Highlights
-
•
Active site characteristics of the β-galactosidase enzyme of Bacillus circulans ATCC 31382.
-
•
Residues essential for catalysis were identified by structure-based alignments.
-
•
Mutational analysis identified two catalytic glutamate amino acid residues.
Abbreviations: DNA, deoxyribonucleic acid; GOS, galacto-oligosaccharides; GH2, glycoside hydrolase family 2; Tm, melting temperatures; o-NPG, ortho-nitrophenyl-β-galactoside; PCR, polymerase chain reaction; SD, standard deviation
Keywords: β-Galactosidase, Galacto-oligosaccharides, Bacillus circulans, Catalytic residue, Mutagenesis, Transglycosylation activity
Abstract
The Bacillus circulans ATCC 31382 β-galactosidase (BgaD) is a retaining-type glycosidase of glycoside hydrolase family 2 (GH2). Its commercial enzyme preparation, Biolacta N5, is used for commercial-scale production of galacto-oligosaccharides (GOS). The BgaD active site and catalytic amino acid residues have not been studied. Using bioinformatic routines we identified two putative catalytic glutamates and two highly conserved active site histidines. The site-directed mutants E447N, E532Q, and H345F, H379F had lost (almost) all catalytic activity. This confirmed their essential role in catalysis, as general acid/base catalyst (E447) and nucleophile (E532), and as transition state stabilizers (H345, H379), respectively.
1. Introduction
The β-galactosidase of Bacillus circulans ATCC 31382 (BgaD) belongs to the family 2 of glycoside hydrolases (GH2) [1,2]; its physiological role is to hydrolyze lactose into galactose and glucose. Its commercial preparation Biolacta N5 has 4 C-terminally truncated active isoforms, products of proteolysis. Recently, we have cloned the B. circulans bgaD gene and characterized the four similar recombinant isoforms rBgaD-(A-D) and the four Biolacta N5 purified isoforms β-GalA-D. The results showed that the smallest isoforms β-Gal-D and rBgaD-D have the same GOS product profiles as the longer and full-length proteins [3]. The GH2 family members have an active site which is composed of two catalytic residues: a catalytic nucleophile and a general acid/base catalyst. GH2 family enzymes are β-retaining and employ a double displacement mechanism (Fig. 1). First, the catalytic nucleophile attacks the anomeric center of the sugar, generating the galactosyl–enzyme intermediate. Subsequently, this intermediate undergoes hydrolysis or transglycosylation. Both steps require assistance of a general acid/base catalyst [4]. At high lactose concentrations some β-galactosidases switch from a hydrolytic to a synthetic mechanism whereby lactose (instead of water) acts as acceptor for the covalently bound galactose [5]. The newly formed product is a galacto-oligosaccharide (GOS).
Fig. 1.
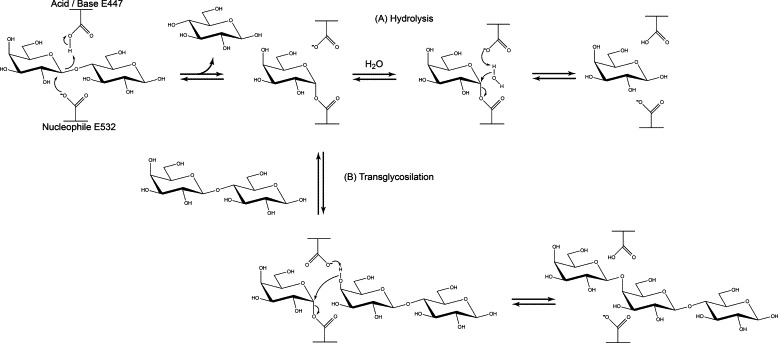
Reaction mechanisms of the B. circulans β-galactosidase enzyme, also showing the catalytic glutamate residues involved, namely E447 as acid/base catalyst, and E532 as nucleophile. Residue H345 forms strong hydrogen bonds with the C3′ hydroxyl group of the galactosyl unit and stabilizes the transition state. The nitrogen of residue H379 is within H-bonding distance of the C3′ hydroxyl group of the galactosyl unit and thereby promotes catalysis by stabilizing the transition state (not visualized in the figure). (A) Hydrolysis reaction, using water as acceptor substrate. (B) Transglycosylation reaction, using lactose as acceptor substrate. Both reactions proceed according to a double-displacement mechanism and use lactose as donor substrates. The glycosidic linkage in lactose is cleaved, resulting in glucose release and covalent binding of galactose to the nucleophile in the active site. A subsequent reaction with water as acceptor substrate results in release of galactose as well (A: hydrolysis). Alternatively, lactose is used as acceptor substrate as well, resulting in release of oligosaccharide products (B: transglycosylation).
