Abstract
AIM: By using comparative genomic hybridization, gain of 3q was found in 45-86% cases of esophageal squamous cell carcinoma (EC-SCC). Chromosome 3q25.3-qter is the minimal common region with several oncogenes found within this region. However, amplification patterns of these genes in EC-SCC have never been reported. The possible association of copy number changes of these genes with pathologic characteristics is still not clear.
METHODS: Real-time quantitative PCR (Q-PCR) was performed to analyze the copy number changes of 13 candidate genes within this region in 60 primary tumors of EC-SCC, and possible association of copy number changes with pathologic characteristics was analyzed by statistics. Immunohistochemistry (IHC) study was also performed on another set of 111 primary tumors of EC-SCC to verify the association between TP63 expression change and lymph node metastasis status.
RESULTS: The average copy numbers (±SE) per haploid genome of individual genes in 60 samples were (from centromere to telomere): SSR3: 4.19 (±0.69); CCNL1: 5.24 (±0.67); SMC4L1: 2.01 (±0.16); EVI1: 2.02 (±0.12); hTERC: 5.28 (±0.54); SKIL: 2.71 (±0.14); EIF5A2: 1.95 (±0.12); ECT2: 9.18 (±1.68); PIK3CA: 8.13 (±1.17); EIF4G1: 1.07 (±0.05); SST: 3.07 (±0.25); TP63: 2.51 (±0.22); TFRC: 2.42 (±0.19). Four clusters of amplification were found: SSR3 and CCLN1 at 3q25.31; hTERC and SKIL at 3q26.2; ECT2 and PIK3CA at 3q26.31-q26.32; and SST, TP63 and TFRC at 3q27.3-q29. Patients with lymph node metastasis had significantly lower copy number of TP63 in the primary tumor than those without lymph node metastasis. IHC study on tissue arrays also showed that patients with lymph node metastasis have significantly lower TP63 staining score in the primary tumor than those without lymph node metastasis.
CONCLUSION: This study showed that different amplification patterns were seen among different genes within 3q25.3-qter in EC-SCC, and several novel candidate oncogenes (SSR3, SMC4L1, ECT2, and SST) were identified. TP63 is amplified in early stage of EC-SCC carcinogenesis but down-regulated in advanced stage of disease.
Keywords: Chromosomal aberration, Comparative genomic hybridization, Esophageal neoplasm, Immunohistochemistry, Quantitative real-time PCR, Tissue array, Tumor protein 63
INTRODUCTION
Esophageal cancer (EC) is one of the most lethal cancers in the world[1] as well as in Taiwan (http://www.doh.gov.tw/dohenglish/Upload/Statistics/S02/9110-eng.xls). In most Asian countries, squamous cell carcinoma (EC-SCC) is the most frequent histological subtype of EC. By using modern molecular cytogenetic study such as comparative genomic hybridization (CGH)[2] and spectral karyotyping (SKY)[3], several recurrent chromosomal aberrations of EC-SCC were identified. Among them, chromosome 3q is especially important, because 45-86% cases of EC-SCC were found to have amplification in this region, with minimal overlapping region over 3q25.3-qter[4-8]. Gain of chromosome 3q was also reported in many other tumors, such as cancer of the lung[9], ovary[10], and cervix[11,12], and head and neck squamous cell carcinoma (HNSCC)[13,14].
The size of 3q25.3-qter is about 40 MB, containing about 367 known genes within this region, not to mention the unknown ones. Several candidate oncogenes were located in this chromosomal region, such as cyclin L1 (CCNL1), at 3q25; human telomerase RNA component (hTERC), at 3q26; phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA), at 3q26; Ski-like protein (SKIL), at 3q26; ecotropic viral integration site 1 (EVI1), at 3q26; eukaryotic translation initiation factor 5A2 (EIF5A2), at 3q26; eukaryotic translation initiation factor 4gamma1 (EIF4G1), at 3q27; tumor protein 63 (TP63), at 3q28; and transferrin receptor (TFRC), at 3q29 (GeneCardsTM, http://bioinformatics.weizmann.ac.il/cards/). However, amplification patterns of these genes in EC-SCC have never been reported.
