Abstract
AIM: Colorectal cancers result from the accumulation of several distinct genetic alterations. This study was to investigate the frequency and prognostic value of loss of heterozygosity (LOH) and microsatellite instability (MSI) at 14 genetic loci located near or within regions containing important genes implicated in colorectal tumorigenesis.
METHODS: We studied colorectal cancers with corresponding normal mucosae in 207 patients (139 males and 68 females, mean age at the time of tumor resection 66.2±12.4 years, range 22-88 years). There were 37 right-sided colonic tumors, 85 left-sided colonic tumors and 85 rectal tumors. The distribution of tumor staging was stage I in 25, stage II in 73, stage III in 68, and stage IV in 41. We analyzed the LOH and MSI of HPC1, hMSH2, hMLH1, APC, MET, P53, NH23-H1, DCC, BAT25, BAT26, D17S250, MYCL1 and D8S254 with fluorescent polymerase chain reaction and denatured gel electrophoresis. High-frequency LOH was determined to be greater than three, or more than 50% of the informative marker with LOH. High-frequency MSI (MSI-H) was determined as more than four markers with instability (>30%). Correlations of LOH and MSI with clinical outcomes and pathological features were analyzed and compared.
RESULTS: The occurrence of MSI-H was 7.25%, located predominantly in the right colons (7/15) and had a higher frequency of poor differentiation (6/15) and mucin production (7/15). LOH in at least one genetic locus occurred in 78.7% of the tumors and was significantly associated with disease progression. Of the 166 potentially cured patients, 45 developed tumor recurrence within 36 mo of follow-up. Clinicopathological factors affecting 3-year disease-free survival (DFS) were TNM staging, grade of differentiation, preoperative CEA level, and high LOH status. Patients with high LOH tumors had a significantly lower DFS (50%) compared with patients with low LOH tumors (84%). Of the patients developing subsequent tumor recurrence, the number and percentage of LOH were 2.97 and 46.8% respectively, similar to the stage IV disease patients. TNM staging had the most significant impact on DFS, followed by high LOH status.
CONCLUSION: Clinical manifestations of LOH and MSI are different in colorectal cancer patients. High-frequency LOH is associated with high metastatic potential of colorectal cancers.
Keywords: Colorectal Cancer, Loss of Heterozygosity, Prognosis
INTRODUCTION
Cumulated genetic alterations could affect proto-oncogenes such as Ki-ras and tumor suppressor genes, including APC, DCC and TP53[1,2]. The common event that inactivates tumor suppressor genes is the loss of one allele frequently associated with an inactivating point mutation in the remaining wild-type allele. This process is referred to as loss of heterozygosity (LOH)[3].
In another mutational pathway, colorectal cancers display increased rates of intragenic mutation characterized by generalized instability of short, tandemly repeated DNA sequences known as microsatellites[4]. High frequency of microsatellite instability (MSI-H) has been found in more than 90% of tumors in patients with hereditary nonpolyposis colorectal cancer[5,6]. The presence of MSI could be the result of defective DNA mismatch repair genes that were misaligned in repetitive sequences (caused by slippage of polymerases) and left unrepaired. In sporadic cases of colorectal cancer, MSI-H occurred in approximately 15% of tumors[4,5,7].
Previous studies have demonstrated that tumors with more alleles lost have a considerably worse prognosis[8,9]. In contrast, the presence of MSI-H carries significant prognostic implications for colorectal cancer patients[10,11]. In this study, we evaluated 207 colorectal cancer patients. LOH and MSI at 14 markers were detected by microsatellite analysis to evaluate the clinical manifestations of LOH and MSI in sporadic colorectal cancers.
MATERIALS AND METHODS
Patients and clinicopathological data
A total of 207 patients with colorectal adenocarcinoma who underwent resection in Taipei Veterans General Hospital from January 1999 to December 1999 were enrolled in this study. Patients were excluded if they received preoperative chemotherapy and/or radiotherapy, and had evidence of hereditary non-polyposis colorectal cancer according to the criteria of Amsterdam or familial adenomatous polyposis, a malignant tumor outside the colon within the previous five years, or died from surgical complications. Clinical data were recorded prospectively and stored in computerized files. The database included (1) patients’ names, gender, age, family history, and major medical problems, (2) location, size, gross appearance, tumor-node-metastasis (TNM) stage, differentiation, and relevant pathologic prognostic features, and (3) types of operations, complications, recurrence of disease, and follow-up conditions. After operation, patients were followed up every 3 mo in the first 2 years, every 6 mo between 2 and 5 years, and annually thereafter. All patients were followed up for at least 3 years after operation or until death.