The B. circulans β-galactosidase (monomeric protein, 1737 amino acids, metal independent, catalytic residues unknown [6]) has high transgalactosylation activity and is applied at industrial scale to produce GOS [6,7]. The enzyme shares 22% sequence identity with the Escherichia coli β-galactosidase (tetrahomomeric protein, 1024 amino acids, metal-ion dependent, 2 Glu catalytic residues) which has been well characterized [8–15]. Whereas the B. circulans β-galactosidase activity is metal independent [6], the E. coli β-galactosidase requires both Mg2+ and Na+ for maximal activity: the sodium ion directly ligands the lactose 6-hydroxyl and the magnesium ion may contribute to the role of the general acid/base catalyst [16]. In contrast to B. circulans β-galactosidase, the E. coli enzyme is a poor producer of GOS [16]. In the E. coli β-galactosidase enzyme the amino acid residues E461 and E537 are respectively the catalytic proton donor and the nucleophile [14]. Moreover, residue H358 was identified as a transition state stabilizer following the observation that it forms strong hydrogen bonds with the C3′ hydroxyl group of the galactosyl unit [10]. Another study showed that the nitrogen of residue H392 is within H-bonding distance of the C3′ hydroxyl group of the galactosyl unit and thereby promotes catalysis by stabilizing the transition state [9]. Here we characterized site-directed mutants of the two putative catalytic glutamates in B. circulans β-galactosidase and two conserved histidines in the active site. This is the first report on mutagenesis of the putative Glu catalytic residues of B. circulans β-galactosidase, showing that a change in these residues results in loss of β-galactosidase activity.
2. Materials and methods
2.1. General
B. circulans ATCC 31382, obtained from LGC Standards (Wesel, Germany), was grown at 30 °C in 3% (w/v) beef extract and 5% (w/v) peptone. E. coli strain TOP10F (Invitrogen, Carlsbad, CA) was used for cloning purposes and grown at 37 °C in Luria–Bertani medium. For plasmid selection the appropriate antibiotic was added at the following concentrations: 50 μg ml−1 for kanamycin and 100 μg ml−1 for ampicillin. E. coli BL21 (DE3) grown at 30 °C was used for high-level expression of β-galactosidases of strain ATCC 31382. Plasmids pZErO-2 and pET15b (both Invitrogen) were used for cloning and expression purposes, respectively. Protein concentrations were determined using the Bradford reagent (Bio-Rad, Munich, Germany) with bovine serum albumin as standard.
2.2. DNA preparation and manipulation
Genomic DNA of B. circulans ATCC 31382 was isolated from an overnight grown nutrient broth culture using the GenElute Bacterial Genomic DNA kit (Sigma) according to the recommendations of the manufacturer. Preparation of plasmid DNA cultures of E. coli was performed using the GenElute Plasmid Miniprep Kit (Sigma) according to the recommendations of the manufacturer. DNA restriction, ligations, and transformation were performed according to protocols described by Sambrook et al. [17].
