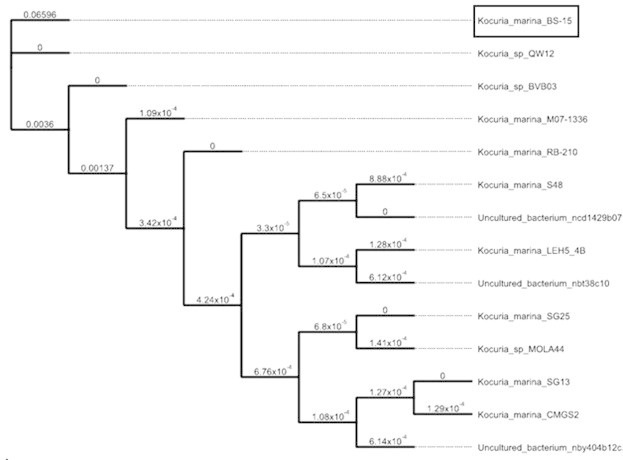
Graphical phylogenetic tree analysis of Kocuria marina BS-15 based on 16S rRNA gene sequence data compare with other sp.
3.6. Purification of K. marina BS-15 yielding biosurfactan
Thin layer chromatography (TLC) data revealed a single spot with Rf value of 0.65 under UV detection. Based on the Rf value, the spot was concluded as a lipid moiety containing the compound of lipopeptide (Fig. 3). This preliminary result suggests that the partially purified biosurfactant produced by K. marina BS-15 should contain a lipopeptide. Anyanwu et al. (2011) confirmed in their study the TLC data with the Rf value of 0.68 and 0.70 after iodine treatment as lipopeptide. Our earlier study Donio et al. (2013) also confirmed that the biosurfactant extracted from halophilic Bacillus BS-3 had the Rf value of 0.68 as lipopetide type. Study conducted by Vater et al., 2002 also substantiated one surfactant with the Rf values of 0.62 as lipopeptide.
Figure 3.
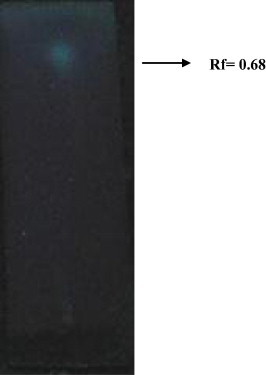
Thin layer chromatography analysis of Kocuria marina BS-15 biosurfactants.
3.7. Partial characterization of biosurfactant
The FTIR analysis (Fig. 4) revealed that the peak at 491 cm−1 is due to C–I (Carbon–Iodine) bond. The peak at 605 cm−1 and the peaks at 651 and 671 cm−1 confirm the presence of C-Br. The peaks at 2084 and 3147 cm−1 are due to the presence of cumulated system R2C N N in the sample. Also, an absorption band at 993 cm−1 showed stretching mode of the RCH CH2 indicating the presence of alkenes. The stretch, 3429 and 2360 cm−1 denoted as the N–H group. The transmittance around at 1400 cm−1 referred to the aliphatic chain of the C–H group. An intense stretching peak 1159, 1537 and 1626 cm−1 indicates the presence of RNO2 groups. The availability of all these functional groups firmly substantiated that the biosurfactant is a peptide nature. Our previous work (Donio et al., 2013a) also confirmed that, the lipopeptide type of biosurfactant isolated from halophilic Bacillus sp. BS-3 had the IR stretch of 3429 cm−1 donated N–H group and the other peak at 2084 cm−1 corresponds to cumulated system like R2C N N in the sample.
Figure 4.
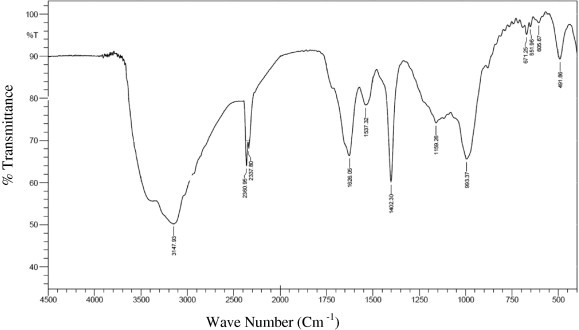
FT-IR spectrum of the partially purified biosurfactant produced by the halophilic bacteria Kocuria marina BS-15.
