Abstract
Cotton is an important crop and its production is affected by various disease pathogens. Monopartite begomovirus associated betasatellites cause Cotton leaf curl disease (CLCuD) in Northern India. In order to access the occurrence and genetic variability of Cotton leaf curl betasatellites, an extensive field survey was conducted in states of Rajasthan, Punjab and Haryana. We selected the betasatellite sequence for analysis as they are reported as important for disease severity and sequence variability. Based on the field observations, the disease incidence ranged from 30% to 80% during the survey. Full genome and DNA β were amplified from various samples while no amplicon was obtained in some samples. The nucleotide sequence homology ranged from 90.0% to 98.7% with Cotton leaf curl virus (CLCuV), 55.2–55.5% with Bhendi yellow vein mosaic virus, 55.8% with Okra leaf curl virus and 51.70% with Tomato leaf curl virus isolates. The lowest similarity (47.8%) was found in CLCuV-Sudan isolate. Phylogenetic analysis showed that analyzed isolates formed a close cluster with various CLCuV isolates reported earlier. The analysis results show sequence variation in Cotton leaf curl betasatellite which could be the result of recombination. The results obtained by genome amplification and sequence variability indicate that some new variants are circulating and causing leaf curl disease in Rajasthan, Punjab and Haryana.
Abbreviations: CLCuD, Cotton leaf curl disease; CLCuV, Cotton leaf curl virus; PCR, polymerase chain reaction; SCR, satellite conserved region
Keywords: Cotton leaf curl virus, Betasatellites, Genetic variability, Northern India
1. Introduction
Cotton is an important crop and cotton production is seriously hampered by CLCuD in India and Pakistan (Sattar et al., 2013). The incidence of CLCuD has been reported in almost all the growing belt in North India (Rishi and Chauhan, 1994; Briddon et al., 2001; Sharma and Rishi, 2003; Zaffalon et al., 2011; Rajagopalan et al., 2012). India is an important producer of cotton in the world. In 1990, this disease was observed as an epidemic with approximately 30–40% estimated loss in Multan, Pakistan (Zhou et al., 1998; Briddon and Markham, 2000; Asad et al., 2003). During 1997–98 a sudden increase in CLCuD was reported in Northern India. The variability of CLCuV and betasatellite molecule in Northern India and other regions has been published earlier in various reports (Sanz et al., 1999; Briddon et al., 2001, 2003; Kirthi et al., 2004; Nawaz-ul-Rehman et al., 2012; Sohail et al., 2014). The characteristic symptoms included leaf curling; vein thickening followed by cup formation under the leaves. Interestingly, DNA β sequences of Cotton leaf curl Gezira virus from various geographical locations were found to be very similar. Recently it has been reported that, only one DNA β molecule can interact with four distinct CLCuV and produce typical symptoms (Mansoor et al., 2003a,b). Most of the begomoviruses identified in the Old World were found to be monopartite while the New World begomoviruses have bipartite genomes. Recently, a native monopartite begomovirus infecting cotton has been identified (Melgarejo et al., 2013; Sanchez-Campos et al., 2013). In the Old World most of the monopartite begomovirus were associated with betasatellites (Briddon and Mansoor, 2008; Briddon et al., 2012).
Betasatellites have small (∼1.4 kb), circular, ssDNA genome and their replication and movement fully depend upon a helper virus (Briddon et al., 2003; Mansoor et al., 2003a,b; Leke et al., 2013). The sequences of betasatellites have three major features – a single βC1 gene, adenine rich sequence region and satellite conserved region containing stem-loop structure, which is known for the origin of replication in geminiviruses (Hanley-Bowdoin et al., 1999; Briddon, 2003; Briddon et al., 2003). The function of βC1 gene is mediated by a typical encoded protein. The βC1 gene is known as a pathogenicity determinant, post-transcriptional gene silencing suppressor and mediates virus movement (Cui et al., 2005; Kon et al., 2007; Qazi et al., 2007; Saeed et al., 2007; Amin et al., 2011; Iqbal et al., 2012). The study presented here has analyzed the sequences of Cotton leaf curl betasatellites recently isolated from North India and has identified specific sequence variations among samples collected from three states.