2.3. PCR amplification
PCR was performed with genomic DNA of B. circulans ATCC 31382 as template to prepare the DNA fragment that correspond to isoform β-Gal-D as found in Biolacta N5 using the common sense primer supplemented by the NcoI restriction enzyme site (5′-TATACCATGGGAAACAGTGTGAGCTATGATGG-3′) and anti-sense primer by the BglII site (rBgaD-D, 5′-TTTAGATCTTTATGGCGTTACACGTAAATAC-3′). The obtained PCR product was purified using the GenElute PCR Clean-Up Kit (Sigma), and blunt ligated into the EcoRV site of pZErO-2 followed by a digestion with NcoI and BglII and separated with agarose gel electrophoresis. DNA fragment with the right size was cut out, purified with the GenElute Gel Extraction Kit (Sigma) and ligated into pET-15b to prepare expression plasmid pET-15b-rBgaD-D.
2.4. Bioinformatic analyses
Using T-Coffee [3,18], the B. circulans rBgaD-D amino acid sequence was aligned with seven other family GH2 β-galactosidases (Table 1). This alignment was improved manually with Jalview 2.8 [19] by careful examination with Pymol of a 3D BgaD (a.a. 50–650) model queried using PHYRE [20] which was structurally aligned with crystal structures of Kluyveromyces lactis BgaL, E. coli LacZ and Arthrobacter sp. C2-3 LacZ.
Table 1.
Comparison of the β-galactosidase amino acid sequences of Bacillus circulans ATCC 31382 with other β-galactosidases.
| Enzyme source | Enzyme name | No. a.a.a | Identity (%) | Similarity (%) | Uniprot code | PDB codeb |
|---|---|---|---|---|---|---|
| Bacillus circulans ATCC 31382 | BgaD-A | 1737 | 100 | 100 | E5RWQ2 | |
| Bifidobacterium bifidum DSM 20215 | BIF3 | 1752 | 28 | 43 | Q9F4D5 | |
| Bifidobacterium bifidum NCIMB 41171 | BbgIII | 1935 | 29 | 42 | A4K5H9 | |
| Bifidobacterium bifidum NCIMB 41171 | BbgI | 1291 | 14 | 24 | E4V7B8 | |
| Bifidobacterium bifidum NCIMB 41171 | BbgIV | 1052 | 13 | 23 | Q0ZII7 | |
| Kluyveromyces lactis CBS2359 | BgaL | 1025 | 13 | 22 | P00723 | 3OB8 |
| Escherichia coli K12 | LacZ | 1024 | 13 | 22 | P00722 | 1DP0 |
| Arthrobacter sp. C2-2 | LacZ | 1023 | 13 | 25 | Q8KRF6 | 1YQ2 |
Number of amino acids.
PDB codes used for structural alignment with the 3D model of the rBgaD-cat (a.a. 50–650).
2.5. Site-directed mutagenesis
Site-directed mutagenesis of the B. circulans rbgaD-D gene was carried out with the QuikChange™ site directed mutagenesis kit (Stratagene) and confirmed by sequencing. Pfu Turbo DNA polymerase (Stratagene) was used for all polymerase chain reactions (PCR) (95 °C, 30 s; 55 °C, 1 min; 68 °C, 10 min for 30 cycles) using plasmid pET15b-rBgaD-D as template [3]. PCR products were digested with DpnI to specifically digest the DNA templates. Subsequently, plasmids containing the desired mutation were transformed to E. coli BL21 (DE3) competent cells. For site-directed mutagenesis the following oligonucleotides were used in PCR reactions: 5′-CACGGCGTTTCGATGTTCCATGATTTAGGGG-3′ (H345F); 5′-CCATCAGGGTTACCTTCAACCCGGCATCAC-3′ (H379F); 5′-ATCATGTGGTCGATCGGAAATAACATATATGATACGACCAATGCC-3′ (E447N) and 5′-CTGTACGGCTCGCAGACGTCCTCGG-3′ (E532Q).