The GC–MS analysis (Fig. 5 and Table 4) characterized, two lipid based compounds that had the biosurfactant properties. The peak at the retention time of 12.78 was confirmed as Nonanoic acid with the molecular weight of 148 and formula of C9H18O2. Findings by Kiran et al. (2010a) supported that the surface active compound produced by Nocardiopsis lucentensis MSA04 was characterized by GC–MS analysis as glycolipid with a hydrophobic non-polar hydrocarbon chain (nonanoic acid methyl ester) and hydrophilic sugar, 3-acetyl 2,5 dimethyl furan reported by Kiran et al. (2010a). Lipopeptide families of biosurfactant including nonanoic acid, 9-oxo-, methyl ester and brevifactin were characterized by GC–MS analysis in Brevibacterium aureum MSA13 (Kiran et al., 2010b). The lipid based compound Cyclopropane (molecular weight 42.08; formula C3H6) was also detected with the peak having the retention time of 24.65. Cyclopropane ring-containing lipids, especially in phospholipids and glycolipids, were reported for many bacteria (Mizoguchi et al., 2013). Escherichia coli, whose cell membranes were dominantly composed of cyclopropane ring containing phospholipids and fatty acids (Brown et al., 1997). Two polycyclopropane fatty acid derivatives, FR-900846 (2) and U-106305 (3) were isolated from Streptoverticillium fervens and Streptomyces (Grogan and Cronan, 1997). Dubey et al. (2012) also confirmed the biosurfactant production in K. turfanesis strain-J using curd whey as substrates. The present study revealed that, the search for novel biosurfactants in extremophiles seems to be particularly promising since they have particular adaptations like increased stability in adverse environments and the microbial products are highly stable and important in various fields. These halo bacterial biosurfactants from solar salt works will help to develop more valuable eco friendly pharmacological products to the pharmacological industries.
Figure 5.
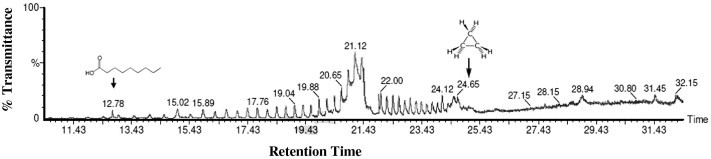
GC analysis of partially purified biosurfactant from halophilic Kocuria marina BS-15.
Table 4.
Biosurfactant characterized from Kocuria marina BS-15 by GC–MS analysis.
| Retention time | Name of the compounds | Molecular formula | Molecular weight | Molecular structure |
|---|---|---|---|---|
| 12.78 | Nonanoic acid | C9H18O2 | 158 | |
| 24.65 | Cyclopropane | C3H6 | 42.08 | 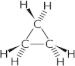 |
Footnotes
Peer review under responsibility of King Saud University.
References
- Anyanwu C.U., Obi S.K.C., Okolo B.N. Lipopeptide biosurfactant production by Serratia marcescens NSK-1 strain isolated from petroleum-contaminated soil. J. Appl. Sci. Res. 2011;7(1):79–87. [Google Scholar]
- Aparna A., Srinikethan G., Smitha H. Production and characterization of biosurfactant produced by a novel Pseudomonas sp. 2B. Colloids Surf. B. 2012;95:23–29. doi: 10.1016/j.colsurfb.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Bodour A.A., Drees K.P., Miller-Maier R.M. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated Arid Southwestern soils. Appl. Environ. Microbiol. 2003;69:3280–3287. doi: 10.1128/AEM.69.6.3280-3287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.L., Ross T., McMeekin T.A., Nichols P.D. Acid habituation of Escherichia coli and the potential role of cyclopropane fatty acids in low pH tolerance. Int. J. Food Microbiol. 1997;37:163–173. doi: 10.1016/s0168-1605(97)00068-8. [DOI] [PubMed] [Google Scholar]
- Cameotra S.S., Makkar R.S. Synthesis of biosurfactants in extreme conditions. Appl. Microbiol. Biotechnol. 1998;50:520–529. doi: 10.1007/s002530051329. [DOI] [PubMed] [Google Scholar]
- Cooper D.G., Goldenberg B.G. Surface-active agents from two Bacillus species. Appl. Environ. Microbiol. 1987;53:224–229. doi: 10.1128/aem.53.2.224-229.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D.G., Zajic J.E. Surface active compounds from microorganisms. Adv. Appl. Microbiol. 1980;26:229–253. [Google Scholar]
- Das K., Mukherjee A.K. Comparison of lipopeptide biosurfactants production by Bacillus subtilis strains in submerged and solid state fermentation systems using a cheap carbon source: some industrial applications of biosurfactants. Process. Biochem. 2007;42:1191–1199. [Google Scholar]