A recently introduced method, real-time quantitative PCR (Q-PCR), could be used to accurately evaluate copy numbers of genes with only minimal amounts of tumor materials with high-throughput capacity[15-17]. In previous study, the accuracy of assessing copy number changes by Q-PCR in comparison with fluorescence in situ hybridization (FISH) had been demonstrated[18]. In this study, Q-PCR was used to study the copy number changes of the afore mentioned nine candidate oncogenes, together with four genes [signal sequence receptor gamma (SSR3) at 3q25.31, structural maintenance of chromosomes 4-like 1 (SMC4L1) at 3q26.1, epithelial cell transforming sequence 2 oncogene (ECT2) at 3q26.31, and somatostatin (SST) at 3q27.3] that were not down-regulated in our preliminary expression profile study, within chromosome 3q25.3-qter region in 60 primary tumors of EC-SCC, and the possible associations of these changes with disease progression were analyzed. Immunohistochemistry (IHC) was also used to verify the association of expression change of TP63 with lymph node metastasis status on another 111 cases of EC-SCC.
MATERIALS AND METHODS
Primary tumors and cell lines of EC-SCC
From 1995 to 1997, 60 ethic Chinese patients with EC-SCC were enrolled for Q-PCR study. All patients underwent primary surgical resection without neoadjuvant chemotherapy or radiotherapy. Only patients with written informed consent were included. Pathological evaluation of depth of tumor invasion, tumor differentiation and lymph node metastasis were done by one of our pathologists (Jung-Ta Chen), and staging and grading of tumor were defined according to the Cancer Staging Manual (5th edition; American Joint Committee on Cancer). Portions of tumor from the paraffin-embedded primary tumor samples, of which at least 70% were tumor cells, were identified under the microscope by one of our pathologists (Jung-Ta Chen) and were cut out for study. The procedure of DNA extraction was modified from a previously described protocol[19]. CGH analysis of part of these patients had been previously reported[8]. DNA extracted from five volunteer donor lymphocytes was used as control. EC-SCC cell lines CE 48T/VGH, CE 81T/VGH, TE6 and TE9, which have been previously characterized by molecular cytogenetics[18], were used to validate the accuracy of Q-PCR.
Fluorescence in situ hybridization (FISH)
FISH was performed using methods as previously suggested[20]. The search for FISH probes covering the 13 genes was done by browsing Ensembl Genome Browser http://www.ensembl.org/ and UCSC Genome Browser http://genome.ucsc.edu for candidate bacterial artificial chromosome (BAC) clones. The resulting clones were then obtained from RPCI-11 BAC library (Table 1)[21]. Their identities were verified by FISH-mapping onto normal lymphocyte metaphases. For each cell line, FISH signals were counted in 10 metaphases, and FISH signals per haploid genome were calculated by using average FISH signals per cell ×23/ average number of chromosomes per cell[18].
Table 1.
Covering BAC clones and position of 13 genes and sequence of Q-PCR primers of LINE1 and 13 genes over 3q25.3-qter.
| Genes | BAC clones | Position | Forward primer | Reverse primer |
| LINE1 | - | - | CCGCTCAACTACATGGAAACTG | GCGTCCCAGAGATTCTGGTATG |
| SSR3 | 304C15 | 3q25.31 | GCCCAGGCATATGAGAGTTGTC | CCAACATGGCAGGGTCAAGT |
| CCNL1 | 6F2 | 3q25.31 | TCATGGCAGTCAACCAACAT | CCATTGTAAGGGCTTTTGGA |
| SMC4L1 | 227J5 | 3q26.1 | GGCAAAGTCCTAAGCAAGGTTGT | TCAACTGGCAAGCTAAGTGGAA |
| EVI1 | 141C22 | 3q26.2 | CATGCATGCTGATTGCAGAAC | CCACCTGCCGCAAAATGGT |
| hTERC | 40O08 | 3q26.2 | CGTAGGCGCCGTGCTTT | TTTTCCGCCCGCTGAA |
| SKIL | 543D10 | 3q26.2 | GCTCGGCATTCCCAAGAAA | CCCCTTCCAACACAGTCTGAGT |
| EIF5A2 | 110O7 | 3q26.2 | GGCTTCCAGCACTTACCCTATG | GACCATGCTTTCCAGTTTTGG |
| ECT2 | 453J16 | 3q26.31 | CCTAACAGCAATCGCAAACG | CTGTCTCTCTTGAAAGCTGAGCAA |
| PIK3CA | 245C23 | 3q26.32 | GGAGGATGCCCAATTTGATG | AACAGTCCATTGGCAGTTGAGA |
| EIF4G1 | 481O2 | 3q27.1 | CCGGTTCCAGAATCTGAGTTTT | GATTCGGAGGGGCAAGCTG |
| SST | 211G3 | 3q27.3 | GATGCCCTGGAACCTGAAGA | GCCGGGTTTGAGTTAGCAGAT |
| TP63 | 313I6 | 3q28 | CCTCGTCCACCAGTCCCTAT | GGAAGGACACGTCGAAACTG |