2. Materials and methods
2.1. Field survey and sample collection
Field survey was conducted during the cotton cropping season from 2008 to 2011 in the major cotton fields of Rajasthan, Punjab and Haryana. Virus infected samples were collected from cotton plants with typical symptoms such as leaf curling, enation and stunting of plant.
2.2. PCR amplification and cloning of betasatellites
Total genomic DNA was isolated using the Cetyl trimethyl ammonium bromide method (Doyle and Doyle, 1990) and polymerase chain reaction (PCR) was conducted by using about 100 ng template DNA and 10 Pico moles of forward and reverse primers. Virus infection was confirmed by PCR using specific coat protein gene forward and reverse primers (CPF-AATTATGTCGAAGCGAGCTGC and CPR-TAATATCAATTCGTTACAGAG). Betasatellites were amplified by specific DNA β primers (βF-GGTACCACTACGCTACGCAGCAGCC and βR-GGTACCTACCCTCCCAGGGGTACAC) designed from the beginning and end of the viral genome from the published sequences. During PCR, Taq DNA polymerase (2.5 units) (MBI Fermentas, USA) 5 μl of 10× buffer, 1 μl of 10 mM dNTPs and 1 μl (10 Pico moles) of forward and reverse primers were used. The final volume was made up to 50 μl using sterile distilled water. The PCR amplified fragments of DNA β were gel eluted and purified by using a QIA quick Gel Extraction Kit (Qiagen, USA) and cloned into pGEMT-easy vector. The positive clones were identified by colony PCR and restriction enzyme digestions.
2.3. Sequence and phylogenetic analyses
The sequencing of clones was performed in DNA sequencer (ABI Prism, Perkin Elmer) at JK AgriGenetics Ltd, Hyderabad. The obtained sequences were analyzed using Bioedit software (version 5.0.9). The full sequence DNA β and associated betasatellites were initially selected for homolog using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST and the sequence showed better scores selected for genetic variability study.
3. Results
3.1. Field survey and sample collection
During the field survey, disease incidence was recorded up to 30–80% in different fields of major cotton growing areas of Rajasthan, Punjab and Haryana. The naturally infected cotton plants showed typical symptoms like leaf curling, vein thickening and enations (Fig. 1). Total three hundred samples were collected from nine locations of North India (Table 1).
Figure 1.

Natural infection of CLCuV in North India.
Table 1.
Locations of samples collected from Northern India.
| Place visited | No. of samples collected | ||
|---|---|---|---|
| Rajasthan | Sadhuwali gaon village | 30 | |
| Sangaria | 30 | ||
| Hanumangarh | 36 | ||
| Punjab | Abohar | 33 | |
| Bhatinda | 39 | ||
| Sangatpura | 30 | ||
| Haryana | Dabwali | 36 | |
| Sirsa | 30 | ||
| Hissar | 36 | ||
| Total | 9 | 300 | |
3.2. PCR amplification and cloning of betasatellites
Virus infection was confirmed by PCR amplification of full DNA-A, coat protein gene and DNA β resulted in an amplicon of about 0.750 and 1.35 kb fragments, respectively from naturally infected samples collected from various locations of North India (Fig. 2). The recombinant clones were confirmed by restriction enzyme digestions and approximately 1.3 kb fragments were released after restriction digestion. The full length clones of DNA β from each state were selected and sequenced. Interestingly, during PCR amplification, some non-desired amplification was also observed with variable sizes (0.6, 1.0 and 1.6 kb-data not shown) and BLAST result showed a close similarity to previously submitted sequences of CLCuV-DNA β.
Figure 2.
