Summary
Aging is accompanied by physiological impairments, which, in insulin-responsive tissues, including the liver, predispose individuals to metabolic disease. However, the molecular mechanisms underlying these changes remain largely unknown. Here, we analyze genome-wide profiles of RNA and chromatin organization in the liver of young (3 months) and old (21 months) mice. Transcriptional changes suggest that de-repression of the nuclear receptors PPARα, PPARγ, and LXRα in aged mouse liver leads to activation of targets regulating lipid synthesis and storage, whereas age-dependent changes in nucleosome occupancy are associated with binding sites for both known regulators (forkhead factors and nuclear receptors) and for novel candidates associated with nuclear lamina (Hdac3 and Srf) implicated to govern metabolic function of aging liver. Winged-helix transcription factor Foxa2 and nuclear receptor co-repressor Hdac3 exhibit reciprocal binding pattern at PPARα targets contributing to gene expression changes that lead to steatosis in aged liver.
Introduction
Aging is associated with increased prevalence of metabolic disease and cancer, reduced capacity for tissue regeneration, and physical decline (Rodriguez et al., 2007; Willis-Martinez et al., 2010). In particular, triglyceride accumulation is the common metabolic phenotype of aging liver and of metabolic syndrome, an age-related disorder that increases the risk of developing diabetes and cardiovascular disease. While mechanisms of age-dependent defects in tissue regeneration have been studied extensively (Jin et al., 2010; Jin et al., 2009), causes of age-onset metabolic impairments remain largely unknown.
Previous studies suggest a role for chromatin and nuclear organization in aging-associated lipid dystrophies. First, changes in chromatin organization mediate age-dependent impairments in several tissues (Chambers et al., 2007; Jin et al., 2010). Furthermore, mutations in lamin A/C (LMNA), a nuclear envelope protein, cause progeria, a premature aging syndrome, and lamin-dependent defects have been connected to physiological human aging (Scaffidi and Misteli, 2006). LMNA is also mutated in partial lipodystrophy (Shackleton et al., 2000), a condition associated with insulin-resistant diabetes and hepatic steatosis. Clinical features of lipodystrophy due to mutations in LMNA closely resemble those in individuals with mutations in PPARG, a nuclear receptor involved in pathogenesis of fatty liver (Gavrilova et al., 2003). Progeria attributed to defects in DNA repair (Schumacher et al., 2009) is modeled in mice by deletion of Ercc1, an enzyme crucial to nucleotide excision repair (Niedernhofer et al., 2006). Ercc1 mutants exhibit upregulation of targets of the hepatic nuclear receptors (PPARα and PPARγ) and hepatic steatosis. Finally, liver-specific loss of Foxa2, a pioneer transcription factor regulating nucleosome dynamics, leads to premature aging, increased hepatic lipogenesis, and age-onset obesity (Bochkis et al., 2013). While the progeroid models described above propose a role for epigenetic conformation in modulating age-dependent metabolic impairments, this hypothesis has not yet been tested in a model of physiological aging.
To study the relation between chromatin organization and age-dependent metabolic impairments in the liver we examined gene expression and genome-wide nucleosome occupancy in livers isolated from young (3 months) and old (21 months) mice (Figure 1A). Age-dependent induction of expression in lipid synthesis and storage genes, similar to metabolic changes seen in progeroid syndromes, is consistent with de-repression of the nuclear receptors PPARα, PPARγ, and LXRα. Analysis of transcription factor binding sites that are overrepresented in regions where nucleosome occupancy changes with age identified established regulators of age-dependent metabolic dysfunction and novel lamina-associated candidates. Winged-helix transcription factor Foxa2 that regulates nucleosome dynamics binds regions of decreased nucleosome occupancy at PPARα targets in old livers. Conversely, binding of nuclear receptor co-repressor Hdac3, detected from motifs found in regions of increased nucleosome occupancy, exhibits a reciprocal pattern. Together, altered Foxa2 and Hdac3 occupancy at PPARα targets contributes to gene expression changes that lead to steatosis in aged liver.
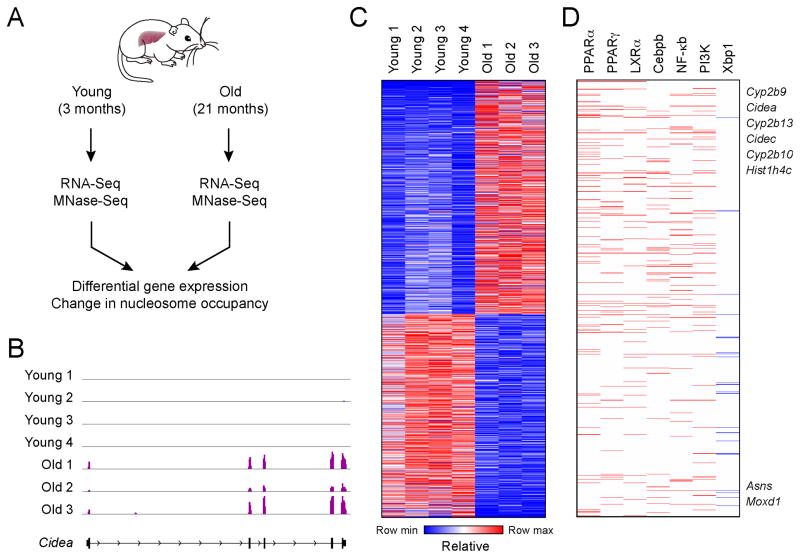
Figure 1. Nuclear receptor-dependent and inflammatory targets are induced during physiological aging in the liver.