| TFRC | 313F11 | 3q29 | CAAAGTGGCACGAGCAGCTG | GCTCTAGAATGAACGGTGGAAG |
LINE1: long interspersed elements; SSR3: signal sequence receptor gamma; CCNL1: cyclin L1; SMC4L1:structural maintenance of chromosomes 4-like 1; EVI1: ecotropic viral integration site 1; hTERC: human telomerase RNA component; SKIL: Ski-like protein; EIF5A2: eukaryotic translation initiation factor 5A2; ECT2: epithelial cell transforming sequence 2 oncogene; PIK3CA: phosphoinositide-3-kinase, catalytic, alpha polypeptide; EIF4G1: eukaryotic translation initiation factor 4gamma1; SST: somatostatin; TP63:tumor protein 63; TFRC: transferrin receptor. BAC clones, clones of bacterial artificial chromosomes covering the genes obtained from RPCI-11 library.
Real-time quantitative PCR (Q-PCR)
All primers were designed with Primer Express 3.0 software (Applied Biosystems Foster City, CA) using default parameters, with modified minimum amplicon length requirements (85 bp). An additional requirement consisted of a maximum GC content of 40% for the five last 3’ end nucleotides. The sequences of the primers are listed in Table 1. PCR reactions were performed as previously described[18]. DNA content was normalized to that of long interspersed elements (LINE1), a repetitive element for which copy numbers per haploid genome are similar both in normal or neoplastic cells[17]. Copy number changes per haploid genome were calculated by using the formula 2(Nt-Nline)-(Tt-Tline), where Nt is the threshold cycle number observed for an experimental primer in the normal DNA sample, Nline is the threshold cycle number observed for a LINE1 primer in the normal DNA sample, and Tt and Tline are the threshold cycle numbers observed for the experimental primer and LINE1 primer in test DNA sample, respectively[17]. For normal cell the copy number of a gene per haploid genome should be one. PCR for each primer set were performed in triplicate, and calculated copy number changes per haploid genome were averaged.
Construction of tissue arrays
Tissue arrays of another 120 EC-SCC tumor samples were constructed using method as previously described[22]. Briefly, the H&E-stained slides of selected tumor samples were examined under a light microscope. The areas of interest were circled with a color pen, and a 16-gauge bone marrow biopsy trephine apparatus was used to punch at the circled areas, extracting a tissue cylinder with 2.0 mm diameter. At least three cylindrical core biopsies were taken from different sites of each tumor. About 40 cylinders (8×5) were carefully transferred with forceps to a recipient metal paraffin block box. The recipient box was then covered with a plastic cassette and then liquid wax was gently poured into the box until it was full. The box was then put on a hot plate for 1 min to homogenize the wax, after which the box was removed from the hot plate and cooled to room temperature slowly. Four-micrometer sections were cut and mounted on silane-coated slides.
Immunohistochemistry (IHC) for TP63
Immunostaining for TP63 was performed on tissue array slides using a mouse anti-human TP63 monoclonal antibody clone: 7JUL (1:30; Novocastra, New Castle Upon Tyne, UK), which recognized all TP63 isoforms, and high temperature antigen unmasking technique (autoclave in pH 8.0 EDTA buffer) as previously described[23]. The bound antibodies were detected using the DAKO Envision system. Positive control (non-neoplastic tonsil tissue) and negative control (replacement of primary antibody by PBS) were included. The slides were independently reviewed by two of the authors (Chin-Chen Pan, Paul Chih-Hsueh Chen) who were blinded to the clinicopathologic data. TP63 immunoreactivity was semi-quantified using a combined intensity and percentage of positive scoring method. Intense nuclear staining was scored as 2, weak as 1, and negative as 0 (Figure 1)[24]. The percentage of cells with each intensity score was estimated. A TP63 staining score was defined as the sum of the percentage of positive cells with each intensity level multiplied by the intensity score [e.g., a case with 30% intense staining and 40% weak staining would be scored as 100 (30×2+40×1)][24].
