Abstract
AIM: To study the influence of redox environment of Escherichia coli (E. coli) cytoplasm on disulfide bond formation of recombinant proteins.
METHODS: Bovine fibroblast growth factor (BbFGF) was selected as a model of simple proteins with a single disulfide bond and free cysteines. Anti-HBsAg single-chain Fv (HBscFv), an artificial multidomain protein, was selected as the model molecule of complex protein with 2 disulfide bonds. A BbFGF-producing plasmid, pJN-BbFGF, and a HBscFv producing-plasmid, pQE-HBscFv, were constructed and transformed into E. coli strains BL21(DE3) and M15[pREP4] respectively. At the same time, both plasmids were transformed into a reductase-deficient host strain, E. coli Origami(DE3). The 4 recombinant E. coli strains were cultured and the target proteins were purified. Solubility and bioactivity of recombinant BbFGF and HBscFv produced in different host strains were analyzed and compared respectively.
RESULTS: All recombinant E. coli strains could efficiently produce target proteins. The level of BbFGF in BL21(DE3) was 15-23% of the total protein, and was 5-10% in Origami (DE3). In addition, 65% of the BbFGF produced in BL21(DE3) formed into inclusion body in the cytoplasm, and all the target proteins became soluble in Origami(DE3). The bioactivity of BbFGF purified from Origami(DE3) was higher than its counterpart from BL21(DE3). The ED50 of BbFGF from Origami(DE3) and BL21(DE3) was 1.6 μg/L and 2.2 μg/L, respectively. Both HBscFv formed into inclusion body in the cytoplasm of M15[pQE-HBscFv] or Origami[pQE-HBscFv]. But the supernatant of Origami[pQE-HBscFv] lysate displayed weak bioactivity and its counterpart from M15[pQE-HBscFv] did not display any bioactivity. The soluble HBscFv in Origami[pQE-HBscFv] was purified to be 1-2 mg/L and its affinity constant was determined to be 2.62×107 mol/L. The yield of native HBscFv refolded from inclusion body in M15[pQE-HBscFv] was 30-35 mg/L and the affinity constant was 1.98×107 mol/L. There was no significant difference between the bioactivity of HBscFvs refolded from the inclusion bodies produced in different host strains.
CONCLUSION: Modification of the redox environment of E. coli cytoplasm can significantly improve the folding of recombinant disulfide-bonded proteins produced in it.
Keywords: Escherichia coli, Recombinant protein expression, Disulfide bond, Solubility, bFGF, HBsAg, Single-chain Fv, Reductase deficient, Protein folding, Affinity constant
INTRODUCTION
Among the systems available for heterologous protein production, Escherichia coli remains the first choice because of its ability to grow rapidly and at high density, its well-characterized genetics and the availability of an increasingly large number of vectors and host strains[1-3]. With a considerable amount of efforts directed at yield during the past 20 years, heterologous proteins could be produced in E. coli with an amazing productivity. At present, one of the mostly focused fields of E. coli system is how to improve the solubility of heterologous proteins in cytoplasm of this bacterium[3]. This problem might be addressed in two approaches. First, heterologous proteins might be fused with chaperone or refoldase, which promotes the proper isomerization or accelerates rate-limiting steps along the folding pathway[4,5]. Plasmids pET32[6] and pET44[7] are the representatives of this kind of vectors. Second, redox environment of E. coli cytoplasm might be altered by genetic engineering, namely, construction of an E. coli mutant with diminished or eliminated reductase system[8]. The effect of this strategy is still not well addressed. To investigate the influence of redox environment of E. coli cytoplasm on the solubility of heterologous proteins, bovine basic fibroblast growth factor (BbFGF) with a single disulfide bond, and human anti-HBsAg single-chain Fv (HBscFv) with 2 disulfide bonds, selected as model molecules of simple and complex proteins, were produced in normal E. coli strains and in E. coli Origami(DE3), a reductase deficient strain. Comparing the solubility and bioactivity of the recombinant proteins produced in different hosts might help us better understand the folding of heterologous proteins, and be a reference for other proteins’ engineering.
