Abstract
AIM: To develop a real-time PCR for detecting hepatitis B virus (HBV) DNA based on TaqMan technology using a new MGB probe.
METHODS: Plasmid containing the sequence of X gene (1414-1744 nt) was constructed as HBV-DNA standard for quantitative analysis. A TaqMan-MGB probe between primers for amplification was designed to detect PCR products. The interested sequence contained in the plasmid and in clinical specimens was quantitatively measured.
RESULTS: The detection limit of the assay for HBV DNA was 1 genome equivalent per reaction. A linear standard curve was obtained between 100 and 109 DNA copies/reaction (r>0.990). None of the negative control samples showed false-positive reactions in duplicate. HBV DNA was detected in 100% (50/50) of HBV patients with HbeAg, and in 72.0% (36/50) with HBsAg, HBeAb and HBcAb. The coefficient of variation for both intra- and inter-experimental variability demonstrated high reproducibility and accuracy.
CONCLUSION: Real-time PCR based on TaqMan-MGB probe technology is an excellent method for detection of HBV DNA.
Keywords: Hepatitis B Virus, DNA, TaqMan-MGB probe, Real-time PCR
INTRODUCTION
Hepatitis B virus (HBV) infection is closely related to the development of acute or chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma[1]. Approximately 300 million individuals are chronically infected with hepatitis B virus in the world. In China, enzyme-linked immunosorbent assay (ELISA) is still a main detection method for HBV infection, but ELISA result can neither efficiently reflect serum viral load or hepatitis activity nor monitor the efficacy of antiviral treatments. Currently, polymerase chain reaction (PCR) assay has been widely used for monitoring HBV load. However, this method is clinically limited by relatively poor reproducibility and accuracy.
It has been reported that real-time PCR is an accurate, sensitive and highly reproducible method for detection of pathogenic microorganisms. Real-time PCR based on TaqMan technology is the prospective method[2-4]. TaqMan-MGB probe[5] is a new class of TaqMan probe, which is conjugated with minor groove binder (MGB) at 3’ end (Figure 1). Compared with TaqMan probe, TaqMan-MGB probe has several advantages, for example, high specificity and sensitivity, remarkable accuracy and reproducibility. In this report, we show our results of a highly sensitive assay for HBV DNA detection by real-time PCR using TaqMan-MGB probe technology.
Figure 1.
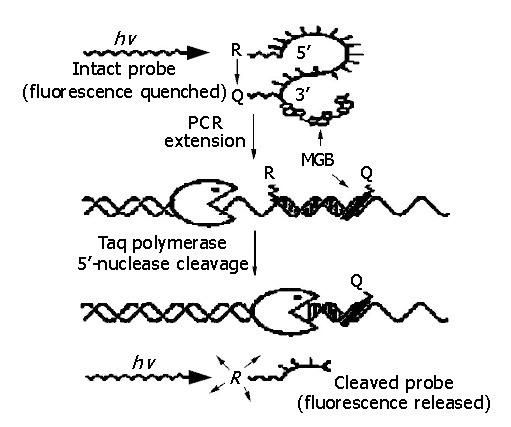
Scheme of real-time PCR using TaqMan-MGB probe.
MATERIALS AND METHODS
Samples
All sera used for this study were obtained from Xijing Hospital of Fourth Military Medical University. Fifty HBV-negative control samples were supplied by National Institute for the Control of Pharmaceuticals and Biological Products. All sera were stored at -20 °C until use.
Serological ELISA assay
Serum HBsAg, HBsAb, HBeAg, HBeAb and HBcAb were measured by commercially available enzyme immunoassays (Sino-American Biotechnology Co.) according to the manufacturer’s instructions.
DNA extraction
Guanidinium thiocyanate (GuSCN)-silica extraction technology is based on the lysing and nuclease-inactivating properties of guanidinium thiocyanate together with the nucleic acid–binding properties of silica particles in the presence of this agent[6]. This nucleic acid extraction method was simplified in practice in our laboratory. Briefly, reaction vessel was prepared by adding 500 μL of lysis buffer [5.25 mol/L GuSCN, 50 mmol/L Tris-HCl pH 6.4, 20 mmol/L EDTA pH 8.0, Triton X-100 13 mL/L] and 40 μL of silica particles (pH 2.0) to an 1.5-mL Eppendorf tube, then 200 μL of serum was added and the vessel was vortexed immediately (for approximately 5 s). After 10 min at room temperature, the vessel was centrifuged for 15 s at 12000 g, and the supernatant was removed by an aspirator. The silica particles were subsequently washed once with ethanol. After disposal of the ethanol, the pellet was dried at 56 °C with an open lid for 10 min. Finally 50 μL TE buffer was added, and the vessel was closed, vortexed, and incubated for 10 min at 56 °C.