Figure 1.
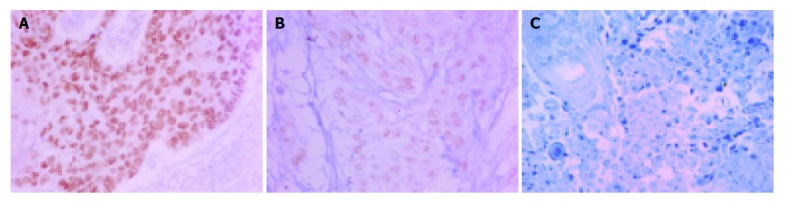
Immunoreactivity for TP63. A: Intense nuclear TP63 staining; B: Weak nuclear TP63 staining; C: Negative staining. (Original magnification: ×400).
Statistical analysis
Statistical comparisons were performed using SPSS 12.0 software. Pearson coefficient of correlation was used to determine the correlation between the copy number changes assessed by FISH and Q-PCR. Student’s t-test was used to test the association between the results of Q-PCR or the TP63 staining score and lymph node metastasis or differentiation status. Kruskal-Wallis test was used to test the association between the results of Q-PCR or the TP63 staining score and primary tumor extent.
RESULTS
Evaluation of Q-PCR accuracy
The accuracy of the assay was tested on 4 EC-SCC cell lines. In Figure 2, copy number changes of 13 genes on these 4 EC-SCC cell lines, assessed by both FISH and Q-PCR, were quite compatible. The Pearson coefficients of correlation for comparing FISH and Q-PCR of 13 genes in 4 cell lines were all >0.9.
Figure 2.
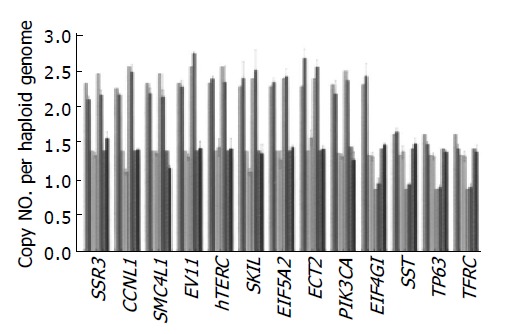
Comparison of copy numbers assessed by FISH (stripped bar) and Q-PCR (±SD) (hollow bar) of 13 genes in 4 EC-SCC cell lines [CE 48T/VGH (pink), CE 81T/VGH (yellow), TE6 (green), TE9 (blue)]. Pearson coefficients of correlation for comparing FISH and Q-PCR of 13 genes in 4 cell lines were all >0.9.
Assessment of copy number changes of candidate genes in primary tumors by Q-PCR
Copy number changes of 13 candidate genes were assessed by Q-PCR in 60 primary EC-SCC tumors. The average copy numbers (±SE) per haploid genome of individual genes in 60 samples were (from centromere to telomere): SSR3: 4.19 (±0.69); CCNL1: 5.24 (±0.67); SMC4L1: 2.01 (±0.16); EVI1: 2.02 (±0.12); hTERC: 5.28 (±0.54); SKIL: 2.71 (±0.14); EIF5A2: 1.95 (±0.12); ECT2: 9.18 (±1.68); PIK3CA: 8.13 (±1.17); EIF4G1: 1.07 (±0.05); SST: 3.07 (±0.25); TP63: 2.51 (±0.22); TFRC: 2.42 (±0.19). Four clusters of amplification could be found: SSR3 and CCLN1 at 3q25.31; hTERC and SKIL at 3q26.2; ECT2 and PIK3CA at 3q26.31-q26.32; and SST, TP63 and TFRC at 3q27.3-q29 (Figure 3).
Figure 3.