BbFGF is a non-glycosylated single-strand polypeptide with a variety of bioactivities[9]. The polypeptide consists of 155 amino acid residues, including 4 cysteines, in which C34 and C101 are linked with disulfide bonds, while C78 and C96 exist freely. It was reported that the majority of recombinant BbFGF forms into inclusion body when it is overproduced in E. coli cytoplasm. Point mutation of C78 and C96 into serine might solve the problem to some extent[10,11], but the primary structure of BbFGF is changed and the bioactivity of the mutant declines, thus becoming an obstacle in drug development[12]. HBscFv[13] is a human recombinant antibody with 4 cysteines participating in disulfide bond formation. It is impossible to minimize the misfolding of recombinant products via point mutation. In addition, there is more uncertainty in the folding process of scFvs, because they are artificial multidomain (VH and VL) molecules[2,14]. It is more difficult to obtain soluble recombinant HBscFv than BbFGF in E. coli cytoplasm.
MATERIALS AND METHODS
Plasmids, bacteria and reagents
pJN-BbFGF, a plasmid constructed from a pET3c derivative, pJN982[15], allows the expression of BbFGF fused in frame to phage10-LacZ leader under control of the T7 promoter. pQE-HBscFv is a HBscFv-producing plasmid derived from pQE-40, in which HBscFv is fused in frame to a 6×His tag downstream of T5 promoter. E. coli BL21(DE3) [F- OmpT hsd SB(rB-,MB-) dcm gal λ(DE3)] and Origami(DE3) [Δara- leu7697 ΔlacX74 ΔphoA PvuII phoR araD139 galE galK rspL F’[lac– (lacIq) pro] gor522::Tn10 (TcR) trxB::kan (DE3)] were purchased from Novagen. E. coli M15[pREP4] with phenotype of Nals, Strs, Rifs, Thi-, lac-, Ara+, Gal+, Mtl-, F-, RecA-, Uvr+, Lon+ was purchased from Qiagen. IPTG was purchased from Promega. Chromatography medium Bio-Rex 70 and Heparin Hyper D were purchased from Bio-Rad and Kronlab respectively. His-Trap HP column and SP-sepharose CL-4B were purchased from Amersham Bioscience. MTT was from Sigma. Rabbit anti-HBscFv antibody was prepared in our laboratory.
Expression of BbFGF
Construction of recombinant E. coli was carried out as previously described[15]. E. coli BL[pJN-BbFGF] or Origami[pJN-BbFGF] was culture to 1.0 A600 U/mL at 37 °C, 220 r/min, in 2×YT medium and expression of BbFGF was induced with 1 mmol/L IPTG for 4 h. The pellet and supernatant of bacteria lysate were collected for analysis or purification.
Purification and bioactivity of BbFGF
Samples containing BbFGF were applied on Bio-Rex70 column. Blank runs were performed to elute any trace of weakly adsorbed ligand and, subsequently, PBS with 0.6 mol/L NaCl was used to elute the protein of interest as the starting material of Heparin Hyper D re-purification. Recombinant BbFGF was eluted by buffer with 1.2 mol/L NaCl and its purity was determined on gel-optical-density scan[16-20]. Bioactivity of recombinant BbFGF with purity over 96% was assayed by MTT method as previously described[18,19].
Expression of HBscFv
HBscFv-producing E. coli strains, M15[pQE-HBscFv] and Origami[pQE-HBscFv] were cultured in 2×YT medium and were induced with 1 mmol/L IPTG as the same condition as BbFGF.
Purification and bioactivity of HBscFv
HBscFv inclusion body from M15[pQE-HBscFv] or Origami[pQE-HBscFv] was lysed in buffer containing 6 mol/L GuHCl, then applied on His-Trap HP column. The fraction of interest was collected and refolded by gradual removal of the denaturing reagent using stepwise dialysis with the introduction of an oxidizing reagent (GSSG) and L-arginine. Soluble HBscFv from Origami[pQE-HBscFv] was preliminarily purified via SP-sepharose CL-4B chromatography, and then applied on His-Trap HP column. Indirect ELISA was performed to determine the HBsAg binding activity of HBscFv products from different host strains[20-22].