(A) Experimental design includes comparing gene expression (RNA-Seq) from livers of young (3 months), middle-aged (12 months), and old (21 months) and nucleosome occupancy (MNase-Seq) of young and old livers. (B) RNA-Seq track view in Integrative Genome Viewer (IGV) of expression of Cidea, a target of nuclear receptors Pparα and Pparγ, not expressed in young healthy livers and highly induced in steatotic older hepatocytes. (C) Heat map of differentially expressed genes in aged livers (FDR 5%, 727 upregulated and 525 downregulated). (D) Regulators of gene expression in heat map in (C). Factors regulating lipid synthesis and storage are nuclear receptors Pparα (p-value 1.1×10−20), Pparγ (p-value 2.7×10−8), and LXRα (p-value 4.2×10−13), and Cebpb (p-value 2.1×10−9). Additional regulators include Nf-κb (p-value 1.8×10−3), PI3K (p-value 6.4×10−6), and Xbp1 (p-value 3.6×10−4). Networks with upregulated and downregulated targets are shown in red and blue, respectively. P values determined by Fisher’s exact test (using Ingenuity Pathway Analysis).
Results
Inflammatory and nuclear receptor target genes are induced in aged liver
To characterize transcriptional changes in aging, we profiled gene expression by RNA-Seq in young (3 months) and old (21 months) male mice (3–4 animals per age group; Figure 1A), and identified 1,252 genes differentially expressed between young and old mice (727 induced with age; 525 repressed; FDR 5% edgeR (Robinson et al., 2010), Experimental Procedures, Table S2). Consistent with previous studies of genetically-induced aging (Niedernhofer et al., 2006), genes induced with age were enriched for ‘Lipid & Fatty Acid Metabolism’ functions (88 of 1,157 pathway genes, P-value = 4.1×10−9, Fisher’s exact test). Among the highly induced genes were Cidea, a target of nuclear receptors PPARα and PPARγ, encoding a lipid-associated protein only detected in fatty and diabetic livers (Gong et al., 2009), related family members Cideb and Cidec, cytochrome p450 detoxification enzymes (Cyp2b9, Cyp2b10, and Cyp2b13) involved in stress response, and histone H4 transcript (Hist1h4c) (Figures 1B–1D). In contrast, mRNA levels of Moxd1, an enzyme in the endoplasmic reticulum (ER) and Asns, an enzyme upregulated by ER stress response, are downregulated in aged hepatocytes.
Genes induced with aging were also enriched for known targets of key transcriptional regulators of lipid homeostasis, including peroxisome proliferator activated receptors (PPARα and PPARγ; p-values 1.1×10−20 and 2.7×10−8, respectively, Ingenuity Pathway Analysis (IPA), Fisher’s exact test), liver X receptor (LXRα, Nr1h3, p-value 4.2×10−13), PGC1α (p-value 1.8×10−4), a co-activator of PPARs, and CEBPB (p-value 2.1×10−9), known to co-regulate PPARγ targets (Lefterova et al., 2008) (Figures 1D, S1A, S1B, Table S3). IPA also identifies gene networks regulated by agonists for PPARα (pirinixic acid/WY-14643 and clofibrate, p-values 9.6×10−20 and 1.8×10−7, respectively) and LXRα (T0901317, p-value 4.2×10−13), suggesting that PPARα and LXRα are ligand-activated in aged liver. In addition, genes activated by inflammatory regulators (NFκB/RELA, IRF3, and TLR4; p-values 1.8×10−3, 2.9×10−2, 2.1×10−3 respectively) are also upregulated in aged mice (Figure 1D, S1A, B, Table S3).
These results are consistent with a model where one or more of these regulators is activated during aging, leading to increased transcription of key lipid metabolism genes. Notably, PPARs are upregulated in progeroid syndromes, LXRα is activated in prematurely aged Foxa2 mutants (Bochkis et al., 2013), and inflammatory signaling is known to be activated by dietary fatty acids and contributes to insulin resistance (Fessler et al., 2009).
Age-dependent changes in nucleosome occupancy are enriched in distal elements
To study the role of chromatin organization in these age-dependent changes in transcription, we next measured genome-wide nucleosome occupancy profiles in livers from young (3 months) and old (21 months) mice. We used MNase digestion of chromatin followed by sequencing (MNase-Seq, Experimental Procedures, (Umlauf et al., 2004)), with two biological replicates for each age group: Replicate 1 with single-end sequencing, and Replicate 2 with paired-end sequencing (Figure 2A). We calculated nucleosome occupancy for each sample separately with the DANPOS package (Chen et al., 2013), with moderately good correlation between replicates (Spearman ρ = 0.78 and 0.76 for young and old replicates, respectively), and occupancy measures substantially higher in the second replicate. Given the differences between the replicates (Figure S3A and above) we calculated changes in nucleosome occupancy between young and old samples separately for each replicate, and used DANPOS (Chen et al., 2013) to identify regions with significant occupancy changes of well-positioned nucleosomes (29,355 and 61,869 regions of loss and 24,979 and 61,540 regions of gain for replicates 1 and 2, respectively) (Figure 2B). Finally, similar pathways enriched for genes with changes in nucleosome occupancy in either replicate (Figure S3B).