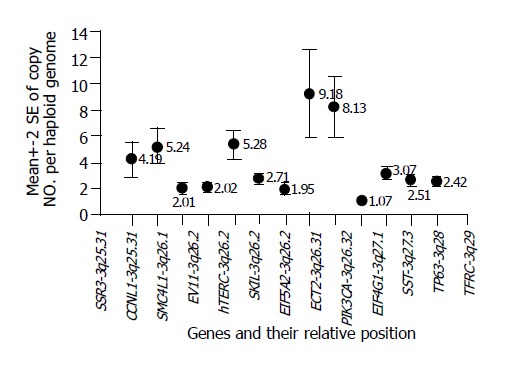
Average copy number changes (indicated by the circles with number listed beside them; crossbar represented 2×standard error) of 13 genes in association of their relative position on chromosome 3q. Four clusters of amplifications could be found: SSR3 and CCLN1 at 3q25.31; hTERC and SKIL at 3q26.2; ECT2 and PIK3CA at 3q26.31-q26.32; and SST, TP63 and TFRC at 3q27.3-q29.
Association of copy number changes of candidate genes in primary tumors with disease staging or differentiation
The association between copy number changes of 13 genes and primary tumor extent, lymph node metastasis or tumor differentiation was analyzed. Patients with lymph node metastasis have significantly lower copy number of TP63 in the primary tumor than those without lymph node metastasis (2.05 vs 3.08; P = 0.017) (Figure 4). There were no significant association between copy number changes of the other 12 genes and lymph node metastasis. Also the copy number changes of all 13 genes had no statistical association with either primary tumor extent or tumor differentiation.
Figure 4.
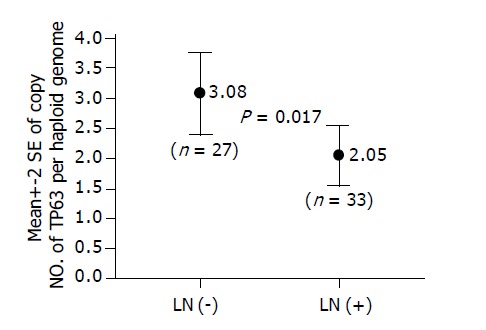
In Q-PCR analysis of 60 EC-SCC tumors, patients with lymph node metastasis [LN (+)] have significantly lower copy number of TP63 in the primary tumor than those without lymph node metastasis [LN (-)] (2.05 vs 3.08; P = 0.017).
Association of TP63 staining score in primary tumors with disease staging or differentiation
Immunostaining of TP63 could be evaluated in 111 cases. The association between TP63 staining score and primary tumor extent, lymph node metastasis or tumor differentiation was analyzed. As in the study of Q-PCR, patients with lymph node metastasis have significantly lower TP63 staining score in the primary tumor than those without lymph node metastasis (51.03 vs 79.62; P = 0.034) (Figure 5). There was no statistical association between TP63 staining score and either primary tumor extent or tumor differentiation.
Figure 5.
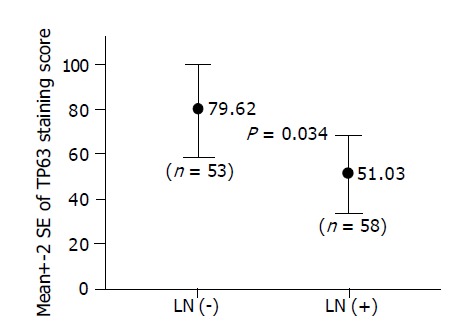
In IHC study of 111 EC-SCC tumors, patients with lymph node metastasis [LN (+)] have significantly lower TP63 staining score in the primary tumor than those without lymph node metastasis [LN (-)] (51.03 vs 79.62; P = 0.034).
DISCUSSION
In this study, the pattern of copy number changes of 13 potential target genes located on 3q25.3-qter in EC-SCC and the association of these changes with disease progression were analyzed. In general, all genes except FIE4G1 had increased copy number. However, four clusters of genes with higher amplification were found: SSR3 and CCNL1 at 3q25.31; hTERC and SKIL at 3q26.2; ECT2 and PIK3CA at 3q26.31-q26.32; and SST, TP63 and TFRC at 3q27.3-q29, with highest peaks over ECT2 and PIK3CA (Figure 3). In previous study, it was demonstrated that copy number of PIK3CA was significantly higher than that of TP63 in EC-SCC[18]. Therefore, in EC-SCC, 3q26.31-q26.32 might be the mostly amplified area within this region.