The affinity constant (KA) of HBscFv products was determined by non-competitive enzyme immunoassay as previously described[23]. In brief, a microtiter plate (Corning Costar) was coated with serially 2-fold diluted HBsAg from 3 to 0.375 mg/L in carbonate buffer at 4 °C overnight. Serially diluted HBscFv (10-7 mol/L) from 1:2 to 1:256 was added to the plate and an indirect ELISA was carried out. According to the sigmoid curve of A versus logarithm of HBscFv concentration (mol/L), the concentrations of HBscFv at A50 were identified and KA was calculated by the formula of KA = (n-1)/( nAb’-Ab).
RESULTS
Construction of recombinant E. coli
Plasmid pJN-BbFGF was transformed into E. coli BL21(DE3) and E. coli Origami(DE3). The recombinant strains were designated as E. coli BL[pJN-BbFGF] and Origami[pJN-BbFGF], respectively. As is shown in Figure 1A, the gene of interest was located at the downstream of T7 promoter and phage10-LacZ leader. T7 RNA polymerase, integrated into the genome of host and induced by IPTG, could start the transcription of target gene, resulting in recombinant BbFGF with the molecule weight of 20 ku. Recombinant E. coli M15[pQE-HBscFv] and Origami[pQE-HBscFv] were constructed by transforming plasmid pQE-HBscFv into E. coli M15[pREP4] and Origami(DE3), respectively. Sketch map of plasmid pQE-HBscFv (Figure 1B) showed that the target gene, controlled by T5 promoter, was located downstream of LacO sequence and sensitive to IPTG induction. Moreover, there was a 6×His tag at N-terminal of HBscFv, which made the recombinant protein could be purified by immobilized metal ion affinity chromatography (IMAC).
Figure 1.
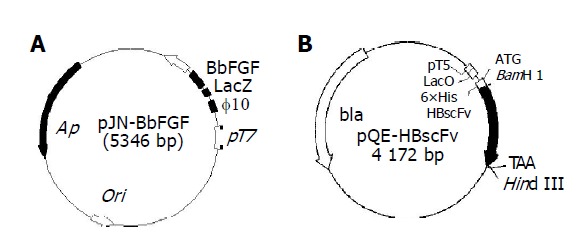
Structure of the expression plasmids pJN-BbFGF (A) and pQE-HBscFv (B).
Expression of BbFGF
SDS-PAGE analysis of induced BL[pJN-BbFGF] is shown in Figure 2A, in which lanes 2 and 3 are supernatant and pellet of the cell lysate, respectively. It could be made out that recombinant BbFGF mainly formed into inclusion body in the cytoplasm of E. coli BL21(DE3). The result of gel-optical-density scan associated with protein quantitative analysis revealed that 65% of the produced BbFGF aggregated into inclusion body. In contrast, soluble BbFGF was obtained when the protein was produced in E. coli Origami(DE3). Cell lysate of induced Origami[pJN-BbFGF] is shown in Figure 2B, in which lanes 2 and 3 are the supernatant and pellet of the cell lysate, respectively. The gel showed that the target protein was included in the supernatant, accounting for 11.6% of the total soluble protein.
Figure 2.
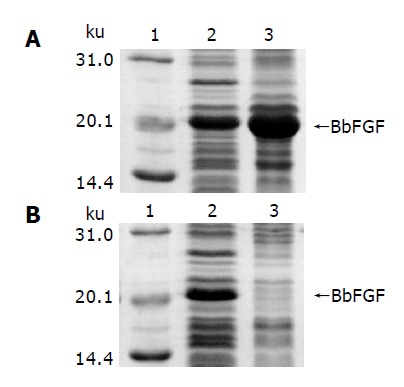
Characteristics of BbFGF in different host bacteria. A: E. coli BL21(DE3), B: E. coli Origami(DE3), lane 1: Molecular weight standard, lane 2: Supernatant of the bacteria lysate, lane 3: Pellet of the bacteria lysate.
Besides, SDS-PAGE analysis also revealed that the yield of BbFGF in Origami(DE3), from 5-10% of the total protein, was significantly lower than that in BL21(DE3), in which the recombinant BbFGF was 15-23% of the total protein.