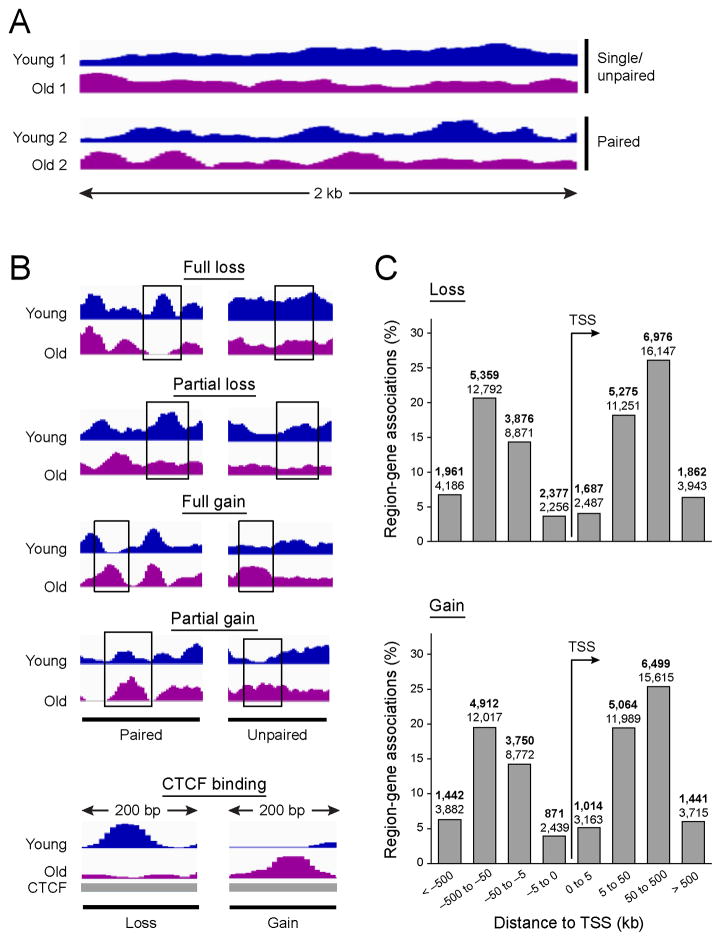
Figure 2. Characterization of regions of age-dependent change in nucleosome occupancy.
(A) MNase-Seq tracks show nucleosome occupancy of young (3M, blue) and old (21M, purple) biological replicates in 2 kb region (top - unpaired, bottom – paired). (B) Examples of loss (full and partial, two top panels) and gain (full and partial, two middle panels) in nucleosome occupancy for paired (left) and unpaired (right) replicates. Examples of loss and gain in nucleosome occupancy overlapping a CTCF binding site (bottom panel). (C) The distribution of age-dependent nucleosome occupancy change regions (both losses and gains) around TSS for replicate 2. Majority of changes occur distally, 50 to 500 kb from TSS Values for replicate 1 in parentheses.
Most age-dependent changes in nucleosome occupancy occur distally, 50 to 500 kb from TSS (Figure 2C). Furthermore, there is an overlap between regions with changes in nucleosome occupancy and those bound the insulator CTCF, or marked with H3K4me1 (10% and 9% of all regions with occupancy change for replicate 1 and 2, respectively) or with H3K9ac (9% and 3% of all regions with occupancy change for replicate 1 and 2, respectively) in young livers, modifications associated with enhancers and active euchromatin, respectively. This suggests that distal enhancers, known to be bound by Foxa factors and nuclear receptors (Bochkis et al., 2012; Lefterova et al., 2008), may be the genomic features most associated with nucleosome occupancy changes.
Foxa2 binds regions of reduced nucleosome occupancy at PPARα targets in aged liver
To identify transcriptional regulators whose binding may be affected by changes in nucleosome positions, we performed both de novo motif discovery (using MEME (Bailey et al., 2009), Experimental Procedures) and positional weight matrix (PWM) scan analysis (using PscanChIP (Zambelli et al., 2013), Experimental Procedures) of 150 bp windows spanning all nucleosomes gained or lost with age for either replicate.
Motifs associated with forkhead transcription factors are the most overrepresented in regions of changed occupancy (both gain and loss) for both replicates. FOXO factors, transducers of insulin signaling, can decompact chromatin (Hatta and Cirillo, 2007), and FOXA2 is known to bind nucleosomal DNA in vivo (Li et al., 2011). Indeed, FOXA2 targets (defined as differentially expressed in 1-year old Foxa2 mutants vs. WT liver (Bochkis et al., 2013)) are particularly differentially expressed between our wild type old vs. young livers (Figure S3C, KS p-value 2.4×10−9).
Motifs for nuclear receptors (RORa p-value 3.1×10−56, LXR p-value 7.6×10−85, PPAR p-value 9.7×10−76) and interferon regulatory factors (IRF p-value 8.1×10−80) were highly enriched in regions of nucleosome occupancy loss (Figure 3A). Protein expression of the lipid metabolism regulators FOXA2 and PPARα is not altered in aged livers, while that of PPARγ is induced, consistent with previous reports of PPARγ induction in fatty liver (Panasyuk et al., 2012) (Figure 3B). Ingenuity Pathway Analysis (IPA) of genes with nucleosome occupancy loss near TSS identified networks regulated by ligand-activated nuclear receptors PPARα, PPARγ, and LXRα. Expression was induced for quarter of these targets (Fold Change >=1.3, Figure 3C).