Tumor tissues
The Institutional Review Board of the Taipei Veterans General Hospital approved the study program. Consents to tissue collection were obtained from all patients. Following removal, specimens were cleaned thoroughly. The tumors were dissected meticulously and samples were collected from 4 different quadrants of the tumor for consideration of intratumoral heterogeneity. The corresponding normal mucosae taken at least 10 cm away from the primary tumor were collected. The fragments of samples were immediately frozen in liquid nitrogen and stored at -70 °C. Sections of cancer tissues and the corresponding normal mucosae were identified, reviewed, and analyzed by a senior gastrointestinal pathologist who did not know the clinical outcome of the patients. The stage of the disease was classified according to the TNM classification of the International Union Against Cancer[12]. There were 139 (67.1%) men and 68 (32.9%) women in the study group. The mean age at the time of tumor resection was 66.2±12.4 years (range 22-88 years). There were 37 (17.8%) right side colonic tumors (cecum to splenic flexure), 85 (41.1%) left side colonic tumors (splenic flexure to sigmoid colon) and 85 (41.1%) rectal tumors. The distribution of tumor staging was stage I in 25 (12.1%), stage II in 73 (35.3%), stage III in 68 (32.9%) and stage IV in 41 (19.8%).
LOH and MSI analysis
High-molecular-weight genomic DNA was prepared from each tumor and the corresponding normal tissue using the QIAamp tissue kit (QIAGEN GmbH, Hilden, Germany). Normal and tumor genomic DNA sample pairs were amplified using the microsatellite instability MSI/LOH assay starter kit (Applied Biosystems, Foster City, CA). According to the international criteria for the determination of MSI[13], five reference markers including D2S123, D5S345, Bat25, Bat26 and D17S250 were enrolled in our microsatellite panel. The chromosomal location of the microsatellite markers and genes surrounding the markers was described in a previous report[14]. In brief, 25 ng template DNA was amplified with fluorescent-labeled primer. PCR was carried out in a GeneAmp PCR System 9, 600 thermal cycler (Applied Biosystems) as follows: a 10-min pre-PCR incubation step at 95 °C, 30 cycles of amplification, each at 96 °C for 10 s, at 55 °C for 30 s, at 70 °C for 3 min, and a final extension at 70 °C for 30 min.
The amplification reactions were pooled and loaded onto a denaturing polyacrylamide gel, and the data were collected with an ABI 377 automated sequencer (Applied Biosystems). At the end of the run, each fluorescent peak was quantitated in terms of size (in base pairs), peak height, and area. Normal and tumor DNA pairs were compared for changes in peak height of each microsatellite marker via GeneScan analysis software (Applied Biosystems).
The LOH index was calculated using the modified method described by Cawkwell[15] for each paired normal and tumor samples. The peak height of two alleles in each tumor was divided by the peak height in normal samples: T1:T2/N1:N2 where T1 and N1 are the peak heights of the tumor and normal samples, respectively for the corresponding allele one, and T2 and N2 for the corresponding allele two. Allele loss, according to the manufacturer’s instructions, occurred with an LOH index of ≤0.67 or ≥1.5. This allele loss also translated to a 33% decrease in peak height of one of the tumor alleles as compared with the normal alleles. The result of LOH analysis of a representative sample is shown in Figure 1. The tumors that exhibited LOH at more than 3 markers, or showed more than 50% of informative markers, were classified as the high-frequency LOH group (LOH-high). Others were classified as the low-frequency LOH group (LOH-low).
Figure 1.
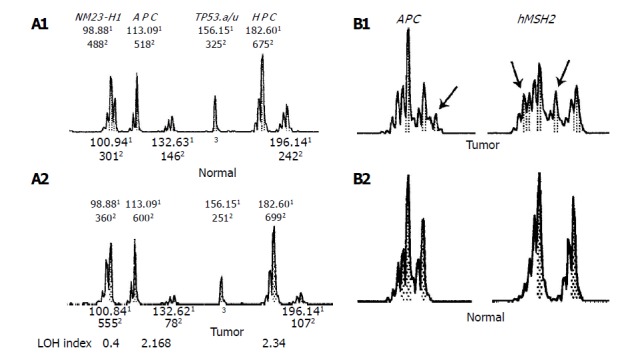
LOH at three genetic loci in a 52-yr-old male with stage IV disease (A) and MSI at two genetic loci in a 47-yr-old male with stage II disease (B) shown as representative results of GeneScan. A: LOH at three genetic loci in a 52-yr-old male with stage IV disease; B: MSI at two genetic loci in a 47-yr-old male with stage II disease. 1Size of the PCR product (in bp). 2Fluorescence intensity of peak. 3Homozygosity. The arrowhead indicated novel alleles.