Purification and bioactivity of recombinant BbFGFs
BbFGFs produced in two recombinant bacteria were purified by cation exchange and heparin affinity chromatography to homology with purity of up to 96%. MTT assay revealed that the specific bioactivity of BbFGF purified from Origami(DE3) was higher than its counterpart from BL21(DE3) (Figure 3). The ED50 of BbFGFs from Origami(DE3) was 1.6 μg/L, and 2.2 μg/L from BL21(DE3).
Figure 3.
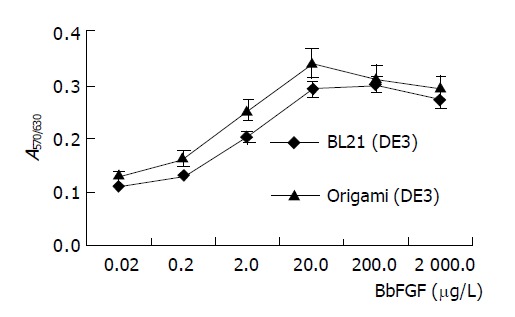
Bioactivity of BbFGFs produced from different host bacteria.
Expression of HBscFv
Induced with IPTG, E. coli M15[pQE-HBscFv] produced a great deal of recombinant HBscFv, which existed in the form of inclusion body in the cytoplasm (Figure 4A). SDS-PAGE analysis of M15[pQE-HBscFv] showed that the 28 ku HBscFv was located in the insoluble fraction of cell lysate. At the same time, recombinant E. coli Origami[pQE-HBscFv] was cultured and induced. The result of SDS-PAGE analysis indicated that HBscFv still formed into inclusion body in the cytoplasm of Origami(DE3). Lane 3 of Figure 4B was the insoluble fraction of Origami[pQE-HBscFv], in which there was an obvious HBscFv-like protein band. There was also an enlarged protein band, amounting 4.7% of the total protein at the corresponding position of HBscFv in the soluble fraction (Lane 2, Figure 4B). To determine the characteristics of this protein band, bioactivity assay was carried out.
Figure 4.
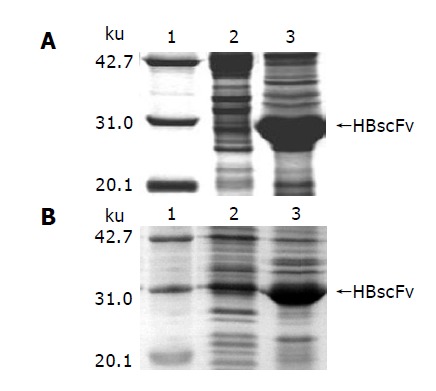
Characteristics of HBscFv in different host bacteria. A: E. coli M15[pREP4], B: E. coli Origami(DE3), lane 1: Molecular weight standard, lane 2: Supernatant of the bacteria lysate, lane 3: Pellet of the bacteria lysate.
Purification and bioactivity of recombinant HBscFvs
The supernatant from the two recombinant E. coli strains was adjusted to identical protein concentration and the activity of HBsAg-binding was determined. There was no detectable bioactivity in the supernatant of M15[pQE-HBscFv] (A450/630 = 0.05, similar to negative control), and weak bioactivity (A450/630 = 0.21) was detectable in that of Origami[pQE-HBscFv]. The hollow columns in Figure 5 show the significant difference of bioactivity between the soluble fractions of the two HBscFv-producing E. coli strains. Two-step purification revealed that the yield of soluble HBscFv from Origami[pQE-HBscFv] was 1-2 mg/L.
Figure 5.
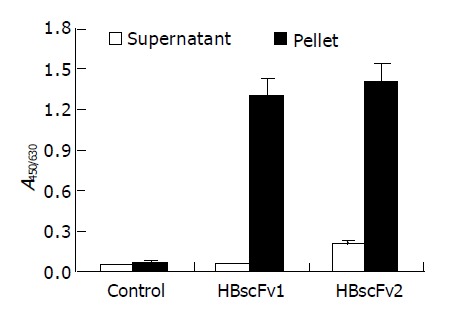
Antigen-binding activity of recombinant HBscFvs produced from different E. coli (mean±SD). HBscFv1: HBscFv produced from M15[pQE-HBscFv], HBscFv2: HBscFv produced from Origami[pQE-HBscFv].