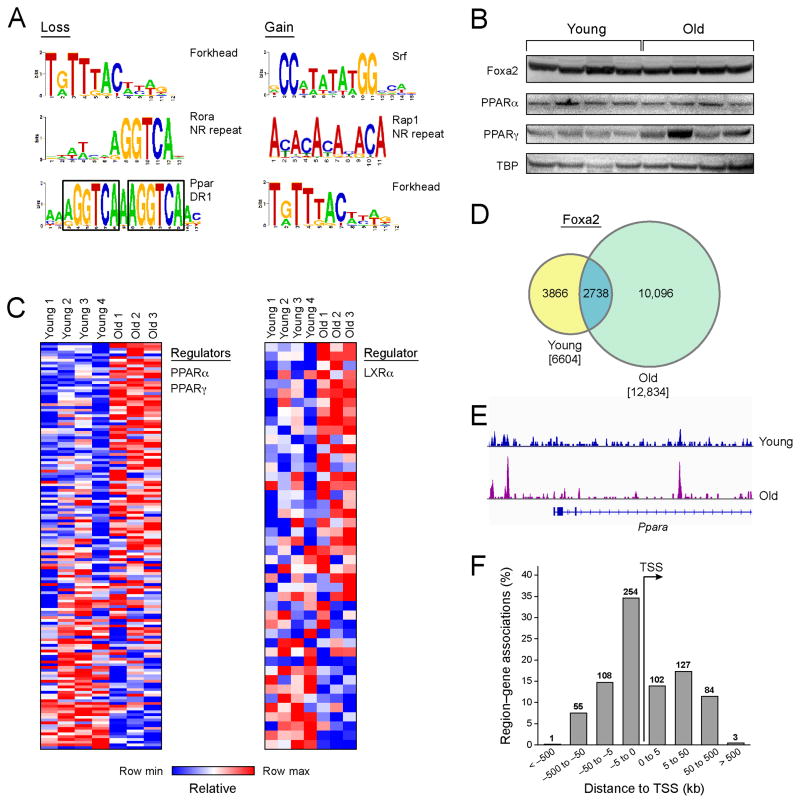
Figure 3. Foxa2 binds PPARα targets in old liver.
(A) PWM scan analysis identifies numerous forkhead PWMs and nuclear receptor motifs (Rora p-value 3.1×10−56, LXR p-value 7.6×10−85, PPAR p-value 9.7×10−76) in regions of age-dependent loss of nucleosome occupancy. PPAR factors bind a direct repeat (DR-1 element). The repeats are enclosed by black rectangles. Forkhead PWMs, as well as matrices for Srf (p-value 3.3×10−45) and Rreb1 (p-value 5.4×10−98), as well as a sequence resembling a telomeric repeat bound by Rap1 (p-value 5.9×10−64) are significantly overrepresented in regions of gain of nucleosome occupancy. (B) Western blot analysis of protein nuclear extracts from 4 young (3 months) and 4 old (21 months) mouse livers with antibodies to FOXA2, PPARα, PPARγ, and TATA box–binding protein (TBP, loading control). (C) Heat maps showing expression of Pparα and Pparγ targets (left panel) and LXRα targets (right panel) with age-dependent nucleosome occupancy loss near transcription start site (TSS). (D) Venn diagram showing the results of genome-wide location analysis for Foxa2 in young and old liver, identifying 6,605 binding sites in young and 12,834 in old, of which 2,738 were called bound by both factors by PeakSeq. (E) ChIP-Seq track view in IGV of increased Foxa2 binding in old liver at the Ppara locus.
Since Foxa2 plays an important role in lipid metabolism in aged liver (Bochkis et al., 2013), we next examined Foxa2 binding in young and old livers using ChIP-Seq. Strikingly, Foxa2 occupies substantially more sites in old hepatocytes (12, 834) than in young ones (6,605; Figure 3D). Additional bound regions in the old livers are found at PPAR target genes, as well as at the Ppara promoter (Figure 3E). The increase in binding is not due to change in FOXA2 expression (Figure 3B).
A subset of 734 Foxa2 sites that are “gained” in old livers correspond to regions of decreased nucleosome occupancy. Interestingly, while the majority of changes in nucleosome occupancy occur distally, these sites are found mostly at promoters (Figure 3F). Genes associated with the binding sites are enriched in functional categories including “hepatic steatosis” (p-value 1.5×10−4), “nuclear import” (p-value 3.8×10−4), and “increased circulating VLDL” (p-value 3.5×10−5). In addition, the PPAR/DR-1 element is enriched at these sites, suggesting that age-dependent Foxa2 binding could enable PPAR binding at these regions. Alternatively, newly-bound FOXA2 at the promoter may interact with PPAR proteins that are bound to existing distal enhancer elements, to activate PPAR targets (Figure 6).
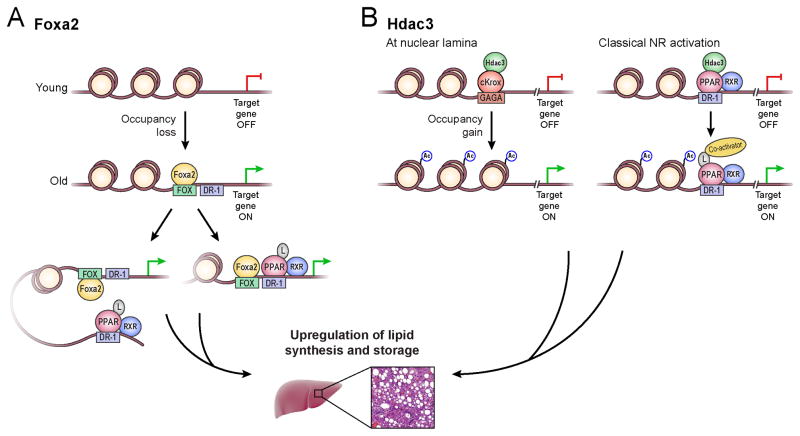
Figure 6. A model relating chromatin changes to development of fatty liver during aging.