Preparation of standard HBV DNA
HBV DNA X region (1414-1744 nt) was amplified (forward primer, 5’-ACGTCCTTTGTYTACGTCCCGT-3’, nucleotides 1414 to 1435; reverse primer, 5’-CCCAACTCCTCCCAGTCYTTAA-3’, nucleotides 1744 to 1723) and cloned into pMD18-T vector. After quantification, recombinant HBV plasmid was serially diluted from 1011 copies per mL to 102 copies per mL. Ten microliters of each diluted recombinant HBV plasmid was used as HBV standard PCR template.
TaqMan-MGB probe
TaqMan-MGB probe was designed as follows: a highly conserved sequence was selected; the first base of 5’ end must not be G; the annealing temperature of TaqMan-MGB probe was set at 10 °C higher than that of PCR primers. The sequence of TaqMan-MGB probe is 5’ FAM-TCAACGACCGACCTTGA-dabcyl-MGB 3’ (Figure 2).
Figure 2.

Location and direction of primers and probe within template.
Optimizing real-time PCR reaction parameters
In regard to the ratio of primer and probe, different data were reported. On the basis of these different reports, the best reaction parameters were determined with high correlation coefficient of standard curve and high sensitivity as criteria.
Real-time PCR reaction was run on DNA Engine Opticon (MJ Research). The fluorescence of 3 to 15 cycles was set up as background. The data were collected at the annealing step (55 °C) of each cycle, and the threshold cycle (Ct) for each sample was calculated by determining the point at which the fluorescence exceeded the threshold limit. The standard curve was calculated automatically by plotting the Ct values against each standard of known concentration and calculation of the linear regression line of this curve.
RESULTS
Recombinant plasmid sequence similarity analysis
Online sequence similarity analysis (http://www.ncbi.nlm.nih.gov/BLAST/) showed that the sequence cloned was 99% (328/331 = 99%) identical to that in HBV DNA X region (1414-1744 nt).
Real-time PCR reaction parameters
Amplification reaction mixture (50 μL) contained 10 μL of DNA, 5 μL of 10 × PCR buffer, 200 μmol/L dATP, 200 μmol/L dCTP, 200 μmol/L dGTP, 500 μmol/L dUTP, 1.5 mmol/L MgCl2, 400 nmol/L forward primer, 400 nmol/L reverse primer, 400 nmol/L TaqMan-MGB probe, 2 U of hot start DNA polymerase (Takara), and 0.5 U of uracil DNA glycosylase (UNG). Thermal cycling conditions were as follows: initial activation of UNG at 50 °C for 2 min followed by inactivation of UNG at 95 °C for 10 min. Subsequently, 60 cycles of amplification were performed at 95 °C for 20 s, at 55 °C for 30 s and at 72 °C for 30 s.
Linear standard curve and detection limit
The detection limit of this assay was 100 DNA copy per reaction. A linear standard curve was obtained between 100 and 109 DNA copies per reaction (r>0.990) (Figures 3, 4).
Figure 3.
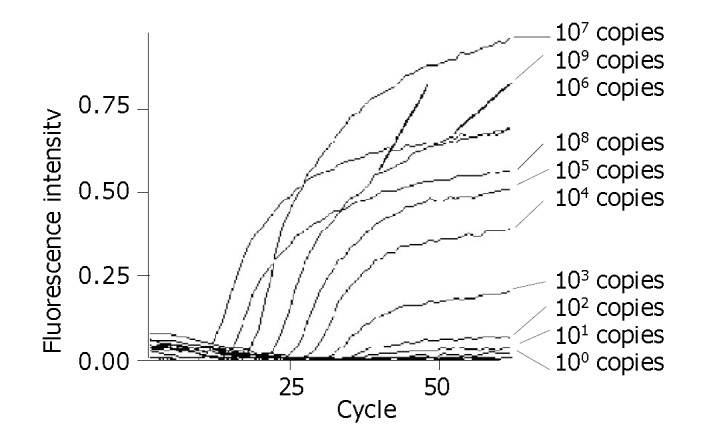
Amplification curves of standard templates of different concentrations.
Figure 4.
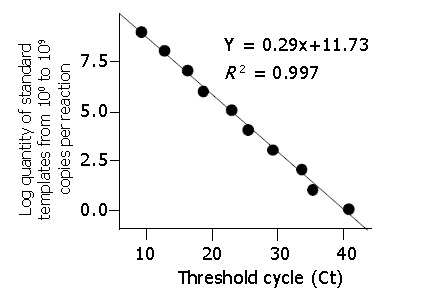
Standard curve for HBV DNA quantification.
Specificity
Fifty HBV-negative control samples and 174 clinical serum samples were analyzed. None of the negative control samples showed false-positive reactions in duplicate. HBV DNA was detected in 100% (50/50) of clinical samples with HBeAg and in 72.0% (36/50) with HBsAg, HBeAb and HBcAb. HBV DNA was detected in 6.9% (2/29) sera with HBsAb, and in 11.1% (5/45) with no HBV serological marker.