2.6. Recombinant protein expression and purification
E. coli expression of the β-galactosidase rBgaD-D and the four mutants were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Thermo) at an optical cell density at 600 nm of 0.6 for 4 h at 30 °C. Cells were harvested, washed with 20 mM Tris–HCl (pH 7.5) (Buffer A) and resuspended in B-PER lysis solution (Thermo). After 10 min incubation at room temperature, cell debris was removed by a centrifugation step and cell free extract with rBgaD-D was loaded on a HiTrap Q 1 ml column (Pharmacia) which was equilibrated with Buffer A. The flow through with rBgaD-D or one of the four mutants was collected, concentrated with an ultrafiltration device (Centricon Plus-20 with 50 kDa molecular cutoff, Millipore, Bedford, MA) and loaded on HiTrap Q 1 ml column (Pharmacia) which was equilibrated with Buffer B (20 mM N-Cyclohexyl-2-aminoethanesulfonic acid (CHES), pH 9.5) and unbound proteins were washed away with 5 column volumes of Buffer B. The elution was carried out with 20 ml of linear salt gradient to Buffer C (1 M NaCl in Buffer B). Fractions of 0.5 ml were collected and analyzed by 8% SDS–PAGE. Recombinant protein containing fractions were pooled and concentrated with an ultrafiltration device (Centricon Plus-20 with 50 kDa molecular cutoff, Millipore, Bedford, MA), loaded on a Superdex 200 column equilibrated with Buffer D (150 mM NaCl in Buffer A), and fractions of 0.5 ml were collected. Fractions with rBgaD-D or one of the four mutants were pooled, concentrated if necessary, and protein concentrations were determined as described.
2.7. Activity measurements
The specific activity of purified (mutant) β-galactosidase enzymes toward lactose was measured by following enzymatically released glucose with the glucose oxidase/peroxidase method (Megazyme). One enzyme activity unit is defined as the amount of enzyme in mg required for the hydrolysis of 1 μmol lactose per min at 40 °C and pH 6.0.
To determine kinetic parameters of (mutant) β-galactosidase, initial reaction rates for the hydrolysis of ortho-nitrophenyl-β-galactoside (o-NPG) with a molar extinction coefficient of 4500 M−1 cm−1 at 420 nm were determined spectrophotometrically (420 nm) at 40 °C and pH 6.0 over a concentration range of 1–38 mM. To prepare the substrate solutions, o-NPG was dissolved in 0.1 M phosphate buffer (pH 6.0) by heating and subsequently kept at 40 °C to prevent precipitation. The reaction was initiated by adding 10 μl of enzyme solution to 190 μl substrate solutions. The Vmax, kcat and KM values, expressed as the mean ± SD of values obtained by triplicate measurements, were determined from Lineweaver–Burk (LB) plots in case of rBgaD-D and mutant E532Q.
2.8. Thermal shift assay
The melting temperature (Tm) (temperature at which 50% of the protein is unfolded) of rGalD-D and the four mutants derived were determined by a fluorescence-based thermal shift assay [21]. Solutions of 5 μl of 250 × SYPRO Orange (Molecular Probes), 2.5 μl of 1 M acetate buffer, pH 6.0, 12.5 μl of Milli-Q and 5 μl of 1 mg/ml protein were mixed, and heated from 25 °C to 95 °C in increments of 0.5 °C/min on a CFX96 real-time system-C1000 Thermal Cycler (Bio-Rad). All measurements were done in triplicate. SYPRO Orange dye interacts with a protein undergoing thermal unfolding, with its fluorescence increasing upon exposure to the protein’s hydrophobic core. The TM for rGalD-D and the four mutants were determined by calculating the first derivative from the melting curve using CFX manager 2.0 software (Bio-Rad).
3. Results and discussion
3.1. Alignment and structural modeling
The active site of β-galactosidase (LacZ) of E. coli is structurally and biochemically well characterized [8–15]. The amino acid residues E461 and E537 were identified as respectively the general acid/base catalyst and the nucleophile. Mutations in these residues, introducing several different residues at these positions, resulted in loss of activity [14,22]. Moreover, residues H357 and H540 were identified as being important for stabilization of the transition state. Mutations in these residues, introducing several different residues at these positions, also resulted in loss of activity [9,10]. Currently, no such information is available about the active site of B. circulans β-galactosidase rBgaD enzyme.