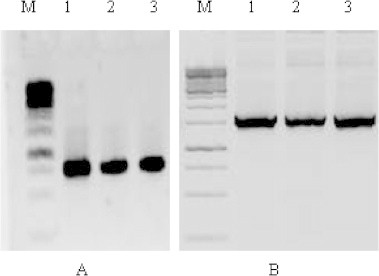
Detection of CLCuV Infection in naturally infected cotton leaves in Northern India. (A and B) M: 1 Kb ladder, 1: Rajasthan, 2: Punjab, 3: Haryana.
In Hanumangarh (Rajasthan), full genome amplification was not observed in any samples and DNA β was amplified in very less samples and no amplicon was observed in two samples. In Sangaria (Rajasthan), full genome was amplified from very less samples and no DNA β was amplified in any sample while three samples did not produce any kind of applicant. In Sadhuwali gaon (Rajasthan), most of the samples produced a full genome and DNA β amplification, and no amplification was observed in one sample.
In Sangatpura and Abohar (Punjab), most of the samples produced the amplicon of DNA-A and DNA β and some non-specific amplicons were also observed with variable sizes (0.6, 1.0 and 1.6 kb) from infected samples. In Bhatinda (Punjab), DNA-A and DNA β were amplified from most of the samples.
In Sirsa and Dabwali (Haryana), DNA-A and DNA β amplification of was observed in many samples and in one sample no amplicon observed. In Hissar (Haryana), no desired amplification was observed while most of the samples produced 0.6 kb fragment.
3.3. Sequence and phylogenetic analysis
Based on the resulted full genome of DNA β sequences, the genetic variability was compared with selected sequences. The resulted sequences consisted of 1371 nucleotides (nt) from Rajasthan 1351 (nt) from Punjab and 1350 (nt) from Haryana and were designated as JK-B5R-(Rajasthan), JK-A2P-(Punjab) and JK-B2H-(Haryana) respectively. During the BLAST search in GenBank, the obtained DNA β sequences showed a higher similarity with various CLCuV isolates, Bhendi yellow vein mosaic virus, Okra leaf curl virus and Tomato leaf curl virus. The comparison revealed sequence homology ranged from 90.0% to 98.7% with CLCuV, 55.2–55.5% with Bhendi yellow vein mosaic virus, 55.8% with Okra leaf curl virus and 51.70% with Tomato leaf curl virus isolates. Interestingly, the lowest (47.8%) similarity was observed with CLCuV-Sudan isolate. Phylogenetic analysis of these isolates showed that the entire three isolates formed a close cluster with the CLCuV isolates previously reported (Table 2), (Fig. 3).
Table 2.
Nucleotide sequence identity matrix of JKB5-R (KP015741) with selected virus isolates.
| Virus isolates | Location | Accession # | Size (nt) | Identity matrix (%) |
|---|---|---|---|---|
| CLCuV-JKA2-P | Punjab | KP015742 | 1351 | 91.5 |
| CLCuV-JK B2H | Haryana | KP015743 | 1350 | 93.0 |
| CLCuV | Pakistan | AM774307 | 1351 | 91.6 |
| CLCuV | Pakistan | FN554724 | 1349 | 91.5 |
| CLCuV | Pakistan | FJ607041 | 1354 | 91.0 |
| CLCuV | Rajasthan | GQ370388 | 1350 | 91.1 |
| CLCuV | Rajasthan | GQ369730 | 1351 | 92.5 |
| CLCuV | Rajasthan | JF502375 | 1419 | 93.3 |
| CLCuV | Punjab | Q191161 | 1350 | 91.4 |
| CLCuV | Punjab | JF502377 | 1351 | 90.5 |
| CLCuV | Punjab | JF502381 | 1415 | 91.7 |
| CLCuV | Haryana | AY795608 | 1415 | 95.6 |
| CLCuV | Haryana | JF502398 | 1354 | 89.1 |
| CLCuV | Haryana | AY744380 | 1374 | 98.1 |
| CLCuV | Lucknow | GQ369731 | 1359 | 90.0 |
| CLCuV | Lucknow | KM065438 | 1371 | 97.0 |
| CLCuV | New Delhi | KM070822 | 1371 | 98.7 |
| CLCuV | Bangalore | AY705381 | 1355 | 82.0 |
| CLCuV | Sudan | AY077797 | 1348 | 47.8 |
| BYVMV | Tamilnadu | AJ308425 | 1352 | 55.2 |
| BYVMV | West Bengal | EF417919 | 1354 | 55.5 |
| BYVMV | Maharashtra | GU233520 | 1358 | 55.3 |
| OLCuV | New Delhi | GQ245761 | 1351 | 55.8 |
| ToLCV | Rajasthan | AY438558 | 1371 | 51.7 |
Figure 3.