HBscFvs from pellets of the two HBscFv-producing E. coli strains were purified and refolded by dialysis. The yield of HBscFv refolded from M15[pQE-HBscFv] was 30-35 mg/L, and 18-22 mg/L from Origami[pQE-HBscFv]. Refolded HBscFvs from different recombinant bacteria were adjusted to the same concentration. Bioactivity assay demonstrated that both HBscFvs exhibited HBsAg-binding activities, and there was no significant difference between the specific bioactivities (blacked column in Figure 5).
Figure 6 is the ELISA curves of HBscFv purified from the supernatant of Origami[pQE-HBscFv]. From the curve, KA of the soluble HBscFv was determined to be 2.62×107 mol/L. Parallel analysis showed that KA of refolded HBscFv from the inclusion body of M15[pQE-HBscFv] was 1.98×107 mol/L.
Figure 6.
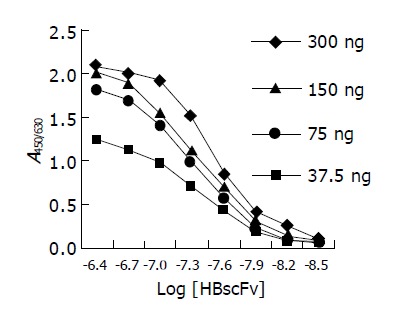
ELISA curves of HBscFv purified from the supernatant of Origami[pQE-HBscFv].
DISCUSSION
Aggregation of recombinant disulfide-bonded proteins in E. coli is a direct consequence of the lack of disulfide bond formation, proceeding from an intermediate present during the unfolding/refolding transition[24]. Under physiological conditions, E. coli cytoplasm is maintained in a reduced state that strongly disfavors the formation of stable disulfide bonds in proteins. Disulfide bonds that form instantaneously in cytoplasm of normal E. coli are reduced by at least 5 proteins, namely, thioredoxins 1 and 2, glutaredoxins 1, 2 and 3[25]. To keep these proteins in reduced situation, thioredoxin reductase and glutathione reductase are necessary. Thus, heterologous proteins can fold correctly when the reductase is fully or partly impaired[26].
As a host strain with inactivated thioredoxin reductase and glutathione reductase, E. coli Origami(DE3) is permissive to the formation of disulfide bonds in the more oxidative cytoplasm. It was reported that alkaline phosphatase can partly fold into enzymatically active structure in this mutated host strain[26]. Results listed above show BbFGF, a simple protein with a single disulfide bond and free cysteines, mainly forming into inclusion body in E. coli BL21(DE3), is synthesized as a soluble product in Origami(DE3). In addition, HBscFv, a complex protein with 2 disulfide bonds, exists mainly as an inclusion body and partially as a soluble protein in this oxidized host strain, suggesting that modification of the redox environment of E. coli cytoplasm may significantly improve the solubility of simple proteins which usually form into inclusion bodies during overproduction. However, it is difficult to obtain a great number of soluble recombinant products for complex proteins with multiple disulfide bonds.
Besides the enhanced solubility, another advantage of E. coli Origami(DE3) that should not be ignored is that recombinant proteins have higher specific bioactivity. Determination of ED50 of BbFGF and KA of HBscFv has proved it. The method of HBscFv refolding applied in this paper resulted in the highly efficient refolding of various denatured scFvs from inclusion bodies and its mechanism is disclosed recently[20,22]. However, the KA of refolded HBscFv still differed significantly from that of soluble HBscFv. Isoelectric point measurement by IEF and disulfide bond determination by MALDI-TOF MS suggested that there are misfolded disulfide bonds in the refolded HBscFv products[27]. Therefore, E. coli Origami(DE3) may be a better choice for the bioactivity-guided application of recombinant proteins.