A protein sequence alignment was made between B. circulans β-galactosidase rBgaD and other β-galactosidases known to be producers of GOS (Table 1). The highest sequence identity was found with the BIF3 and BbgIII enzymes of Bifidobacterium bifidum DSM 20215 and B. bifidum NCIMB 41171, respectively (Table 1). However, also their active site residues have not been identified and characterized yet. The catalytic residues of β-galactosidases of K. lactis, E. coli and Arthrobacter sp. C2-2 were previously identified but their sequence identity (Table 1) was too low for identification of the catalytic residues of B. circulans β-galactosidase. To improve this protein sequence alignment, a 3D protein model was predicted for the catalytic domain of B. circulans rBgaD-50–650 (a.a.) and structurally aligned with crystal structures of K. lactis BgaL, E. coli LacZ and Arthrobacter sp. C2-2 LacZ (Fig. 2). Manual adjustments based on the structural alignment were made and enabled the identification of the rBgaD-D conserved residues E447 and E532 as the putative general acid/base catalyst and the nucleophile, respectively. The two histidines in E. coli LacZ identified as being important for stabilization of the transition state were also conserved in B. circulans with H345 and H379 as their equivalents. Moreover, two conserved glutamates, the putative general acid/base catalyst and the nucleophile for B. bifidum DSM 20215 BIF3 (E479 and E567), B. bifidum NCIMB 41171 BbgI, BbgIII and BbgIV (E589 and E681, E479 and E567, E490 and E564, respectively) which have not been reported before, were identified as well (Fig. 3).
Fig. 2.
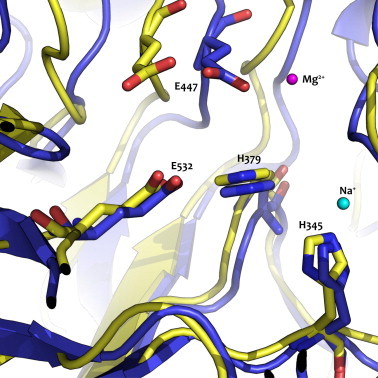
Overlay of the protein model for B. circulans rBgaD-50–650 in yellow and E.coli β-galactosidase crystal structure (PDB: 1DP0) in blue. The two catalytic glutamates (E477, E532) and two conserved histidines (H345, H397) are shown for B. circulans rBgaD-50–650. The magnesium (magenta) and sodium (light blue) ions are required for E.coli β-galactosidase, but not for the β-galactosidase of B. circulans. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.

Multiple sequence alignment (created using T-Coffee and Jalview 2.8) of beta-galactosidase proteins, revealing four conserved blocks. The first two blocks contain the conserved histidine residues and the last two blocks contain the conserved and (putative catalytic) glutamate residues (boxed in grey). Asterisks indicate fully conserved residues. Residues subjected to mutagenesis are shown in bold [9,10,14,22]. BgaD_Bacci, B. circulans ATCC 31382 BgaD; BIF3_Bifbi, Bifidobacterium bifidum DSM 20215 BIF3; BbgIII_Bifbi, Bifidobacterium bifidum NCIMB 41171 BbgIII; BbgI_Bifbi, Bifidobacterium bifidum NCIMB 41171 BbgI; BbgIV_Bifbi, Bifidobacterium bifidum NCIMB 41171 BbgIV; BgaL_Klula, Kluyveromyces lactis CBS2359 BgaL; LacZ_Escco, Escherichia coli K12 LacZ; LacZ_Arthr, Arthrobacter sp. C2–2 LacZ.
3.2. Characterization of site-directed mutants
B. circulans β-galactosidase residues H345, H379, E447 and E532 were changed by site-directed mutagenesis. The activity of the purified enzymes with lactose was determined by measuring glucose release. Three mutants (H345F, H379F and E447N) were inactive with the assay used, whereas the activity of mutant E532Q was reduced at least 340-fold compared to the wild type enzyme (Table 2). This is the first report on mutagenesis of these residues of the B. circulans ATCC 31382 β-galactosidase, showing that when they are changed, enzyme activity is lost (virtually) completely.
Table 2.