CCNL1 [25], hTERC [26-28], SKIL[29], PIK3CA[30-32], TP63 [33-37] and TFRC [38] were reported as potential oncogenes in different diseases . However, SSR3, SMC4L1, ECT2 and SST have never been previously identified as oncogenes. SSR3 is one of the 4 members of translocon-associated protein (TRAP) over endoplasmic reticulum (ER) membrane. TRAP is responsible for the passage of peptide through ER membrane[39]. SMC4L1 is one of the members of structural maintenance of chromosomes (SMC) proteins. The eukaryotic SMC proteins could form heterodimers, which may be involved in chromosome condensation, sister chromatid cohesion, and DNA recombination[40]. ECT2 was identified as an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis[41]. Recent studies have showed that N-terminal truncation of ECT2 may render it as an oncogenic protein, which may cause malignant transformation of cell[42]. SST (somatostatin) and its analogues have been used in the treatment of endocrine tumor. Whether these genes play roles in carcinogenesis of EC-SCC deserve further investigation.
It is very interesting to find that the copy number changes as well as expression of TP63 decrease in more advanced stage of disease in EC-SCC. Heselmeyer et al reported that gain of chromosome 3q could be found in the early dysplasia lesion as well as in the invasive carcinoma of cervical cancer, but at reduced frequency in advanced stage of disease[11,12]. However, this phenomenon has never been reported in EC-SCC.
TP63 gene, one of TP53 gene family, is a well-known oncogene within this region. Overexpression of TP63 was found in most squamous cell carcinoma[33-37]. In a IHC study of EC-SCC, TP63 protein was found highly expressed in carcinoma (50/51), dysplasia (10/11), and even in histologically normal epithelia of esophagus adjacent to the cancerous tissues (45/47)[35]. On the contrary, in the study of TP63 expression in urothelial carcinoma, Koga et al reported that lower TP63 expression was significantly associated with higher Tumor-Node- Metastasis (TNM) stage (P = 0.0004) and lymph-node metastasis (P = 0.013). It was found that cancer cells with lower TP63 staining had higher chance of lower membranous β-catenin expression, which plays a role in cell-cell adherent junctions, and cancer invasion and metastasis could be promoted by reduced membranous β-catenin expression[24]. In a similar study of upper urinary tract urothelial carcinoma, Zigeuner et al found that decreased TP63 immunoreactivity was significantly associated with advanced tumor stages and poor prognosis. They also found that cases with decreased TP63 immunoreactivity had higher chance of TP53 overexpression in comparison with cases with normal TP63 immunoreactivity.
By combining results of this study with other reports, it is very likely that amplification of genes located on 3q25.3-qter may occur in quite early stage in the carcinogenesis of EC-SCC. But some of the genes, such as TP63, may be down-regulated as disease progressed. One possible explanation is that the alterations of genes are no longer necessary for maintenance of cancer cells survival. The other possibility is that down-regulation of these genes may accompany alterations of other genes (such as β-catenin or TP53), which may render cancer cell more invasive or malignant.
In conclusion, this study demonstrated different amplification patterns of different genes within 3q25.3-qter in EC-SCC, with highest amplification over 3q26.31-q26.32. SSR3, SMC4L1, ECT2 and SST were identified as novel candidate oncogenes within this region. TP63 is amplified in early stage of EC-SCC carcinogenesis but down-regulated in advanced stage of disease.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the technical support from National Yang-Ming University Genome Research Center (http://genome.ym.edu.tw/).