Protein folding is a complex process that results in various assistant enzymes or proteins[28]. A great number of folding enhancers, methods and kinetic models[29] are described and applied. What dismays us is that optimizing one factor may improve the folding of certain proteins, but it is impossible to design a universal expression system that adapts to all kinds of heterologous proteins[2,3,30,31]. Therefore, to overproduce a complex protein, we usually have to make “trial-and-error” experiments to select an expression system; and to optimize culture conditions and modify host strains to maximize the yield of native products.
In this study, soluble BbFGF and partially soluble HBscFv were obtained in E. coli Origami(DE3), but the total yield of recombinant products decreased. The expression level of BbFGF in Origami(DE3) was 33-43% of that in BL21(DE3). The yield of soluble HBscFv in Origami(DE3) was 4.5-11.1% of the inclusion bodies, and the total yield of the target proteins in Origami(DE3) was 63-68% of that in M15[pREP4]. Whether the soluble expression is resulted from the lower yield, or from the alteration of the redox environment of cytoplasm is still unknown. We had tried to change the induction condition of BL[pJN-BbFGF] to decrease BbFGF yield, but failed to eliminate inclusion body formation. As the relationship between yield decrease and reductase deficiency is concerned, there might be certain correlation between the two factors. E. coli requires either a functional thioredoxin or a glutaredoxin system to reduce disulfide bonds which appear after each catalytic cycle in the essential enzyme ribonucleotide reductase and perhaps to reduce non-native disulfide bonds in cytoplasmic proteins[25]. Chaperones are able to improve the folding yield of multidomain proteins but markedly delay the folding process both in vivo and in vitro. Based on these finding, we might conclude that correct folding of heterologous proteins in reductase deficient host E. coli Origami(DE3) is at the expense of total yield.
HBscFv is a recombinant human antibody fragment that is more suitable to be produced in mammalian cells or other eukaryotic cells[2,13,32-34], but the technology of large-scale culture of mammalian cells is still in developing. Large scale production of recombinant antibodies efficiently and costly is the last obstacle in antibody engineering. If the partially soluble HBscFv described here could be further optimized in the culture condition[35] and/or the level of molecular chaperones or refoldase (DsbC or other proved solubility enhancer)[36,37], the proportion of soluble products might be greatly increased. That should pose further significance of E. coli on the production of recombinant antibody or other complex pharmaceutical proteins.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30371661 and No. 30400071; and the Natural Science Foundation for Research Team of Guangdong Province, China, No. 2004E039213
Co-correspondence: Yi-Fei Wang
References
- 1.Hua Q, Yang C, Oshima T, Mori H, Shimizu K. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl Environ Microbiol. 2004;70:2354–2366. doi: 10.1128/AEM.70.4.2354-2366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma R, Boleti E, George AJ. Antibody engineering: comparison of bacterial, yeast, insect and mammalian expression systems. J Immunol Methods. 1998;216:165–181. doi: 10.1016/s0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 3.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 4.Choi GH, Lee DH, Min WK, Cho YJ, Kweon DH, Son DH, Park K, Seo JH. Cloning, expression, and characterization of single-chain variable fragment antibody against mycotoxin deoxynivalenol in recombinant Escherichia coli. Protein Expr Purif. 2004;35:84–92. doi: 10.1016/j.pep.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Machida S, Yu Y, Singh SP, Kim JD, Hayashi K, Kawata Y. Overproduction of beta-glucosidase in active form by an Escherichia coli system coexpressing the chaperonin GroEL/ES. FEMS Microbiol Lett. 1998;159:41–46. doi: 10.1111/j.1574-6968.1998.tb12839.x. [DOI] [PubMed] [Google Scholar]
- 6.Mao YF, Yan J. Construction of prokaryotic expression system of ureB gene from a clinical Helicobacter pylori strain and identification of the recombinant protein immunity. World J Gastroenterol. 2004;10:977–984. doi: 10.3748/wjg.v10.i7.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Available from: http: //www.merckbiosciences.com/docs/NDIS/TB330-000.pdf.