Change in nucleosome occupancy in aged liver is associated with upregulation of nuclear receptor targets and development of steatosis. (A) Foxa2 binding leads to nucleosome eviction in older livers. Foxa2 cooperates with ligand-activated PPAR receptors (L: ligand), either interacting with existing PPAR proteins bound to enhancer elements (left) or enabling additional PPAR binding at the promoter (right), leading to upregulation of targets regulating lipid synthesis and storage. (B) Hdac3 regulates hepatic lipid targets in two ways: 1) at the nuclear lamina through GAGA sites bound by cKrox/Hdac3 (left), and 2) by repressing PPAR sites in young but not old livers in a classical mechanism of nuclear receptor (NR) action. Regions with a GAGA motif, bound by cKrox (Zbtb7b) in complex with Hdac3, inhibit expression of lipogenic targets in young livers. Age-dependent gain in nucleosome occupancy at these locations leads to eviction of histone deacytelase Hdac3. Nucleosomes can now be acetylated, leading to active transcription of nuclear receptor targets. In addition, in a classical model of nuclear receptor activation, Hdac3 binds unliganded PPARα in young livers and is evicted upon agonist stimulation in old livers. A co-activator with histone acetyltransferase activity is recruited to the PPAR complex. Nearby nucleosomes are acetylated and gene expression of the targets is turned on. The reciprocal binding pattern of Foxa2 and Hdac3 at loci encoding PPARα targets contributes to dysregulation of hepatic lipid homeostasis during aging.
Increased nucleosome occupancy suggests a role for cKrox-Hdac3 in age-dependent dysfunction
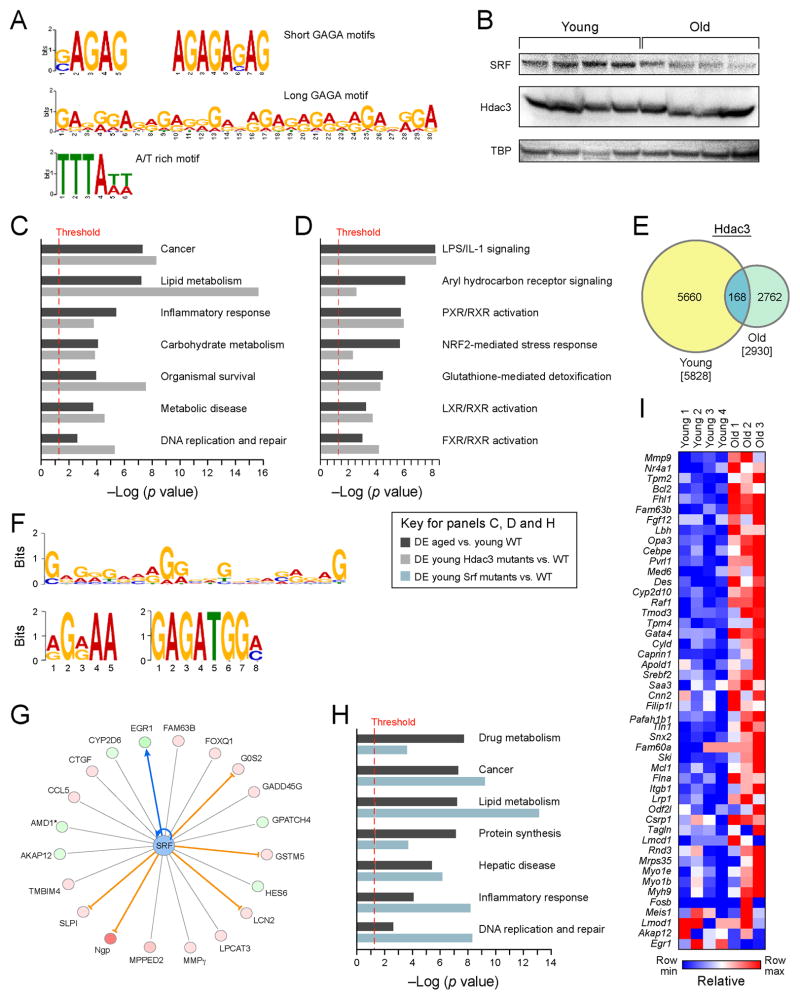
Regions associated with increased nucleosome occupancy were enriched, among other elements, with a motif bound by Srf (p-value 3.3×10−45), a factor that interacts with the nuclear lamina (Swift et al., 2013) (Figure 3A) and with novel motifs identified as a GAGA repeat (short motif p-value 1.8×10−111, long motif p-value 2.3×10−8) (Figure 4A). The short GAGA motif is frequent along the genome (background frequency 0.416), but the prevalence of this sequence in the regions of age-dependent occupancy gain was higher (frequency 0.527). The GAGA repeat is associated with lamina-associated domains (LADs) (Lund et al., 2013) and is bound by the transcriptional repressor cKrox (Zbtb7b) in a complex with Hdac3. While SRF protein levels decrease, expression of HDAC3 is not altered in older livers (Figure 4B). Hdac3 can act as a co-repressor of nuclear receptors regulating hepatic lipid metabolism genes (Knutson et al., 2008; Sun et al., 2012). Furthermore, the Ncor complex, an activating cofactor for Hdac3 (Guenther et al., 2001), is predicted to be repressed in our aged livers based on the expression of its known targets (Ingenuity Pathway Analysis, Experimental Procedures). Taken together, this suggests that Hdac3 activity may also be reduced in older animals, resulting in induction of its targets by nuclear receptors.