Tumor samples that exhibited novel allele peaks compared with the corresponding normal samples were classified as MSI at that marker. Such markers were considered uninformative for the LOH study. The pattern of MSI of a representative sample is shown in Figure 2. The analysis was performed twice if the data were controversial. According to the international criteria for the determination of MSI[13], MSI in at least 4 or more loci was defined as high microsatellite instability (MSI-H); otherwise it was considered microsatellite stability (MSS).
Figure 2.

The number and percentage of LOH. Panel A: The number of LOH; Panel; B: The percentage of LOH.
Statistical analysis
The statistical end point in our analysis was the occurrence of metastasis or death. The group distribution of each clinicopathological characteristic according to the presence or absence of LOH or MSI was compared using the χ2 test. The numerical value was compared with the Student’s t test. Statistical significance was defined as P<0.05 (SPSS for Windows version 10.0). The variables that resulted in P values <0.1 were entered into the Cox proportional hazards model for the determination of the independent prognostic factor for colorectal cancer.
RESULTS
Markers with detectable heterozygous alleles were defined as informative. One hundred and sixty-three tumors (78.8%) exhibited LOH in at least one microsatellite marker. LOH in more than 3 markers (LOH-high) was noted in 93 tumors (44.9%). The highest frequency of LOH in the tumor suppressor gene was TP53.alu (65%), followed by DCC (64.3%), D8S254 (51.7%) and APC (47.8%) (Table 1, Figure 1A). There was no difference in the distribution of gender, age, preoperative CEA level and location of tumors between the LOH-high and LOH-low groups. Considering the pathological variables, there was a linear trend between TNM staging and the LOH-high tumors (P<0.001). The mean number and percentage of LOH in stage I tumors were 1.52% and 22.8%, respectively, with an increase to 3.02% and 48%, respectively, in stage IV tumors (Figure 2). The other pathological variables such as lymph-vascular invasion, mucin component, invasive pattern of tumors, and grade of differentiation showed no significant association with the LOH-high tumors (Table 2).
Table 1.
Frequency of LOH and MSI of tumor suppressor genes in sporadic colorectal cancers.
| Marker | Informative /Total (%) | LOH/Informative (%) | MSI No./Total (%) |
| NM23-H1 | 125/207 (60.4) | 43/125 (31.6) | 4/164 (2.4) |
| APC | 121/207 (58.5) | 56/121 (47.8) | 10/151 (6.6)1 |
| Tp53.alu | 103/207 (49.8) | 67/103 (65.0) | 1/140 (0.7) |
| HPC1 | 127/207 (61.4) | 15/127 (11.8) | 6/192 (3.1) |
| D8S254 | 89/207 (43.0) | 46/89 (51.7) | 5/161 (3.1) |
| DCC | 98/207 (47.3) | 63/98 (64.3) | 3/144 (2.1) |
| hMSH2 | 153/207 (73.9) | 24/153 (15.7) | 19/183 (10.4)1 |
| hMLH1 | 99/207 (47.8) | 26/99 (26.3) | 11/181 (6.1) |
| MET | 91/207 (44.0) | 27/91 (29.7) | 7/180 (3.9) |
| Tp53.pcr15 | 80/207 (38.6) | 31/80 (38.8) | 5/174 (2.9) |
| MYCL1 | 110/207 (53.1) | 38/110 (34.5) | 5/169 (3.0) |
| Bat-25 | 30/207 (14.5) | 3/30 (10) | 22/204 (9.8)1 |
| Bat-26 | 41/207 (19.8) | 4/41 (4.4) | 15/203 (7.4)1 |
| Mfd15 | 33/207 (15.9) | 2/33 (7.2) | 9/205 (4.4)1 |
Reference markers according to the international criteria for the determination of MSI[13].
Table 2.
Comparison between clinico-pathological features of loss of heterozygosity-high and loss of heterozygosity-low in colorectal cancer, n (%).