Footnotes
Supported by the National Microarray and Gene Expression Analysis Core Facility of the National Research Program for Genomic Medicine at National Yang-Ming University (http://www.ym.edu.tw/microarray), and annual project Grant From National Science Council (Grant NO. NSC 92-2314-B-075-055), Taiwan, China
Assistant Editor Guo SY Edited by Gabbe M
References
- 1.Wobst A, Audisio RA, Colleoni M, Geraghty JG. Oesophageal cancer treatment: studies, strategies and facts. Ann Oncol. 1998;9:951–962. doi: 10.1023/A:1008273110272. [DOI] [PubMed] [Google Scholar]
- 2.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 3.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, et al. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 4.Du Plessis L, Dietzsch E, Van Gele M, Van Roy N, Van Helden P, Parker MI, Mugwanya DK, De Groot M, Marx MP, Kotze MJ, et al. Mapping of novel regions of DNA gain and loss by comparative genomic hybridization in esophageal carcinoma in the Black and Colored populations of South Africa. Cancer Res. 1999;59:1877–1883. [PubMed] [Google Scholar]
- 5.Pack SD, Karkera JD, Zhuang Z, Pak ED, Balan KV, Hwu P, Park WS, Pham T, Ault DO, Glaser M, et al. Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosomes Cancer. 1999;25:160–168. doi: 10.1002/(sici)1098-2264(199906)25:2<160::aid-gcc12>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 6.Tada K, Oka M, Tangoku A, Hayashi H, Oga A, Sasaki K. Gains of 8q23-qter and 20q and loss of 11q22-qter in esophageal squamous cell carcinoma associated with lymph node metastasis. Cancer. 2000;88:268–273. doi: 10.1002/(sici)1097-0142(20000115)88:2<268::aid-cncr4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 7.Ueno T, Tangoku A, Yoshino S, Abe T, Toshimitsu H, Furuya T, Kawauchi S, Oga A, Oka M, Sasaki K. Gain of 5p15 detected by comparative genomic hybridization as an independent marker of poor prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2002;8:526–533. [PubMed] [Google Scholar]
- 8.Yen CC, Chen YJ, Chen JT, Hsia JY, Chen PM, Liu JH, Fan FS, Chiou TJ, Wang WS, Lin CH. Comparative genomic hybridization of esophageal squamous cell carcinoma: correlations between chromosomal aberrations and disease progression/prognosis. Cancer. 2001;92:2769–2777. doi: 10.1002/1097-0142(20011201)92:11<2769::aid-cncr10118>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Lu YJ, Dong XY, Shipley J, Zhang RG, Cheng SJ. Chromosome 3 imbalances are the most frequent aberration found in non-small cell lung carcinoma. Lung Cancer. 1999;23:61–66. doi: 10.1016/s0169-5002(98)00093-2. [DOI] [PubMed] [Google Scholar]
- 10.Arnold N, Hagele L, Walz L, Schempp W, Pfisterer J, Bauknecht T, Kiechle M. Overrepresentation of 3q and 8q material and loss of 18q material are recurrent findings in advanced human ovarian cancer. Genes Chromosomes Cancer. 1996;16:46–54. doi: 10.1002/(SICI)1098-2264(199605)16:1<46::AID-GCC7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Heselmeyer K, Macville M, Schröck E, Blegen H, Hellström AC, Shah K, Auer G, Ried T. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–240. [PubMed] [Google Scholar]
- 12.Heselmeyer K, Schröck E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Guan X, Guo Y, Sham J, Deng M, Liang Q, Li H, Zhang H, Zhou H, Trent J. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30:254–260. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1086>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 14.Gain of DNA copy number on chromosomes 3q26-qter and 5p14-pter is a frequent finding in head and neck squamous cell carcinomas. Int J Mol Med. 1998;2:173–179. doi: 10.3892/ijmm.2.2.173. [DOI] [PubMed] [Google Scholar]
- 15.Bièche I, Olivi M, Champème MH, Vidaud D, Lidereau R, Vidaud M. Novel approach to quantitative polymerase chain reaction using real-time detection: application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. doi: 10.1002/(sici)1097-0215(19981123)78:5<661::aid-ijc22>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 16.De Preter K, Speleman F, Combaret V, Lunec J, Laureys G, Eussen BH, Francotte N, Board J, Pearson AD, De Paepe A, et al. Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod Pathol. 2002;15:159–166. doi: 10.1038/modpathol.3880508. [DOI] [PubMed] [Google Scholar]