Kinetic parameters for the Bacillus circulans ATCC 31382 β-galactosidase rGalD-D and the E532Q site-directed mutant.a All parameters were determined at 40 °C and pH 6.0. One enzyme activity unit is defined as the amount of enzyme in mg required for the hydrolysis of 1 μmol lactose per min at 40 °C and pH 6.0. Hydrolysis of oNPG (oNP release at 420 nm) was used to determine the kinetic parameters Vmax (μmol min−1 mg−1 of protein), KM (mM) and kcat (s−1). Subscripts high and low indicate parameters determined from the high and low substrate regions in biphasic Lineweaver–Burk plots, respectively. All measurements were done in triplo.
| Enzyme | rGalD-D | E532Q |
|---|---|---|
| Lactose hydrolysis units min−1 mg−1 | 158.5 ± 5.4 | 0.47 ± 0.02 |
| oNPG hydrolysis kinetic parameters | ||
| Vmax,high | 277.7 ± 2.7 | 1.0 ± 0.1 |
| Km,high | 45.9 ± 2.7 | 43.6 ± 0.1 |
| kcat,high | 424.1 ± 4.1 | 1.6 ± 0.1 |
| Vmax,low | 35.8 ± 9.3 | 0.2 ± 0.01 |
| Km,low | 1.5 ± 0.9 | 1.9 ± 0.3 |
| kcat,low | 56.3 ± 11.9 | 0.2 ± 0.01 |
H379F, H345F and E447N site-directed mutants were inactive.
Kinetic properties could only be determined for wild type and site-directed mutant E532Q (Table 2). Both wild type and E532Q mutant showed biphasic Lineweaver–Burk (LB) plots for which kinetic parameters could be determined separately. This resulted in two sets of kinetic parameters (Vmax, KM and kcat): one for the lower and one for the higher substrate concentration regions, similar to what was shown previously for rGalD-D [3]. Such biphasic LB plots have been reported for hydrolytic enzymes that show transglycosylation activity at higher substrate concentrations [2,6]. The kcat,high for the E532Q mutant had decreased 270-fold, with an almost unchanged Km,high. A decreased kcat,low value (250-fold) also was determined for the E532Q mutant, whereas its Km,low increased about 30% (from 1.5 to 1.9 mM). The assay sensitivity could not be increased enough to determine kinetic parameters for the three other mutants.
On basis of this data we conclude that E447 and E532 of B. circulans β-galactosidase act as general acid/base catalyst and nucleophile, respectively. The data also shows that H345 and H379 are essential for activity of the B. circulans β-galactosidase. This strongly suggests that these two residues are important for transition state stabilization as was shown for the similar residues in the E. coli β-galactosidase [9,10].
3.3. Melting temperatures (Tm) determined with the thermal shift assay
Mutations in the catalytic residues and two histidines of the E. coli β-galactosidase caused minor, if any, structural changes in the protein [9,10,15,22]. To determine whether mutations in the catalytic residues and two histidines of the B. circulans ATCC 31382 β-galactosidase had caused structural changes; we recorded protein melting curves for rGalD-D and mutant enzymes derived and determined the melting temperatures (Tm). All 5 enzymes showed typical melting curve profiles (Fig. 4). The Tm values obtained are shown in Table 3. Introduction of mutations H345F and H379F caused a decrease of 9 °C in Tm values whereas mutation E447N lowered the Tm by 3.5 °C. Interestingly, the E532Q mutant enzyme showed 2 distinct melting temperatures, the first one is 10 °C lower than the Tm value for rGalD-D, the second one being equal to the Tm value for rGalD-D. This observation was only made for mutant E532Q and may be explained by successive melting of independent protein domains caused by the mutation E532Q. From these results we conclude that the mutant proteins were properly folded, however, introduction of point mutants in and close to the active site somewhat decreased the thermal stability of the proteins.
Fig. 4.