| Features | Total | LOH-high | LOH-low | P |
| Number of cases | 207 | 93 (44.9) | 114 (55.1) | |
| Age (yr) | 66.2±12.4 | 67.2±12.4 | 65.4±12.3 | 0.2921 |
| Gender (male/female) | 139/68 | 61/32 | 78/36 | 0.7782 |
| Location of tumor | ||||
| Right colon | 37 (17.9) | 16 (17.2) | 21 (18.4) | 0.5443 |
| Left colon | 85 (41.1) | 42 (45.2) | 43 (37.7) | |
| Rectum | 85 (41.1) | 35 (37.6) | 50 (43.9) | |
| TNM stage | ||||
| I | 25 (12.1) | 7 (7.5) | 18 (15.8) | <0.0014 |
| II | 73 (35.3) | 22 (23.7) | 51 (44.7) | |
| III | 68 (32.9) | 37 (39.8) | 31 (27.2) | |
| IV | 41 (19.8) | 27 (29.0) | 14 (12.3) | |
| Invasive pattern | ||||
| Expanding | 52 (25.1) | 20 (21.5) | 32 (28.1) | 0.2862 |
| Infiltrative | 155 (74.9) | 73 (78.5) | 82 (71.9) | |
| Grade | ||||
| Well-differentiated | 5 (2.4) | 2 (2.2) | 3 (2.6) | 0.2633 |
| Moderate differentiated | 182 (87.9) | 81 (87.0) | 101 (88.6) | |
| Poorly differentiated | 20 (9.7) | 10 (10.8) | 10 (8.8) | |
| Lymphovascular invasion | ||||
| Yes | 48 (23.2) | 27 (56.3) | 21 (43.7) | 0.1022 |
| No | 159 (76.8) | 66 (41.5) | 93 (58.5) | |
| Mucin production | ||||
| Yes | 29 (14.0) | 9 (9.7) | 20 (17.5) | 0.1552 |
| No | 178 (86.0) | 84 (90.3) | 94 (82.5) |
Student t test,
Fisher exact test,
Pearson χ2 test,
χ2 test with linear-by-linear association.
In a total of 2, 451 MSI analyses, excluding LOH markers, 122 markers had MSI (4.97%, Figure 1B). Fifty-five tumors (26.6%) exhibited MSI in at least one genetic locus. MSI in at least 4 or more loci (MSI-H) was noted in 15 tumors (7.25%). The distribution of MSI at different markers is shown in Table 1. The highest frequency of MSI was hMSH2 (10.4%), followed by Bat25 (9.8%), and Bat26 (7.4%). The information on the clinicopathological features of MSI-H and MSS colorectal cancers is shown in Table 3. Seven (46.7%) of the 15 MSI-H cancers were located in the right colon, a significantly greater number than that of the MSS cancers (15.6%). Poor tumor differentiation was found in 40% of MSI-H cancers (6/15), significantly higher than that found in MSS cancers (7.3%, 14/192). High mucin component of tumors was found in 46.7% of MSI-H cancers (7/15), significantly higher than that found in MSS cancers (22/192). There was no difference in the distribution of gender, TNM stage, invasive pattern of tumors, and lymph-vascular invasion between the MSI-H and MSS cancers.
Table 3.
Comparison between clinico-pathological features of MSI-H and MSS in colorectal cancer, n (%).
| Features | Total | MSI-H | MSS | P |
| Number of cases (%) | 207 | 15 (7.25) | 192 (92.7) | |
| Age (yr) | 66.2±12.4 | 69.7±14.8 | 65.9±12.2 | 0.2481 |
| Gender(male/female) | 139/68 | 8/7 | 131/61 | 0.3692 |
| Location of tumor | ||||
| Right colon | 37 (17.9) | 7 (46.7) | 30 (15.6) | 0.0053 |
| Left colon | 85 (41.1) | 2 (13.3) | 83 (43.2) | |
| Rectum | 85 (41.1) | 6 (40.0) | 79 (41.2) | |
| TNM stage | ||||
| I | 25 (14.2) | 1 (6.7) | 24 (12.5) | 0.7804 |
| II | 73 (36.8) | 6 (40.0) | 67 (34.9) | |
| III | 68 (31.6) | 6 (40.0) | 62 (32.3) | |
| IV | 41 (17.4) | 2 (13.3) | 39 (20.3) | |
| Invasive pattern | ||||
| Expanding | 52 (25.1) | 4 (26.7) | 48 (25.0) | 1.02 |
| Infiltrative | 155 (74.9) | 11 (73.3) | 144 (75.0) | |
| Grade of differentiation | ||||
| Well | 5 (2.4) | 0 | 5 (2.6) | <0.0013 |
| Moderate | 182 (87.9) | 9 (60.0) | 173 (90.1) | |
| Poor | 20 (9.7) | 6 (40.0) | 14 (7.3) | |
| Lymphovascular invasion | ||||
| Yes | 48 (23.2) | 1 (6.7) | 47 (24.5) | 0.2092 |
| No | 159 (76.8) | 14 (93.3) | 145 (75.5) | |
| Mucin component | ||||
| Yes | 29 (14.0) | 7 (46.7) | 22 (11.5) | 0.0012 |
| No | 178 (86.0) | 8 (53.3) | 170 (88.5) |
Student t test,
Fisher exact test,
Pearson χ2 test,
χ2 test with linear-by-linear association.