- 8.Derman AI, Prinz WA, Belin D, Beckwith J. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science. 1993;262:1744–1747. doi: 10.1126/science.8259521. [DOI] [PubMed] [Google Scholar]
- 9.Shin EY, Woo KN, Lee CS, Koo SH, Kim YG, Kim WJ, Bae CD, Chang SI, Kim EG. Basic fibroblast growth factor stimulates activation of Rac1 through a p85 betaPIX phosphorylation-dependent pathway. J Biol Chem. 2004;279:1994–2004. doi: 10.1074/jbc.M307330200. [DOI] [PubMed] [Google Scholar]
- 10.Fox GM, Schiffer SG, Rohde MF, Tsai LB, Banks AR, Arakawa T. Production, biological activity, and structure of recombinant basic fibroblast growth factor and an analog with cysteine replaced by serine. J Biol Chem. 1988;263:18452–18458. [PubMed] [Google Scholar]
- 11.Rinas U, Tsai LB, Lyons D, Fox GM, Stearns G, Fieschko J, Fenton D, Bailey JE. Cysteine to serine substitutions in basic fibroblast growth factor: effect on inclusion body formation and proteolytic susceptibility during in vitro refolding. Biotechnology (N Y) 1992;10:435–440. doi: 10.1038/nbt0492-435. [DOI] [PubMed] [Google Scholar]
- 12.McCafferty J, Glover DR. Engineering therapeutic proteins. Curr Opin Struct Biol. 2000;10:417–420. doi: 10.1016/s0959-440x(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 13.Xiong S, Ren XR, Tang YH, Su KY, Yu ZY, Luo Y, Wang YF, Li JX. Expression of a human single-chain Fv antibody against HBsAg in Pichia pastoris. ShengWu GongCheng XueBao. 2003;19:19–23. [PubMed] [Google Scholar]
- 14.Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 15.Xiong S, Lin J, Hong A, Liu JS, Zhang L, Li ZY, Yao RH. Construction and application of a pET3c derivative. Yaowu Shengwu Jishu. 2001;8:4–7. [Google Scholar]
- 16.Sheng Z, Chang SB, Chirico WJ. Expression and purification of a biologically active basic fibroblast growth factor fusion protein. Protein Expr Purif. 2003;27:267–271. doi: 10.1016/s1046-5928(02)00601-0. [DOI] [PubMed] [Google Scholar]
- 17.Garke G, Deckwer WD, Anspach FB. Preparative two-step purification of recombinant human basic fibroblast growth factor from high-cell-density cultivation of Escherichia coli. J Chromatogr B Biomed Sci Appl. 2000;737:25–38. doi: 10.1016/s0378-4347(99)00394-1. [DOI] [PubMed] [Google Scholar]
- 18.Knoerzer W, Binder HP, Schneider K, Gruss P, McCarthy JE, Risau W. Expression of synthetic genes encoding bovine and human basic fibroblast growth factors (bFGFs) in Escherichia coli. Gene. 1989;75:21–30. doi: 10.1016/0378-1119(89)90379-x. [DOI] [PubMed] [Google Scholar]
- 19.Kajio T, Kawahara K, Kato K. Quantitative colorimetric assay for basic fibroblast growth factor using bovine endothelial cells and heparin. J Pharmacol Toxicol Methods. 1992;28:9–14. doi: 10.1016/1056-8719(92)90059-a. [DOI] [PubMed] [Google Scholar]
- 20.Tsumoto K, Shinoki K, Kondo H, Uchikawa M, Juji T, Kumagai I. Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent--application to a human single-chain Fv fragment. J Immunol Methods. 1998;219:119–129. doi: 10.1016/s0022-1759(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 21.Sánchez L, Ayala M, Freyre F, Pedroso I, Bell H, Falcón V, Gavilondo JV. High cytoplasmic expression in E. coli, purification, and in vitro refolding of a single chain Fv antibody fragment against the hepatitis B surface antigen. J Biotechnol. 1999;72:13–20. doi: 10.1016/s0168-1656(99)00036-x. [DOI] [PubMed] [Google Scholar]
- 22.Umetsu M, Tsumoto K, Hara M, Ashish K, Goda S, Adschiri T, Kumagai I. How additives influence the refolding of immunoglobulin-folded proteins in a stepwise dialysis system. Spectroscopic evidence for highly efficient refolding of a single-chain Fv fragment. J Biol Chem. 2003;278:8979–8987. doi: 10.1074/jbc.M212247200. [DOI] [PubMed] [Google Scholar]