- 17.Wang TL, Maierhofer C, Speicher MR, Lengauer C, Vogelstein B, Kinzler KW, Velculescu VE. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen CC, Chen YJ, Lu KH, Hsia JY, Chen JT, Hu CP, Chen PM, Liu JH, Chiou TJ, Wang WS, et al. Genotypic analysis of esophageal squamous cell carcinoma by molecular cytogenetics and real-time quantitative polymerase chain reaction. Int J Oncol. 2003;23:871–881. [PubMed] [Google Scholar]
- 19.Isola J, DeVries S, Chu L, Ghazvini S, Waldman F. Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol. 1994;145:1301–1308. [PMC free article] [PubMed] [Google Scholar]
- 20.Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan CC, Chen PC, Chiang H. KIT (CD117) is frequently overexpressed in thymic carcinomas but is absent in thymomas. J Pathol. 2004;202:375–381. doi: 10.1002/path.1514. [DOI] [PubMed] [Google Scholar]
- 23.Pan CC, Chen PC, Chou TY, Chiang H. Expression of calretinin and other mesothelioma-related markers in thymic carcinoma and thymoma. Hum Pathol. 2003;34:1155–1162. doi: 10.1053/j.humpath.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Koga F, Kawakami S, Fujii Y, Saito K, Ohtsuka Y, Iwai A, Ando N, Takizawa T, Kageyama Y, Kihara K. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003;9:5501–5507. [PubMed] [Google Scholar]
- 25.Redon R, Hussenet T, Bour G, Caulee K, Jost B, Muller D, Abecassis J, du Manoir S. Amplicon mapping and transcriptional analysis pinpoint cyclin L as a candidate oncogene in head and neck cancer. Cancer Res. 2002;62:6211–6217. [PubMed] [Google Scholar]
- 26.Soder AI, Hoare SF, Muir S, Going JJ, Parkinson EK, Keith WN. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- 27.Hiyama T, Yokozaki H, Kitadai Y, Haruma K, Yasui W, Kajiyama G, Tahara E. Overexpression of human telomerase RNA is an early event in oesophageal carcinogenesis. Virchows Arch. 1999;434:483–487. doi: 10.1007/s004280050372. [DOI] [PubMed] [Google Scholar]
- 28.Yokoi S, Yasui K, Iizasa T, Imoto I, Fujisawa T, Inazawa J. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin Cancer Res. 2003;9:4705–4713. [PubMed] [Google Scholar]
- 29.Imoto I, Pimkhaokham A, Fukuda Y, Yang ZQ, Shimada Y, Nomura N, Hirai H, Imamura M, Inazawa J. SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus. Biochem Biophys Res Commun. 2001;286:559–565. doi: 10.1006/bbrc.2001.5428. [DOI] [PubMed] [Google Scholar]
- 30.Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, du Manoir S. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001;61:4122–4129. [PubMed] [Google Scholar]
- 31.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, Liu JM, Yang DM, Yang WK, Shen CY. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 32.Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, Dreyer T, Stahl U. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198:335–342. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- 33.Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hibi K, Nakayama H, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. AIS overexpression in advanced esophageal cancer. Clin Cancer Res. 2001;7:469–472. [PubMed] [Google Scholar]
- 35.Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL, Wei F, Chen BS, Huang XP, Han YS, et al. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer. 2002;102:580–583. doi: 10.1002/ijc.10739. [DOI] [PubMed] [Google Scholar]
- 36.Glickman JN, Yang A, Shahsafaei A, McKeon F, Odze RD. Expression of p53-related protein p63 in the gastrointestinal tract and in esophageal metaplastic and neoplastic disorders. Hum Pathol. 2001;32:1157–1165. doi: 10.1053/hupa.2001.28951. [DOI] [PubMed] [Google Scholar]
- 37.Wang TY, Chen BF, Yang YC, Chen H, Wang Y, Cviko A, Quade BJ, Sun D, Yang A, McKeon FD, et al. Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: a study of 250 cases. Hum Pathol. 2001;32:479–486. doi: 10.1053/hupa.2001.24324. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka N, Sugihara K, Odajima T, Mimura M, Kimijima Y, Ichinose S. Oral squamous cell carcinoma: electron microscopic and immunohistochemical characteristics. Med Electron Microsc. 2002;35:127–138. doi: 10.1007/s007950200016. [DOI] [PubMed] [Google Scholar]
- 39.Hartmann E, Görlich D, Kostka S, Otto A, Kraft R, Knespel S, Bürger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375–381. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 40.Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- 41.Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, Chen X, Lee CC, Lorenzi MV, Ohara N, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]