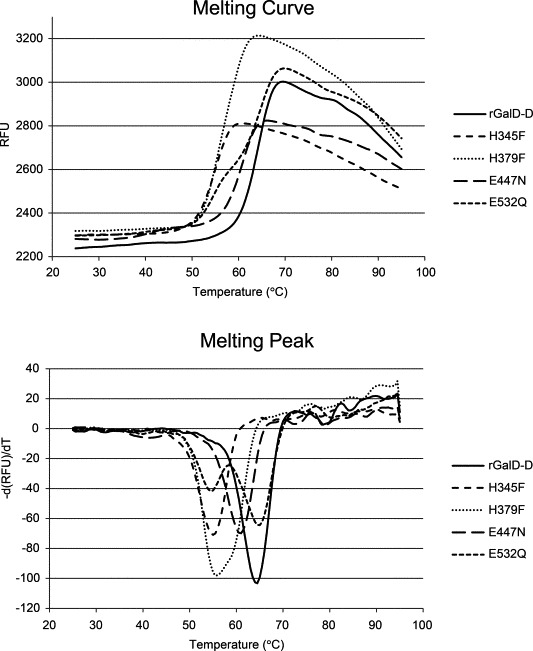
(Top) Melting curves showing protein unfolding by increasing temperatures, accompanied by increase in SYPRO Orange fluorescence. The melting temperature (Tm) values were determined by calculating the first derivative from the melting curve with CFX manager 2.0 software (Bio-Rad). (Bottom) The X-axis value (T) that corresponds with the lowest Y-axis value (−d(RFU)/dT) in the resulting curve resembles Tm. In the case of E532Q two melting temperatures were found.
Table 3.
Melting temperatures (Tm) determined with the thermal shift assay. The melting temperature (Tm) represents the temperature at which 50% of the protein is unfolded [21].
| Enzyme | Tm (°C) | Tm (°C) |
|---|---|---|
| rGalD-D | 64.3 ± 0.3 | – |
| rGalD-D H379F | 55.7 ± 0.3 | – |
| rGalD-D H345F | 55.2 ± 0.3 | – |
| rGalD-D E447N | 60.8 ± 0.3 | – |
| rGalD-D E532Q | 54.7 ± 0.3 | 64.8 ± 0.3 |
In conclusion, active site residues in the B. circulans β-galactosidase essential for catalysis and activity have been identified. Such information is of prime importance for our future attempts to change enzyme properties. Our current work provides a firm basis for enzyme engineering to further investigate the reaction mechanism.
Author contributions
JBB designed the experiments, conducted the experiments and wrote the paper, BJHK wrote the paper, LD designed the experiments and wrote the paper.
Acknowledgements
This project was jointly financed by the European Union, European Regional Development Fund and The Ministry of Economic Affairs, Agriculture and Innovation, Peaks in the Delta, the Municipality of Groningen, the Provinces of Groningen, Fryslân and Drenthe as well as the Dutch Carbohydrate Competence Center (CCC WP[6a]).
References
- 1.Lombard V., Golaconda R.H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song J., Imanaka H., Imamura K., Minoda M., Katase T., Hoshi Y., Yamaguchi S., Nakanishi K. Cloning and expression of a beta-galactosidase gene of Bacillus circulans. Biosci. Biotechnol. Biochem. 2011;75:1194–1197. doi: 10.1271/bbb.110014. [DOI] [PubMed] [Google Scholar]
- 3.J.B. Bultema, S.S. van Leeuwen, J.P. Kamerling, L. Dijkhuizen, Characterization of the 4 (recombinant) beta-galactosidase isoforms of Bacillus circulans ATCC 31382 and the commercial Biolacta N5 preparation, and their galacto-oligosaccharide products, in preparation.