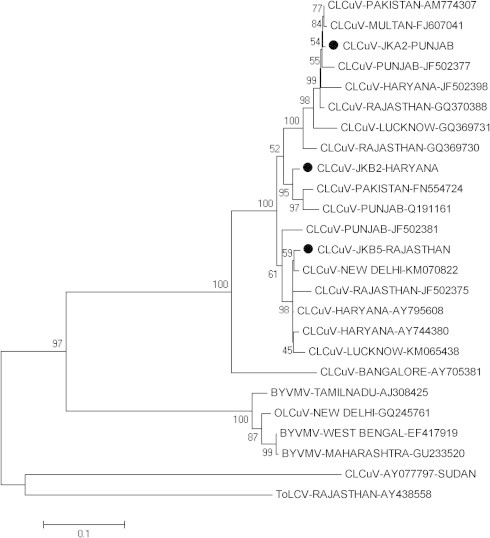
Phylogenetic tree of CLCuV isolated from Northern India. Each isolate is indicated by GenBank accession number.
4. Discussion
This paper reports the occurrence, identification and genetic variability of Cotton leaf curl betasatellites in Northern India. The incidence of the CLCuV was confirmed by PCR in naturally infected leaf samples. Cotton is an important crop and its production is affected by CLCuV for more than 25 years. Globally, cotton is being grown in warmer parts. CLCuV is the major cause of disease in cotton plants in Northern India. In Asia and Africa, begomovirus–betasatellite complexes are involved in causing CLCuD with different etiologies and symptoms. Various efforts have been made and some resistance sources have been identified against the resistance (Sattar et al., 2013). Deletion studies with Ageratum yellow vein betasatellite showed that the sequences between the satellite conserved region and the A-rich region are important for the trans-replication of the betasatellites (Saunders et al., 2008). The satellites conserved region (SCR) does contain a nonanucleotide sequence, which is presumably required for the helper virus-encoded Rep to initiate satellite DNA replication, although this has not yet been proven experimentally. The position of the SCR surrounding the origin of replication, may suggest a role in interaction with host factors involved in DNA replication. It has been reported that when trans-replicated in planta by Cabbage leaf curl virus, a virus that is not associated with betasatellites, mutations occur in the SCR and the A-rich region sequences, and levels of betasatellites DNA are higher, suggesting that the sequence changes improve trans-replication (Nawaz-ul-Rehman et al., 2009). The position of the SCR in betasatellites is analogous to the position of the common region of bipartite begomoviruses (Brown et al., 2012).
In this study, during PCR amplification, some non specific amplicons were also obtained and it is expected that maybe some mutations have occurred at specific priming sites. Some diversities in India (Cotton leaf curl Burewala virus and Cotton leaf curl Rajasthan virus) have been identified (Rajagopalan et al., 2012) and some diversities in Sindh (Cotton leaf curl Gezira virus, Cotton leaf curl Kokhran virus Cotton leaf curl Shahdadpur virus, and Cotton leaf curl Multan virus) have been already identified (Amrao et al., 2010; Tahir et al., 2011; Azhar et al., 2012; Briddon et al., 2012; Sohail et al., 2014). There are some published reports about the co-evolution evidence of begomovirus betasatellites with their cognate viral DNA-A and their genetic change effects on CLCuD (Mansoor et al., 2003a,b; Zhou et al., 2003; Briddon et al., 2014). On the basis of the above reports our findings are strongly supported and it is expected that mutation has taken place at the PCR priming sites resulting in more non specific amplicons of variable sizes. If this is the case, then it is likely that the diversity of the satellites is limited by the virus, which in turn is limited by the cotton variety. The reason for this is unclear, but could be due to the founder effect, with sequence changes selected for due to a requirement other than resistance breaking, spreading throughout a region. Although it is possible that each of these distinct CLCuB types/strains has arisen independently by recombination (inserting Tomato leaf curl betasatellites sequence and possibly also sequences of unknown origin into the SCR and also between the SCR and A-rich region), a far more plausible explanation is that after an initial (large) insertion, the sequence of the recombinant fragment was sequentially reduced by recombination with Cotton leaf curl betasatellites (Sohail et al., 2014).