Of the 166 potentially cured patients, 45 developed tumor recurrence within a median of 36 mo of follow-up. The sites of tumor recurrence were: liver (20), lung (18), bone (5), peritoneum (5), remote nodes (4), and other sites (3). As shown in Table 4, 5 and Figure 3, the clinicopathological factors affecting the 3-year DFS were TNM staging, grade of differentiation, preoperative CEA level, and high LOH status. Using multivariate analysis, we identified that TNM staging showed the most significant impact on DFS [hazard ratio (HR) = 3.31; 95% confidence interval (CI) = 1.82-6.01], followed by high LOH status (HR = 3.18; 95% CI = 1.68-5.98) and CEA level (HR = 2.30; 95% CI = 1.21-4.37). Of the respective markers, LOH of Tp53.alu, D8S254, DCC, and Tp53.pcr15 had a significant impact on DFS, as shown in Table 6.
Table 4.
Correlation of the clinico-pathological factors with three-year disease-free survival in potentially cured colorectal cancer patients (stage I, II, III patients).
| Variables | Number of cases | 3-yr disease -free survival | P1 |
| Gender | |||
| Male | 116 | 70 | 0.614 |
| Female | 50 | 75 | |
| Preoperative CEA level | |||
| <5 ng/mL | 80 | 82 | 0.003 |
| >5 ng/mL | 86 | 63 | |
| TNM stage | |||
| I | 25 | 95 | <0.001 |
| II | 73 | 84 | |
| III | 68 | 47 | |
| Lymphovascular permeation | |||
| No | 129 | 73 | 0.091 |
| Yes | 37 | 63 | |
| Invasive pattern of tumor | |||
| Expansive | 45 | 73 | 0.517 |
| Infiltrative | 121 | 68 | |
| Grade of differentiation | |||
| Well | 3 | 100 | 0.004 |
| Moderate | 146 | 75 | |
| Poor | 17 | 48 | |
| Mucinous component | |||
| <50% | 143 | 72 | 0.642 |
| >50% | 23 | 68 | |
| Microsatellite instability | |||
| MSI-H | 13 | 69 | 0.91 |
| MSS | 153 | 71 | |
| LOH status | |||
| Low | 100 | 84 | <0.001 |
| High | 66 | 50 |
1Log-rank test
Table 5.
Multivariate analysis1.
| Factors | Hazard ratio | 95% CI | P |
| LOH (High vs Low) | 3.18 | 1.68-5.98 | <0.001 |
| TNM | 3.31 | 1.82-6.01 | <0.001 |
| CEA level | |||
| >5 ng/mL vs <5 ng/mL | 2.3 | 1.21-4.37 | 0.011 |
| Grade of differentiation | 1.12 | 0.45-2.77 | 0.791 |
| Lymphovascular invasion | 1 | 0.49-2.01 | 0.981 |
1The P value results from the hypothesis that the relative risk as determined by a multivariate binary logistic regression analysis equaled 1.0.
Figure 3.
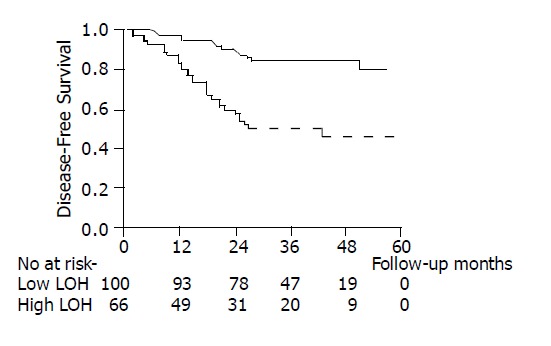
Comparison of the clinicopathological factors with 3-yr survival in colorectal cancer patients. ─ : Low LOH group;-----: High LOH group.
Table 6.
Informative markers that influenced three-year DFS in potentially cured colorectal cancer patients (stage I, II, III patients).
| Markers | Number of cases | 3-yr disease-free survival | P1 |
| Nm23-H1 | |||
| Non-LOH | 68 | 75 | 0.266 |
| LOH | 28 | 62 | |
| APC | |||
| Non-LOH | 54 | 75 | 0.881 |
| LOH | 45 | 72 | |
| Tp53.alu | |||
| Non-LOH | 29 | 92 | <0.001 |
| LOH | 55 | 51 | |
| HPC1 | |||
| Non-LOH | 87 | 75 | 0.743 |
| LOH | 12 | 70 | |
| D8S254 | |||
| Non-LOH | 39 | 75 | 0.03 |
| LOH | 31 | 52 | |
| DCC | |||
| Non-LOH | 27 | 79 | 0.042 |
| LOH | 45 | 55 | |
| MSH2 | |||
| Non-LOH | 107 | 73 | 0.972 |
| LOH | 15 | 75 | |
| MLH1 | |||
| Non-LOH | 62 | 66 | 0.287 |
| LOH | 19 | 57 | |
| Met ONC | |||
| Non-LOH | 50 | 72 | 0.35 |
| LOH | 16 | 64 | |
| Tp53.pcr15 | |||
| Non-LOH | 42 | 76 | <0.001 |
| LOH | 20 | 33 | |
| MYCL1 | |||
| Non-LOH | 59 | 71 | 0.421 |
| LOH | 29 | 65 |
1Log-rank test; The markers that had frequency of informative tumors lower than 30% were excluded in the survival analysis.