- 23.Beatty JD, Beatty BG, Vlahos WG. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 24.Martineau P, Betton JM. In vitro folding and thermodynamic stability of an antibody fragment selected in vivo for high expression levels in Escherichia coli cytoplasm. J Mol Biol. 1999;292:921–929. doi: 10.1006/jmbi.1999.3105. [DOI] [PubMed] [Google Scholar]
- 25.Prinz WA, Aslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 26.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong S, Wang YF, Qian CW, Luo Y, Chen WY, Rao GR, Su KY, Yu ZY. Isoelectric point determination and structure modeling of a single-chain Fv against HBsAg. Huanan Ligong Daxue Xuebao. 2004;31:73–76. [Google Scholar]
- 28.Masip L, Pan JL, Haldar S, Penner-Hahn JE, DeLisa MP, Georgiou G, Bardwell JC, Collet JF. An engineered pathway for the formation of protein disulfide bonds. Science. 2004;303:1185–1189. doi: 10.1126/science.1092612. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann F, Posten C, Rinas U. Kinetic model of in vivo folding and inclusion body formation in recombinant Escherichia coli. Biotechnol Bioeng. 2001;72:315–322. doi: 10.1002/1097-0290(20010205)72:3<315::aid-bit8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 30.Lamark T, Ingebrigtsen M, Bjørnstad C, Melkko T, Mollnes TE, Nielsen EW. Expression of active human C1 inhibitor serpin domain in Escherichia coli. Protein Expr Purif. 2001;22:349–358. doi: 10.1006/prep.2001.1445. [DOI] [PubMed] [Google Scholar]
- 31.Schwalbach G, Sibler AP, Choulier L, Deryckère F, Weiss E. Production of fluorescent single-chain antibody fragments in Escherichia coli. Protein Expr Purif. 2000;18:121–132. doi: 10.1006/prep.1999.1185. [DOI] [PubMed] [Google Scholar]
- 32.Park SS, Ryu CJ, Kang YJ, Kashmiri SV, Hong HJ. Generation and characterization of a novel tetravalent bispecific antibody that binds to hepatitis B virus surface antigens. Mol Immunol. 2000;37:1123–1130. doi: 10.1016/s0161-5890(01)00027-x. [DOI] [PubMed] [Google Scholar]
- 33.Ramírez N, Ayala M, Lorenzo D, Palenzuela D, Herrera L, Doreste V, Pérez M, Gavilond JV, Oramas P. Expression of a single-chain Fv antibody fragment specific for the hepatitis B surface antigen in transgenic tobacco plants. Transgenic Res. 2002;11:61–64. doi: 10.1023/a:1013967705337. [DOI] [PubMed] [Google Scholar]
- 34.Mayfield SP, Franklin SE, Lerner RA. Expression and assembly of a fully active antibody in algae. Proc Natl Acad Sci USA. 2003;100:438–442. doi: 10.1073/pnas.0237108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ito M, Nagata K, Kato Y, Oda Y, Yamagoe S, Suzuki K, Tanokura M. Expression, oxidative refolding, and characterization of six-histidine-tagged recombinant human LECT2, a 16-kDa chemotactic protein with three disulfide bonds. Protein Expr Purif. 2003;27:272–278. doi: 10.1016/s1046-5928(02)00634-4. [DOI] [PubMed] [Google Scholar]
- 36.Kurokawa Y, Yanagi H, Yura T. Overproduction of bacterial protein disulfide isomerase (DsbC) and its modulator (DsbD) markedly enhances periplasmic production of human nerve growth factor in Escherichia coli. J Biol Chem. 2001;276:14393–14399. doi: 10.1074/jbc.M100132200. [DOI] [PubMed] [Google Scholar]
- 37.Jurado P, Ritz D, Beckwith J, de Lorenzo V, Fernández LA. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J Mol Biol. 2002;320:1–10. doi: 10.1016/S0022-2836(02)00405-9. [DOI] [PubMed] [Google Scholar]