Figure 4. Novel regulators associated with nuclear lamina are implicated in mediating age-dependent dysfunction in the liver.
(A) GAGA repeat motif (short motif p-value 1.8×10−111, long motif p-value 2.3×10−8), bound by a transcriptional repressor cKrox (Zbtb7b) at the nuclear lamina, and an A/T rich sequence associated with lamina-associated domains (LADs), are enriched in regions of gain of nucleosome occupancy. (B) Western blot analysis of protein nuclear extracts from 4 young (3 months) and 4 old (21 months) mouse livers with antibodies to SRF, HDAC3, and TATA box–binding protein (TBP, loading control). Comparison of overrepresented biological functions (C) and pathways (D) between differentially expressed genes in aged wildtype livers and young liver-specific Hdac3 mutants (K-S p-value 2.2×10−16). (E) ) Venn diagram showing the results of genome-wide location analysis for Hdac3 in young and old liver, identifying 5,828 binding sites in young and 2,930 in old, of which 168 were called bound by both factors by PeakSeq. (F) GAGA repeat motif (short motif p-value 7.0×10−44, long motif p-value 3.6×10−13) is also enriched in regions bound by Hdac3 in young liver. (G) IPA Analysis of Srf-dependent network of genes differentially expressed in older livers suggests that Srf activity is downregulated with aging. (blue line represents activation, orange line – repression, grey line – association with expression change). (H) Comparison of overrepresented biological functions in aged wildtype livers and young liver-specific Srf mutants (I) Heat map of Srf target genes with age-dependent gain in nucleosome occupancy (replicate 2, pattern similar for replicate 1).
To test this hypothesis, we first compared the genes differentially expressed between (young) Hdac3 liver-specific mutants vs. WT (Knutson et al., 2008) with the genes differentially expressed between aged and young livers. We found that the Hdac3-dependent genes are particularly differentially expressed in old vs. young livers (compared to the background of all old vs. young differentially expressed genes; K-S p-value 2.2×10−16). In addition, genes from the same biological functions and pathways were affected in both young Hdac3 mutant mice and differentially expressed genes in aged livers (Figure 4C, 4D).
Next, we used ChIP-Seq to measure Hdac3 binding in young and old livers and found markedly less occupied regions in old livers (5,828 in young, 2,930 in old; Figure 4E), especially at PPAR-dependent genes. PPAR response element (DR-1) was significantly overrepresented in the sequences bound by Hdac3 in young but not old livers (p-value 1.4×10−31). While the overlap between Hdac3 sites bound in young livers and regions of increased nucleosome occupancy is small (152 regions), the GAGA motif is also enriched at sites bound by Hdac3 in the young livers (short motif p-value 7.0×10−44, long motif p-value 3.6×10−13) (Figure 4F), suggesting the overlap could be greater with deeper coverage of MNase-Seq data and more statistically significant nucleosome occupancy changes calls.
Sites for Serum Response Factor (SRF) were also enriched in regions with age-dependent gain of nucleosome occupancy (p-value <6×10−74, PScanChIP analysis for overrepresented PWMs). Furthermore, SRF-dependent genes were differentially expressed in older livers (IPA analysis, Experimental Procedures, Figure 4G), similar gene functions are enriched in genes differentially expressed in liver-specific Srf mutants (vs. WT) or in old liver (vs. young) (Figure 4H), and most (36 out of 48) Srf target genes with age-dependent gain in nucleosome occupancy near the TSS are induced in aged livers (Figure 4I). Taken together, these data suggest a down-regulation of Srf activity with aging, consistent with reduced SRF protein levels (Figure 4B), leading to de-repression of its gene targets. Srf activity has also been shown to be repressed during senescence by nuclear exclusion (Ding et al., 2001).
Foxa2 and Hdac3 exhibit reciprocal binding pattern at PPARα targets
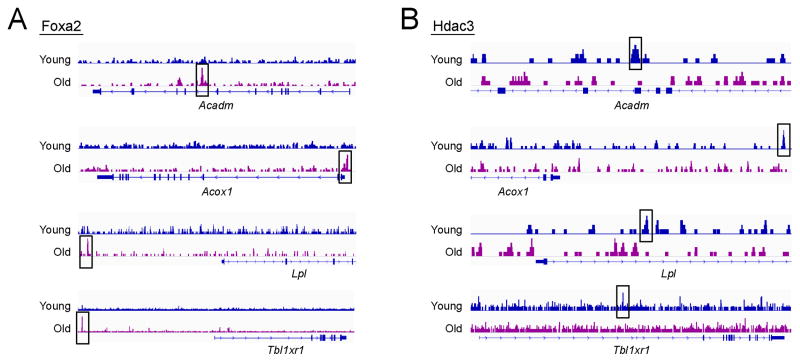
Strikingly, we found that Foxa2, a pioneer factor that binds the forkhead motif, occupies substantially more regions in old livers (6,605 in young, 12,834 in old), whereas lamina-associated nuclear receptor co-repressor Hdac3 binds markedly less regions (5,828 in young, 2,930 in old). These differential binding sites for both factors are found at functional PPARα target genes (Rakhshandehroo et al., 2010) (Figure 5) and contain the PPAR response element. This is consistent with a model where Foxa2 binding likely leads to nucleosome eviction in older livers. Foxa2 cooperates with PPAR receptors, either enabling PPAR binding at the promoter or interacting with existing PPAR proteins bound to enhancer elements, leading to upregulation of targets involved in lipid synthesis and storage (Figure 6A). Hdac3 may regulate hepatic lipid targets in either of two ways: (1) through GAGA sites bound by cKrox/Hdac3; or (2) by repressing PPAR sites in young but not old livers (Figure 6B). Together, the reciprocal binding pattern of Foxa2 and Hdac3 contributes to gene expression changes leading to steatosis in aged liver.