The study presented shows that the Cotton leaf curl betasatellites in Northern India are evolving quite rapidly, although the forces driving this are unclear. A better understanding of virus–satellite interactions is urgently required. In addition the results have highlighted a high gene flow between the three regions-Rajasthan, Punjab and Haryana, in India. A better understanding of the movement of the pathogen might be useful in future efforts to control the disease; particularly should there be further epidemics.
Conflict of interest
The authors confirm that there is no conflict of interest for the information presented in this manuscript.
Acknowledgements
Authors extend their thanks to Mr. SK Gupta (CEO, JK AgriGenetics Ltd. Hyderabad, India) for providing funds, necessary facilities, guidance, valuable suggestions and continuous encouragement.
Footnotes
Peer review under responsibility of King Saud University.
References
- Amin I., Hussain K., Akbergenov R., Yadav J.S., Qazi J., Mansoor S., Hohn T., Fauquet C.M., Briddon R.W. Suppressors of RNA silencing encoded by the components of the Cotton leaf curl begomovirus–betasatellite complex. Mol. Plant Microbe Interact. 2011;24:973–983. doi: 10.1094/MPMI-01-11-0001. [DOI] [PubMed] [Google Scholar]
- Amrao L., Amin I., Shahid M.S., Briddon R.W., Mansoor S. Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Res. 2010;152:153–163. doi: 10.1016/j.virusres.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Asad S., Haris W.A., Bashir A., Zafar Y., Malik K.A., Malik N.N., Lichtenstein C.P. Transgenic tobacco expressing geminiviral RNAs are resistant to the serious viral pathogen causing Cotton leaf curl disease. Arch. Virol. 2003;148:2341–2352. doi: 10.1007/s00705-003-0179-5. [DOI] [PubMed] [Google Scholar]
- Azhar M.T., Akhtar S., Mansoor S. Cotton leaf curl Multan betasatellites strains cloned from Gossypium barbadense further supports selection due to host resistance. Virus Genes. 2012;45:402–405. doi: 10.1007/s11262-012-0766-1. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Mansoor S., Bedford I.D., Pinner M.S., Saunders K., Stanley J., Zafar Y., Malik K.A., Markham P.G. Identification of DNA components required for induction of Cotton leaf curl disease. Virology. 2001;285:234–243. doi: 10.1006/viro.2001.0949. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Markham P.G. Cotton leaf curl virus disease. Virus Res. 2000;71:151–159. doi: 10.1016/s0168-1702(00)00195-7. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Mansoor S. Beta ssDNA satellites. In: Mahy B.W.J., van Regenmortel M.H.V., editors. Encyclopedia of Virology. Academic Press; Oxford, UK: 2008. pp. 314–321. [Google Scholar]
- Briddon R.W. Cotton leaf curl disease, a multicomponent begomovirus complex. Mol. Plant Pathol. 2003;4:427–434. doi: 10.1046/j.1364-3703.2003.00188.x. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Akbar F., Iqbal Z., Amrao L., Amin I., Saeed M., Mansoor S. Effects of genetic changes to the begomovirus/betasatellite complex causing Cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2014;186:114–119. doi: 10.1016/j.virusres.2013.12.008. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Bull S.E., Amin I., Idris A.M., Mansoor S., Bedford I.D., Dhawan P., Rishi N., Siwatch S.S., Abdel-Salam A.M., Brown J.K., Zafar Y., Markham P.G. Diversity of DNA beta, a satellite molecule associated with some monopartite begomoviruses. Virology. 2003;312:106–121. doi: 10.1016/s0042-6822(03)00200-9. [DOI] [PubMed] [Google Scholar]