- Das P., Mukherjee S., Sen R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J. Appl. Microbiol. 2008;04:675–684. doi: 10.1111/j.1365-2672.2007.03701.x. [DOI] [PubMed] [Google Scholar]
- Donio, M.B.S., Ronica, F.A., Thanga Viji, V., Velmurugan, S., Adlin Jenifer, J., Michaelbabu, M., Dhar, P., Citarasu, T., 2013. Halomonas sp. BS4, A Biosurfactant Producing Halophilic Bacterium isolated From Solar Salt Works in India and their Biomedical Importance, Springer Plus, Berlin, 2, p. 149. [DOI] [PMC free article] [PubMed]
- Donio M.B.S., Ronica S.F.A., Thanga Viji V., Velmurugan S., Adlin Jenifer J., Michaelbabu M., Citarasu T. Isolation and characterization of halophilic Bacillus sp. BS3 able to produce pharmacologically important biosurfactants. Asia Pac.. J. Trop. Med. 2013:876–883. doi: 10.1016/S1995-7645(13)60156-X. [DOI] [PubMed] [Google Scholar]
- Dubey K.V., Charde P.N., Meshram S.U., Yadav S.K., Singh S., Juwarkar A.A. Potential of new microbial isolates for biosurfactant production using combinations of distillery waste with other industrial wastes. J. Petrol. Environ. Biotechnol. 2012;S1:002. doi: 10.4172/2157-7463.S1-002. [DOI] [Google Scholar]
- Grogan D.W., Cronan J.E., Jr. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 1997;61(4):429–441. doi: 10.1128/mmbr.61.4.429-441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J.G., Krieg N.R., Sheath P.H.A., Staley J.T., Williams S.T. 9th ed. The Williams & Wilkins Co.; Baltimore, Md: 1994. ‘Bergey’s manual of Determinative bacteriology. [Google Scholar]
- Jain D.K., Collins-Thompson D.L., Lee H., Trevors J.T. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods. 1991;13:271–279. [Google Scholar]
- Jones T.J., Esposito M.F. An assay evaluation of the methylene blue method for the detection of anionic surfactan in urine. J. Anal. Toxicol. 2000;24:323–327. doi: 10.1093/jat/24.5.323. [DOI] [PubMed] [Google Scholar]
- Karanth N.G.K., Deo P.G., Veenanadig N.K. Production of biosurfactants and their importance. Curr. Sci. 1999;77:116–125. [Google Scholar]
- Kaur C., Kaur I., Raichand R., Bora T.C., Mayilraj S. Description of a novel actinobacterium Kocuria assamensis sp. nov., isolated from a water sample collected from the river Brahmaputra, Assam, India. Antonie Van Leeuwenhoek. 2011;99:721–726. doi: 10.1007/s10482-010-9547-9. [DOI] [PubMed] [Google Scholar]
- Kemp W. third ed. Palgrave Published; New York: 1991. Organic Spectroscopy. pp. 243–269. [Google Scholar]
- Kim S.B., Nedashkovskaya O.I., Mikhailov V.V., Han S.K., Kim K.O., Rhee M.S., Bae K.S. Kocuria marina sp. nov, a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2004;54:1617–1620. doi: 10.1099/ijs.0.02742-0. [DOI] [PubMed] [Google Scholar]
- Kiran G.S., Thomas T.A., Selvin J. Production of a new glycolipid biosurfactant from marine Nocardiopsis lucentensis MSA04 in solid-state cultivation. Colloids Surf. B. 2010;78:8–16. doi: 10.1016/j.colsurfb.2010.01.028. [DOI] [PubMed] [Google Scholar]
- Kiran G., Anto Thomas T., Selvin J., Sabarathnam B., Lipton A.P. Optimization and characterization of a new lipopeptide biosurfactant produced by marine Brevibacterium aureum MSA13 in solid state culture. Bioresour. Technol. 2010;101:2389–2396. doi: 10.1016/j.biortech.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Kim S.H., Jeong H.S., Oh S.H., Kim H.R., Kim Y.H., Lee J.N., Kook J.K., Kho W.G., Bae K., Shin J.H. Two cases of peritonitis caused by Kocuria marina in patients undergoing continuous ambulatory peritoneal dialysis. Clin. Microbiol. 2009;47(10):3376–3378. doi: 10.1128/JCM.00847-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R., Schinner F. Bioremediation (natural attenuation and biostimulation) of diesel-oil contaminated soil in alpine glacier skiing area. Appl. Environ. Microbiol. 2001;67:3127–3133. doi: 10.1128/AEM.67.7.3127-3133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayilraj S., Kroppenstedt R.M., Suresh K., Saini H.S. Kocuria himachalensis sp. nov., an actinobacterium isolated from the Indian Himalayas. Int. J. Syst. Evol. Microbiol. 2006;56:1971–1975. doi: 10.1099/ijs.0.63915-0. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Tsukatani Y., Harada J., Takasaki S., Yoshitomi T., Tamiaki H. Cyclopropane-ring formation in the acyl groups of chlorosome glycolipids is crucial for acid resistance of green bacterial antenna systems. Bioorg. Med. Chem. 2013;21(13):3689–3694. doi: 10.1016/j.bmc.2013.04.030. [DOI] [PubMed] [Google Scholar]