DISCUSSION
Our approach consisted of examining 14 microsatellite markers located near or within regions containing important genes implicated in the complex process of colorectal tumorigenesis. The use of automated assessment of loss of heterozygosity and microsatellite instability with fluorescent-PCR was described by Cawkwell[15] and could be adapted to the analysis of both LOH and MSI, two important mechanisms of colorectal carcinogenesis. In our results, the frequency of MSI exhibited a trend to cluster in hMSH2, BAT25 and BAT26 loci, but not in markers in regions displaying high levels of allelic loss (e.g., p53 and DCC). The higher susceptibility of mononucleotide repeats to instability in MSI tumors has been reported by Ionov[4] and Dietmaier[16]. The microsatellite markers with a low frequency MSI might provide prognostic information about LOH. The present study demonstrated a significant inverse correlation between the two molecular events (LOH vs MSI). The tumors with MSI-H phenotypes had a lower frequency of LOH than those with MSS phenotype.
In the present series of patients with resected primary colorectal carcinomas, the frequency of MSI-H phenotypes was 7.25%, lower than in previous reports[7,17]. This may be attributed to several markers having a low-frequency MSI. If only considering five reference markers[13] as the current definition of MSI-H, the frequency of MSI-H could be as high as 11.1% of tumors. Previous reports indicated that MSI-H colorectal cancers were associated with distinct clinicopathological features, including poor differentiation, extracellular mucin accumulation, younger age at onset, and proximal tumor location[18-20]. Our MSI-H tumors displayed similar clinicopathological findings.
Loss of heterozygosity, i.e., loss of one allele at a constitutional heterozygous locus indicates the probability of loss of a tumor suppressor gene, which might promote neoplastic progression[21,22]. The frequency of an informative tumor for each microsatellite marker varied in the present study (range 14.5-73.9%). Analysis of a single marker may be biased by non-informative tumors, especially in the marker in which the frequency of an informative tumor was less than 50%. Therefore, we defined the LOH-high group as LOH at more than 3 markers, or more than 50% of informative markers for considering both the number and percentage of LOH. In our results, high-frequency LOH could be an independent prognostic factor affecting tumor recurrence. Of the 166 potentially cured patients, those with a high LOH tumor showed a DFS of 50 %, which was poorer than those with low LOH tumors (84%). Of the 45 patients who developed tumor recurrence, the mean number of LOH was 2.97 and the percentage of LOH in the primary tumor was 46.8%, which were similar to those tumors that already had distant metastasis at operation. These findings imply that high LOH tumors behave comparably to those stage IV diseases.
Metastasis is a complicated multistep process. The cells must detach and acquire motility, invade through the basement membrane and extracellular matrix, gain access to and survive in the vascular system, adhere to and immigrate into the target organ, and then proliferate in the new site[23]. It is reasonable to believe that multiple genes control the complicated process. Previous reports have hypothesized that accumulation of various genetic abnormalities is more important in tumorigenesis than the sequence of genetic alterations[2,24].
Additionally, a previous report demonstrated that p53 and DCC were the most common mutated tumor suppressor genes in colorectal cancer, and occurred mainly at the late phase of carcinogenesis[1]. In our study, the frequency of LOH was the highest at p53 (65.0%) followed by DCC (64.3%), D8S254 (51.7%) and APC (47.8%). There were four markers associated with metastatic potential, including p53, DCC, and a tumor suppressor gene located in the 8p22. In our series, the incidence of LOH of p53 was similar to a previous report[24]. The p53 gene has been thought to modulate two discrete functions, both in response to stress (particularly DNA damage), cell cycle arrest, and apoptosis[25,26]. The loss of p53 in the final stages of colon cancer not only allows the cells to divide at an unrestrained rate, but this rapid replication further promotes the development of even more genetic mutations because the cell cycle is not interrupted when errors occur. Several authors have found that mutations of p53 or allelic loss of the p53 was correlated with poor outcomes[27-29]. Our study yielded a similar finding. The incidence of allele loss of DCC in colorectal cancer has been reported to be from 33%[15] to 75%[29-31]. According to previous reports, LOH or decreased DCC expression was associated with liver and lymph node metastasis[29,31]. In our study, LOH of DCC was significantly associated with lymph node metastasis (data not shown). The DCC protein was a transmembrane glycoprotein similar to the neural cell adhesion molecules[32]. LOH of DCC could thus lead to impaired contacts between cells, thereby contributing to tumor growth and invasion. The prognostic value of LOH of DCC has been confirmed in several previous studies[31,33,34]. Our observations also showed that LOH of DCC was significantly associated with metastasis potential.