- Briddon R.W., Ghabrial S., Lin N.S., Palukaitis P., Scholthof K.B.G., Vetten H.J. Satellites and other virus-dependent nucleic acids. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy – Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Inc.; London, UK; Waltham, MA/San Diego, CA, USA: 2012. pp. 1209–1219. [Google Scholar]
- Brown J.K., Fauquet C.M., Briddon R.W., Zerbini M., Moriones E., Navas-Castillo J. Geminiviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy – Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Inc.; London UK; Waltham, MA/San Diego, CA, USA: 2012. pp. 351–373. [Google Scholar]
- Cui X., Li G., Wang D., Hu D., Zhou X. A begomovirus DNA β-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005;79:10764–10775. doi: 10.1128/JVI.79.16.10764-10775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Hanley-Bowdoin L., Settlage S.B., Orozco B.M., Nagar S., Robertson D. Geminviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999;18:71–106. [PubMed] [Google Scholar]
- Iqbal Z., Sattar M.N., Kvarnheden A., Mansoor S., Briddon R.W. Effects of the mutation of selected genes of Cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of Cotton leaf curl Multan betasatellite. Virus Res. 2012;169:107–116. doi: 10.1016/j.virusres.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Kirthi N., Priyadarshini C.G.P., Sharma P., Maiya S.P., Hemalatha V., Sivaraman P., Dhawan P., Rishi N., Savithri H.S. Genetic variability of begomoviruses associated with Cotton leaf curl disease originating from India. Arch. Virol. 2004;149:2047–2057. doi: 10.1007/s00705-004-0352-5. [DOI] [PubMed] [Google Scholar]
- Kon T., Sharma P., Ikegami M. Suppressor of RNA silencing encoded by the monopartite Tomato leaf curl Java begomovirus. Arch. Virol. 2007;152:1273–1282. doi: 10.1007/s00705-007-0957-6. [DOI] [PubMed] [Google Scholar]
- Leke W.N., Sattar M.N., Ngane E.B., Ngeve J.M., Kvarnheden A., Brown J.K. Molecular characterization of begomoviruses and DNA satellites associated with Okra leaf curl disease in Cameroon. Virus Res. 2013;174:116–125. doi: 10.1016/j.virusres.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Mansoor S., Amin I., Iram S., Hussain M., Zafar Y., Malik K.A., Briddon R.W. The breakdown of resistance in cotton to Cotton leaf curl disease in Pakistan. Plant. Pathol. 2003;52:784. [Google Scholar]
- Mansoor S., Briddon R.W., Bull S.E., Bedford I.D., Bashir A., Hussain M., Saeed M., Zafar Y., Malik K.A., Fauquet C., Markham P.G. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA beta. Arch. Virol. 2003;148:1969–1986. doi: 10.1007/s00705-003-0149-y. [DOI] [PubMed] [Google Scholar]
- Melgarejo T.A., Kon T., Rojas M.R., Paz-Carrasco L., Zerbini F.M., Gilbertson R.L. Characterization of a New World monopartite begomovirus causing leaf curl disease of Tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J. Virol. 2013;87:5397–5413. doi: 10.1128/JVI.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz-ul-Rehman M.S., Briddon R.W., Fauquet C.M. A melting pot of Old World begomoviruses and their satellites infecting a collection of Gossypium species in Pakistan. PLoS One. 2012;7:e40050. doi: 10.1371/journal.pone.0040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz-ul-Rehman M.S., Mansoor S., Briddon R.W., Fauquet C.M. Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 2009;83:9347–9355. doi: 10.1128/JVI.00795-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi J., Amin I., Mansoor S., Iqbal M.J., Briddon R.W. Contribution of the satellite encoded gene βC1 to Cotton leaf curl disease symptoms. Virus Res. 2007;128:135–139. doi: 10.1016/j.virusres.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Rajagopalan P.A., Naik A., Katturi P., Kurulekar M., Kankanallu R.S., Anandalakshmi R. Dominance of resistance-breaking Cotton leaf curl Burewala virus (CLCuBuV) in northwestern India. Arch. Virol. 2012;157:855–868. doi: 10.1007/s00705-012-1225-y. [DOI] [PubMed] [Google Scholar]