Figure 5. Foxa2 and Hdac3 exhibit reciprocal binding pattern at PPARα targets.
Chip-Seq track view of (A) Foxa2 and (B) Hdac3 binding in young and old livers at the loci encoding Ppara targets.
Discussion
Here, we used an unbiased approach to find candidate regulators that affect age-dependent metabolic dysfunction. Since nucleosomes and transcription factors compete for DNA binding (Workman and Kingston, 1992), mapping genome-wide nucleosome composition and tracking changes in nucleosome occupancy in aged mice in vivo allowed us to test for differences in transcription factor binding that are responsible for downstream gene regulation governing age-dependent phenotypes.
Motifs bound by forkhead transcription factors and nuclear receptors are significantly overrepresented in regions of age-dependent loss of nucleosome occupancy. We have examined binding of Foxa2 in young and old livers, and it is likely that other Fox factors, especially Foxa1 and Foxa3 and members of the Foxo subfamily, could play a role in this process and that possibility should be explored further. While nucleosome occupancy dynamics observed in aged livers associates with distal enhancers, elements bound by forkhead transcription factors and nuclear receptors in young livers (Bochkis et al., 2012) (Lefterova et al., 2008), we find that most Foxa2 sites that are bound only in old livers and correspond to regions of decreased nucleosome occupancy are found near the promoters. These sites are also enriched for the PPAR/DR-1 element, suggesting that additional Foxa2 binding might enhance accessibility and enable recruitment of PPAR factors to these elements (Figure 6A). We also observe upregulation of PPAR-dependent gene expression for genes with a nucleosome loss at the promoter. A recent study has challenged the classical model of nuclear-receptor-dependent gene regulation, reporting that LXRα and PPARα binding to their target loci in the liver is largely ligand-dependent, with the agonists enabling the receptors to occupy less accessible sites (Boergesen et al., 2012). Two additional reports involving progesterone receptor (PR) and estrogen receptor (ER) showed that nucleosome occupancy observed in unstimulated cells is significantly depleted upon hormone activation (Ballare et al., 2013; Tropberger et al., 2013), allowing for nuclear receptor binding. Our findings are consistent with this revised model and suggest that nucleosome dynamics may mediate ligand-dependent activation of “metabolic” nuclear receptors.
While Foxa2 binding sites are also enriched for the PPAR/DR-1 element, we cannot pinpoint which PPAR receptor (PPARα, PPARγ, or PPARδ) binds these sites and in which physiological condition. PPARα mediates the hepatic fasting response, and binding of this factor should also be examined in the fasted state. Hence, binding of PPAR receptors should be explored in young and old livers to determine the relationship between the factors and their roles in aged livers.
We find that shifts in hepatic gene expression in physiological aging mirror differences observed in progeroid conditions. Changes in nucleosome occupancy are associated with our inferred de-repression of nuclear receptors regulating hepatic lipid metabolism, leading to fatty liver (Figure 6). Examining changes in nucleosome occupancy in vivo highlighted novel lamina-associated regulators, Hdac3 and Srf, whose role in age-dependent metabolic dysfunction should be explored further.
Histone deacetylases related to Hdac3, Hdac1, and Sirt1, are known to play important roles in aging liver (Jin et al., 2011; Willis-Martinez et al., 2010). Liver-specific deletion of Hdac3 leads to fatty liver, a phenotype associated with aging, due to de-repression of nuclear hormone receptor-dependent gene expression (Sun et al., 2012) (Knutson et al., 2008). Hdac3 mutant livers also exhibit upregulation of mTOR signaling similar to a model of premature aging due to hepatocyte-specific ablation of Foxa2 (Bochkis et al., 2013). Deletion of Hdac3 also impacts DNA repair and reduces heterochromatin content, as observed in aging nuclei (Bhaskara et al., 2010). Loss of Hdac3 binding and transcriptional de-repression of targets is observed in adipocytes in a mouse model of progeria (Karakasilioti et al., 2013). Hence, it is likely that Hdac3 is a pivotal regulator of epigenetic and metabolic changes during chronological aging.
The second candidate, Srf, regulates liver proliferation, hepatic lipid metabolism, and growth hormone/Igf-1 signaling crucial to longevity (Sun et al., 2009). Transcription factors, including Hif1a, Hsf1, and Xbp1, that govern different stress responses, similar to Srf, affect gene expression during aging (Henis-Korenblit et al., 2010; Hsu et al., 2003; Kang et al., 2005). Loss of Srf in the liver also alters mRNA levels of histone proteins and chromatin regulators, similar to changes seen in aged livers. A recent study reported that lamin A regulates Srf mRNA levels and Srf-dependent gene transcription (Swift et al., 2013), providing another link to aging.