Another putative tumor suppressor gene located in the short arm of chromosome 8 had a high frequency of LOH in our study, as shown in a previous report[35]. Several studies showed that this novel tumor suppressor gene locus was related to the stage progression of colorectal cancer[36,37], similar to our findings.
In the multivariate analysis, TNM stage was the most significantly independent prognostic factor for colorectal cancer, followed by high-frequency LOH and preoperative CEA level. The outcome of patients with colorectal cancer was determined by the presence or absence of metastasis at the time of operation. We further analyzed the prognostic impact of high LOH status on early state tumors (stage I or II). Of the 29 stage I or II tumor patients with high-frequency LOH, 9 patients (32.1%), developed distant metastasis within three years after surgery, which was significantly higher than those with low-frequency LOH (8.7%, P = 0.037). This finding implies that early stage tumors with LOH-high have a stronger metastatic potential.
In conclusion, genetic change in both MSI and LOH phenotypes may occur in sporadic colorectal tumors, as established by microsatellite analysis. Variation of genetic phenotypes is related to different clinical manifestations, an important finding that may be useful in identifying subgroups of patients with different prognostic risks.
Footnotes
Supported by grants from the Veterans General Hospital-Taipei, VGH90348 and VGH910305
Edited by Wang XL Proofread by Zhu LH
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton SR. Molecular genetics of colorectal carcinoma. Cancer. 1992;70:1216–1221. doi: 10.1002/1097-0142(19920901)70:3+<1216::aid-cncr2820701505>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 3.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 4.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 5.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 6.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomäki P, Chadwick RB, Kääriäinen H, Eskelinen M, Järvinen H, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338:1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 8.Kern SE, Fearon ER, Tersmette KW, Enterline JP, Leppert M, Nakamura Y, White R, Vogelstein B, Hamilton SR. Clinical and pathological associations with allelic loss in colorectal carcinoma [corrected] JAMA. 1989;261:3099–3103. doi: 10.1001/jama.261.21.3099. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 10.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 11.Lukish JR, Muro K, DeNobile J, Katz R, Williams J, Cruess DF, Drucker W, Kirsch I, Hamilton SR. Prognostic significance of DNA replication errors in young patients with colorectal cancer. Ann Surg. 1998;227:51–56. doi: 10.1097/00000658-199801000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobin LH, Wittekind C. UICC TNM classification of malignant tumors. 5th ed. New York: Wiley Liss; 1997. pp. 66–69. [Google Scholar]
- 13.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 14.Lin JK, Chang SC, Yang YC, Li AF. Loss of heterozygosity and DNA aneuploidy in colorectal adenocarcinoma. Ann Surg Oncol. 2003;10:1086–1094. doi: 10.1245/aso.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Cawkwell L, Lewis FA, Quirke P. Frequency of allele loss of DCC, p53, RBI, WT1, NF1, NM23 and APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer. 1994;70:813–818. doi: 10.1038/bjc.1994.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Rüschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 17.Speicher MR. Microsatellite instability in human cancer. Oncol Res. 1995;7:267–275. [PubMed] [Google Scholar]
- 18.Risio M, Reato G, di Celle PF, Fizzotti M, Rossini FP, Foà R. Microsatellite instability is associated with the histological features of the tumor in nonfamilial colorectal cancer. Cancer Res. 1996;56:5470–5474. [PubMed] [Google Scholar]