- Morikawa M., Ito M., Imanaka T. Isolation of a new surfactin producer Bacillus pumilus A-I, and cloning and nucleotide sequence of the regulator gene, psf-1. J. Ferment. Bioeng. 1992;74:255–261. [Google Scholar]
- Nitschke M., Pastore G.M. Production and properties of a surfactant obtained from Bacillus subtilis grown on cassava waste water. Bioresour. Technol. 2006;97:336–341. doi: 10.1016/j.biortech.2005.02.044. [DOI] [PubMed] [Google Scholar]
- Pastewski S., Hallmann E., Medrzycka K. Physiochemical aspects of application of surfactants and biosurfactants in soil remediation. Environ. Eng. Sci. 2006;23:579–588. [Google Scholar]
- Post F.J., Collins N.F. A preliminary investigation of the membrane lipid of Halobacterium halobium as a food additive. J. Food Biochem. 1982;6:25–38. [Google Scholar]
- Prieto L.M., Michelon M., Burkert J.F.M., Kalil S.J., Burkert C.A.V. The production of rhamnolipid by a Pseudomonas aeruginosa strain isolated from a southern coastal zone in Brazil. Chemosphere. 2008;71:1781–1785. doi: 10.1016/j.chemosphere.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Rodrigues L.R., Teixeira J.A., van der Mei H.C., Oliveira R. Physicochemical and funtional characterization of a biosurfactant produced by Lactococcus lactis 53. Colloids Surf. B. 2006;49:79–86. doi: 10.1016/j.colsurfb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Satpute S.K., Bhawsar B.D., Dhakephalkar P.K., Chopade B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Ind. J. Mar. Sci. 2008;37:243–250. [Google Scholar]
- Shavandi M., Mohebali G., Haddadi A., Shakarami H., Nuhi A. Emulsification potential of a newly isolated biosurfactant- producing bacterium, Rhodoccus sp. strain TA6. Colloids Surf. B. 2011;82:477–482. doi: 10.1016/j.colsurfb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Siegmund I., Wagner F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991;5(4):265–268. [Google Scholar]
- Singh A., Van Hamme J.D., Ward O.P. Surfactants in microbiology and biotechnology: part 2. Application aspects. Biotechnol. Adv. 2007;25:99–121. doi: 10.1016/j.biotechadv.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Sneath P.H.A., Sokal R.R. Freeman; San Francisco: 1973. Numerical Taxonomy. [Google Scholar]
- Stackebrandt E., Koch C., Gvozdiak O., Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Bacteriol. 1995;45:682–692. doi: 10.1099/00207713-45-4-682. [DOI] [PubMed] [Google Scholar]
- Syldatk C., Wagner F. In: Biosurfactants and Biotechnology. Kosaric N., Cairns W.L., Gray N.C.C., editors. Marcel Dekker, Inc.; New York: 1987. Production of biosurfactants; pp. 89–120. [Google Scholar]
- Tang S.K., Wang Y., Lou K., Mao P.H., Xu L.H., Jiang C.L., Kim C.J., Li W.J. Kocuria halotolerans sp. nov., an actinobacterium isolated from a saline soil in China. Int. J. Syst. Evol. Microbiol. 2009;59:1316–1320. doi: 10.1099/ijs.0.006627-0. [DOI] [PubMed] [Google Scholar]
- Tvrzova L., Schumann P., Sedlacek I., Pacova Z., Sproer C., Verbarg S., Kroppenstedt R.M. Reclassification of strain CCM 132, previously classified as Kocuria varians, as Kocuria carniphila sp. nov. Int. J. Syst. Evol. Microbiol. 2005;55:139–142. doi: 10.1099/ijs.0.63304-0. [DOI] [PubMed] [Google Scholar]
- Vater J., Kablitz B., Wilde C., Franke P., Mehta N., Cameotra S.S. Matrix-assisted laser desorption ionization-time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 2002;68(12):6210–6219. doi: 10.1128/AEM.68.12.6210-6219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov M.M., Timmis K.N., Wray V., Fredrickson H.L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 1995;61(5):1706–1713. doi: 10.1128/aem.61.5.1706-1713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]