- 4.Juers D.H., Matthews B.W., Huber R.E. LacZ beta-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012;21:1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park A.R., Oh D.K. Galacto-oligosaccharide production using microbial beta-galactosidase: current state and perspectives. Appl. Microbiol. Biotechnol. 2010;85:1279–1286. doi: 10.1007/s00253-009-2356-2. [DOI] [PubMed] [Google Scholar]
- 6.Song J., Abe K., Imanaka H., Imamura K., Minoda M., Yamaguchi S., Nakanishi K. Causes of the production of multiple forms of beta-galactosidase by Bacillus circulans. Biosci. Biotechnol. Biochem. 2011;75:268–278. doi: 10.1271/bbb.100574. [DOI] [PubMed] [Google Scholar]
- 7.Warmerdam A., Paudel E., Jia W., Boom R.M., Janssen A.E. Characterization of beta-galactosidase isoforms from Bacillus circulans and their contribution to GOS production. Appl. Biochem. Biotechnol. 2013;170:340–358. doi: 10.1007/s12010-013-0181-7. [DOI] [PubMed] [Google Scholar]
- 8.Lo S., Dugdale M.L., Jeerh N., Ku T., Roth N.J., Huber R.E. Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg2+ is not important for structure and indicate that the main role of Mg2+ is to mediate optimization of active site chemistry. Protein J. 2010;29:26–31. doi: 10.1007/s10930-009-9216-x. [DOI] [PubMed] [Google Scholar]
- 9.Huber R.E., Hlede I.Y., Roth N.J., McKenzie K.C., Ghumman K.K. His-391 of beta-galactosidase (Escherichia coli) promotes catalyses by strong interactions with the transition state. Biochem. Cell Biol. 2001;79:183–193. doi: 10.1139/o00-101. [DOI] [PubMed] [Google Scholar]
- 10.Roth N.J., Rob B., Huber R.E. His-357 of beta-galactosidase (Escherichia coli) interacts with the C3 hydroxyl in the transition state and helps to mediate catalysis. Biochemistry. 1998;37:10099–10107. doi: 10.1021/bi972796t. [DOI] [PubMed] [Google Scholar]
- 11.Roth N.J., Huber R.E. Glu-416 of beta-galactosidase (Escherichia coli) is a Mg2+ ligand and beta-galactosidases with substitutions for Glu-416 are inactivated, rather than activated, by Mg2+ Biochem. Biophys. Res. Commun. 1996;219:111–115. doi: 10.1006/bbrc.1996.0190. [DOI] [PubMed] [Google Scholar]
- 12.Juers D.H., Heightman T.D., Vasella A., McCarter J.D., Mackenzie L., Withers S.G., Matthews B.W. A structural view of the action of Escherichia coli (lacZ) beta-galactosidase. Biochemistry. 2001;40:14781–14794. doi: 10.1021/bi011727i. [DOI] [PubMed] [Google Scholar]
- 13.Juers D.H., Rob B., Dugdale M.L., Rahimzadeh N., Giang C., Lee M., Matthews B.W., Huber R.E. Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli) Protein Sci. 2009;18:1281–1292. doi: 10.1002/pro.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebler J.C., Aebersold R., Withers S.G. Glu-537, not Glu-461, is the nucleophile in the active site of (lac Z) beta-galactosidase from Escherichia coli. J. Biol. Chem. 1992;267:11126–11130. [PubMed] [Google Scholar]
- 15.Juers D.H., Jacobson R.H., Wigley D., Zhang X.J., Huber R.E., Tronrud D.E., Matthews B.W. High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation. Protein Sci. 2000;9:1685–1699. doi: 10.1110/ps.9.9.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber R.E., Kurz G., Wallenfels K. A quantitation of the factors which affect the hydrolase and transgalactosylase activities of beta-galactosidase (E. coli) on lactose. Biochemistry. 1976;15:1994–2001. doi: 10.1021/bi00654a029. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Frisch E.J., Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 18.Di T.P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley L.A., Sternberg M.J. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 21.Giuliani S.E., Frank A.M., Collart F.R. Functional assignment of solute-binding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry. 2008;47:13974–13984. doi: 10.1021/bi801648r. [DOI] [PubMed] [Google Scholar]
- 22.Martinez-Bilbao M., Gaunt M.T., Huber R.E. E461H-beta-galactosidase (Escherichia coli): altered divalent metal specificity and slow but reversible metal inactivation. Biochemistry. 1995;34:13437–13442. doi: 10.1021/bi00041a022. [DOI] [PubMed] [Google Scholar]