- 19.Jass JR, Do KA, Simms LA, Iino H, Wynter C, Pillay SP, Searle J, Radford-Smith G, Young J, Leggett B. Morphology of sporadic colorectal cancer with DNA replication errors. Gut. 1998;42:673–679. doi: 10.1136/gut.42.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forster S, Sattler HP, Hack M, Romanakis K, Rohde V, Seitz G, Wullich B. Microsatellite instability in sporadic carcinomas of the proximal colon: association with diploid DNA content, negative protein expression of p53, and distinct histomorphologic features. Surgery. 1998;123:13–18. [PubMed] [Google Scholar]
- 21.Lasko D, Cavenee W, Nordenskjöld M. Loss of constitutional heterozygosity in human cancer. Annu Rev Genet. 1991;25:281–314. doi: 10.1146/annurev.ge.25.120191.001433. [DOI] [PubMed] [Google Scholar]
- 22.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 23.Fidler IJ. 7th Jan Waldenström Lecture. The biology of human cancer metastasis. Acta Oncol. 1991;30:668–675. doi: 10.3109/02841869109092438. [DOI] [PubMed] [Google Scholar]
- 24.Fearon ER, Jones PA. Progressing toward a molecular description of colorectal cancer development. FASEB J. 1992;6:2783–2790. doi: 10.1096/fasebj.6.10.1321771. [DOI] [PubMed] [Google Scholar]
- 25.Bates S, Hickman ES, Vousden KH. Reversal of p53-induced cell-cycle arrest. Mol Carcinog. 1999;24:7–14. doi: 10.1002/(sici)1098-2744(199901)24:1<7::aid-mc2>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Kastan M. On the TRAIL from p53 to apoptosis? Nat Genet. 1997;17:130–131. doi: 10.1038/ng1097-130. [DOI] [PubMed] [Google Scholar]
- 27.Campo E, Miquel R, Jares P, Bosch F, Juan M, Leone A, Vives J, Cardesa A, Yague J. Prognostic significance of the loss of heterozygosity of Nm23-H1 and p53 genes in human colorectal carcinomas. Cancer. 1994;73:2913–2921. doi: 10.1002/1097-0142(19940615)73:12<2913::aid-cncr2820731207>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Hamelin R, Laurent-Puig P, Olschwang S, Jego N, Asselain B, Remvikos Y, Girodet J, Salmon RJ, Thomas G. Association of p53 mutations with short survival in colorectal cancer. Gastroenterology. 1994;106:42–48. doi: 10.1016/s0016-5085(94)94217-x. [DOI] [PubMed] [Google Scholar]
- 29.Ookawa K, Sakamoto M, Hirohashi S, Yoshida Y, Sugimura T, Terada M, Yokota J. Concordant p53 and DCC alterations and allelic losses on chromosomes 13q and 14q associated with liver metastases of colorectal carcinoma. Int J Cancer. 1993;53:382–387. doi: 10.1002/ijc.2910530307. [DOI] [PubMed] [Google Scholar]
- 30.Barletta C, Scillato F, Sega FM, Mannella E. Genetic alteration in gastrointestinal cancer. A molecular and cytogenetic study. Anticancer Res. 1993;13:2325–2329. [PubMed] [Google Scholar]
- 31.Jen J, Kim H, Piantadosi S, Liu ZF, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–221. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 32.Cho KR, Fearon ER. DCC: linking tumour suppressor genes and altered cell surface interactions in cancer? Eur J Cancer. 1995;31A:1055–1060. doi: 10.1016/0959-8049(95)00128-6. [DOI] [PubMed] [Google Scholar]
- 33.Font A, Abad A, Monzó M, Sanchez JJ, Guillot M, Manzano JL, Piñol M, Ojanguren I, Rosell R. Prognostic value of K-ras mutations and allelic imbalance on chromosome 18q in patients with resected colorectal cancer. Dis Colon Rectum. 2001;44:549–557. doi: 10.1007/BF02234328. [DOI] [PubMed] [Google Scholar]
- 34.Ogunbiyi OA, Goodfellow PJ, Herfarth K, Gagliardi G, Swanson PE, Birnbaum EH, Read TE, Fleshman JW, Kodner IJ, Moley JF. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–433. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara Y, Ohata H, Emi M, Okui K, Koyama K, Tsuchiya E, Nakajima T, Monden M, Mori T, Kurimasa A. A 3-Mb physical map of the chromosome region 8p21.3-p22, including a 600-kb region commonly deleted in human hepatocellular carcinoma, colorectal cancer, and non-small cell lung cancer. Genes Chromosomes Cancer. 1994;10:7–14. doi: 10.1002/gcc.2870100103. [DOI] [PubMed] [Google Scholar]
- 36.Fujiwara Y, Emi M, Ohata H, Kato Y, Nakajima T, Mori T, Nakamura Y. Evidence for the presence of two tumor suppressor genes on chromosome 8p for colorectal carcinoma. Cancer Res. 1993;53:1172–1174. [PubMed] [Google Scholar]
- 37.Yaremko ML, Wasylyshyn ML, Paulus KL, Michelassi F, Westbrook CA. Deletion mapping reveals two regions of chromosome 8 allele loss in colorectal carcinomas. Genes Chromosomes Cancer. 1994;10:1–6. doi: 10.1002/gcc.2870100102. [DOI] [PubMed] [Google Scholar]