Accuracy study
Three different loads of sera were assayed 10 times on one plate simultaneously. Inter-coefficients of variation (%CV) were 6.1%, 4.2% and 3.4% respectively. Twelve sera of different loads were measured 3 times on three different plates. Mean coefficient of variation (%CV) was 7.1%.
DISCUSSION
HBV DNA monitoring in serum has become an important tool to identify individuals with high viral replication, to monitor patients on therapy, and to predict whether antiviral therapy is successful. For example, with the introduction of new antiviral agents like lamivudine, close monitoring of patients has become increasingly important due to the occurrence of antiviral drug-resistant virus strains or the presence of flares after withdrawal of antiviral therapy[7].
DNA polymerase assay has been widely used for monitoring antiviral treatments. In this assay, levels of DNA polymerase are quantified in vitro by incorporation of radiolabeled deoxynucleotide monophosphates, which is difficult to standardize among laboratories. Therefore, the assay has been replaced by a simpler and more accurate HBV DNA quantification system that uses a branched-DNA probe. However, this method is not sensitive enough to monitor serum virus levels in patients undergoing antiviral treatment.
The main merits of real-time PCR are as follows. First, single-tube operation prevents contamination effectively. Second, all steps are accomplished automatically by computer except for sample preparation. Third, false positive rate is low. Fourth, PCR reaction is monitored timely. And the last quantitative test result is absolutely true.
Besides the merits mentioned above, TaqMan-MGB probe has several other advantages. On the one hand, conjugated MGB can improve the melting temperature of probe, thus increasing probe specificity. In addition, it permits shorter probes to be used (usually 13 to 18 nt), which can facilitate probe design especially in AT rich region. On the other hand, shorter probes make fluorescence and quencher closer in space, thus fluorescence quenching more efficiently and increasing sensitivity and accuracy.
Because of these advantages mentioned above, theoretically, real-time PCR based on TaqMan-MGB probe technology should have more remarkable detection performance than that based on common probes, and we have proved it in practice. The detection limit of the assay based on TaqMan-MGB probe could reach 100 DNA copies per reaction, while that based on TaqMan probe could not. Additionally, the assay based on TaqMan-MGB probe has a wider linear detection range. A linear standard curve was obtained between 100 and 109 DNA copies per reaction (r>0.990). While the linear detection range of that based on TaqMan probe was usually from 101 to 108 DNA copies per reaction. Fernandez et al[8] drew a similar conclusion while quantifying HHV-7 genome by real-time PCR using MGB probe technology.
HBV DNA extraction from serum is a key step for real-time PCR. Silica- guanidinium thiocyanate (GuSCN) protocol is a well-known classical method, but its extraction procedure is too complicated and time-consuming, and not suitable for rapid clinical detection; thus, we modified it in practice. Simplified method needs less lysis buffer, which is relatively expensive, and makes the whole extraction procedure accomplished within 40 min.
HBV DNA was detected in 100% (50/50) of clinical samples with HBeAg. This result confirms that HBeAg is a real HBV replication marker. In accordance with previous reports, HBV DNA was detected in a few of sera with HBsAb and sera with no HBV serological marker. This shows that determining HBV replication only by detection of HBV marker is not enough, and real-time PCR should be used simultaneously.
In summary, a highly sensitive, less laborious and quantitative real-time PCR assay is developed based on TaqMan-MGB probe for the detection of HBV DNA.
Footnotes
Edited by Chen WW and Wang XL Proofread by Zhu LH
References
- 1.Bai YJ, Zhao JR, Lv GT, Zhang WH, Wang Y, Yan XJ. Rapid and high throughput detection of HBV YMDD mutants with fluorescence polarization. World J Gastroenterol. 2003;9:2344–2347. doi: 10.3748/wjg.v9.i10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pas SD, Fries E, De Man RA, Osterhaus AD, Niesters HG. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J Clin Microbiol. 2000;38:2897–2901. doi: 10.1128/jcm.38.8.2897-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidmann M, Meyer-König U, Hufert FT. Rapid detection of herpes simplex virus and varicella-zoster virus infections by real-time PCR. J Clin Microbiol. 2003;41:1565–1568. doi: 10.1128/JCM.41.4.1565-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3'-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barcena Marugan R, Cid Gomez L, Lopez Serrano P. Use of adefovir in the treatment of the chronic hepatitis B virus infection with resistance to lamivudine. Transplant Proc. 2003;35:1841–1843. doi: 10.1016/s0041-1345(03)00686-9. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez C, Boutolleau D, Manichanh C, Mangeney N, Agut H, Gautheret-Dejean A. Quantitation of HHV-7 genome by real-time polymerase chain reaction assay using MGB probe technology. J Virol Methods. 2002;106:11–16. doi: 10.1016/s0166-0934(02)00131-3. [DOI] [PubMed] [Google Scholar]