- Rishi N., Chauhan M.S. Appearances of leaf curl disease of cotton in Northern India. J. Cotton Res. Dev. 1994;8:179–180. [Google Scholar]
- Saeed M., Zafar Y., Randles J.W., Rezaian M.A. A monopartite begomovirus-associated DNA β satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007;88:2881–2889. doi: 10.1099/vir.0.83049-0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Campos S., Martínez-Ayala A., Márquez-Martín B., Aragón-Caballero L., Navas-Castillo J., Moriones E. Fulfilling Koch’s postulates confirms the monopartite nature of Tomato leaf deformation virus, a begomovirus native to the New World. Virus Res. 2013;173:286–293. doi: 10.1016/j.virusres.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Sanz A.I., Fraile A., Gallego J.M., Malpica J.M., Garcia-Arenal F. Genetic variability of natural populations of Cotton leaf curl geminivirus, a single-stranded DNA virus. J. Mol. Evol. 1999;49:672–681. doi: 10.1007/pl00006588. [DOI] [PubMed] [Google Scholar]
- Sattar M.N., Kvarnheden A., Saeed M., Briddon R.W. Cotton leaf curl disease. An emerging threat to cotton production worldwide. J. Gen. Virol. 2013;94:695–710. doi: 10.1099/vir.0.049627-0. [DOI] [PubMed] [Google Scholar]
- Saunders K., Briddon R.W., Stanley J. Replication promiscuity of DNA-β satellites associated with monopartite begomoviruses; deletion mutagenesis of the Ageratum yellow vein virus DNA-β satellite localizes sequences involved in replication. J. Gen. Virol. 2008;89:3165–3172. doi: 10.1099/vir.0.2008/003848-0. [DOI] [PubMed] [Google Scholar]
- Sharma P., Rishi N. Host range and vector relationships of Cotton leaf curl virus from Northern India. Indian Phytopathol. 2003;56:496–499. [Google Scholar]
- Sohail A., Tahir M.N., Baloch G.R., Javaid S., Khan A.Q., Amin I., Briddon R.W., Mansoor S., Baloch G.R. Regional changes in the sequence of Cotton leaf curl Multan betasatellite. Viruses. 2014;6:2186–2203. doi: 10.3390/v6052186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir M.N., Amin I., Briddon R.W., Mansoor S. The merging of two dynasties-identification of an African Cotton leaf curl disease-associated begomovirus with cotton in Pakistan. PLoS One. 2011;6:e20366. doi: 10.1371/journal.pone.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffalon V., Mukherjee S., Reddy V., Thompson J., Tepfer M. A survey of geminiviruses and associated satellite DNAs in the cotton-growing areas of northwestern India. Arch. Virol. 2011;157:483–495. doi: 10.1007/s00705-011-1201-y. [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu Y., Robinson D.J., Harrison B.D. Four DNA-A variants among Pakistani isolates of Cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from Okra. J. Gen. Virol. 1998;79(4):915–923. doi: 10.1099/0022-1317-79-4-915. [DOI] [PubMed] [Google Scholar]
- Zhou X., Xie Y., Tao X., Zhang Z., Li Z., Fauquet C.M. Characterization of DNA beta associated with begomoviruses in China and evidence for co-evolution with their cognate viral DNA-A. J. Gen. Virol. 2003;84:237–247. doi: 10.1099/vir.0.18608-0. [DOI] [PubMed] [Google Scholar]