Notably, ‘Nuclear lumen’ genes, including a number of histone transcripts, were highly overrepresented in targets changed in older livers. Histone expression has been reported to decline in a number of aging paradigms (Feser et al., 2010) (Celona et al., 2011) (Liu et al., 2013). In contrast, we found that whereas some histone transcripts are downregulated with age, others are upregulated (Figures S2A–S2C). Downregulated histone H2 transcripts included replication-dependent (Hist2h2aa & Hist1h2b) and replication-independent genes (H2afx). H2afx is the principal chromatin component involved in DNA repair and reduced levels of this histone could explain defects in DNA repair in aged livers. Histone variants differ in stability and DNA binding and play distinct functions in the nucleus (Talbert and Henikoff, 2010). Changing composition of histone variants in aged tissues in vivo could impact gene regulation and should be investigated further.
Premature aging, due to either mutation in lamin A or defects in DNA repair, is associated with dysregulation of lipid homeostasis and upregulation of PPAR-dependent gene expression (Niedernhofer et al., 2006; Savage, 2009). We find that similar pathways, also implicated in metabolic syndrome, are perturbed in chronologically aged livers. We suggest a relationship between lamina-associated factors and age-dependent dysregulation of hepatic lipid metabolism. Whether lamina-dependent mechanisms could mediate age-onset degeneration in other tissues remains to be explored.
Experimental Procedures
Young and old mice
Male mice (C57BL6) were purchased from the National Institute of Aging (NIA) aged rodent colony (Charles River Laboratories). Four young (3 months), two middle-aged (12 months) and three old (21 months) mice were used for RNA studies. Two biological replicates of young and old mice were used for microccocal nuclease digestion and sequencing. Two biological replicates of young and old mice were used for chromatin immunoprecipitation and sequencing. Animals were housed for a week after arrival to acclimate to the light-dark cycle at the MIT facility before tissue harvest. All animal work was approved by MIT’s Committee on Animal Care (CAC protocol number Regev-0612-058-15).
RNA isolation and profiling
Liver RNA isolation and quantitative real-time PCR (QPCR) was performed as previously described (Bochkis et al., 2013). Total RNA (2 μg) was enriched for mRNA (Dynabeads mRNA direct kit, Life Technologies) used for strand-specific library preparation. mRNA was fragmented (Ambion fragmentation buffer, 70°C for 3 min), dephosphorylated, and concentrated (RNA Clean & Concentrator columns, Zymo Research). Ligation with RNA adapters was followed by first strand cDNA synthesis (AffinityScript reverse transcriptase enzyme). RNA was degraded (EDTA & NaOH) while adapters containing barcodes were ligated to cDNA, which subsequently was amplified for 10 cycles of PCR (KAPA HiFi DNA Polymerase, Kapa Biosystems). Libraries were sequenced on an Illumina HiSeq 2500 (31 bp paired-end reads; Table S1).
Microccocal nuclease digestion and sequencing
Digestion was performed on snap-frozen liver, as previously described (Umlauf et al., 2004) with a few modifications. Briefly, 100 mg of tissue was pulverized (Covaris cryoPrep impactor), and nuclei were purified using a sucrose gradient. The nuclei were digested with MNase (Roche catalog number 10107921001) at 37°C for 12 minutes. Several titrations of MNase enzyme were tested. Samples that were well-digested, with most of the input as a mononucleosome (150 bp) and a slight band of a dinucleosome (300 bp) visible on a BioAnalyzer trace, to prevent over-digestion, were used for subsequent library preparation. Libraries (starting material 10 ng) were made as previously described (Bochkis et al., 2012) and sequenced on Illumina HiSeq 2000 instrument (36 bp single-end, 25 bp paired-end, read summary in Table S4).
Chromatin immunoprecipitation and ChIP-Seq
Snap-frozen mouse liver (100 mg) from wild type mice was used to prepare chromatin. ChIP and ChIP-Seq were performed as reported previously described (Bochkis et al., 2012). A slight modification involved using multiplex adapters for sequencing and Kapa HiFi DNA polymerase (Kapa Biosystems) for PCR amplification. Foxa2-specific rabbit antiserum (Seven Hills Bioreagents, WRAB-1200) and rabbit antibody to Hdac3 (Abcam, ab7030) were used for immunoprecipitation. Libraries were sequenced on an Illumina HiSeq 2500 with 60bp single-end reads (Table S6).
Western blot analysis
Nuclear extracts preparation and protein immunoblot analysis were performed as reported previously (Bochkis et al., 2008). The primary antibodies used were rabbit antibody to Foxa2 (Seven Hills Bioreagents, WRAB-1200, 1:5,000), rabbit antibody to Hdac3 (Abcam, ab7030, 1:5,000), rabbit antibody to PPARα (Santa Cruz, sc-9000, 1:100), rabbit antibody to PPARγ (Santa Cruz, sc-7196, 1:100), rabbit antibody to SRF (Santa Cruz, sc-335, 1:200), and rabbit antibody to TBP (Santa Cruz, sc-273, 1:100).
RNA-seq data analysis
RNA-Seq reads were aligned using TopHat (Trapnell et al., 2009) to mouse genome build mm9 (library and alignment statistics in Table S1). Expression levels were calculated using RSEM (Li and Dewey, 2011). Differential expression analysis of RNA-seq (p-value <0.05) was performed in R using EdgeR package (Robinson et al., 2010), with a Benjamini-Hochberg FDR of 5%.
Functional analysis
Ingenuity curates information gathered from the literature, as well as genomic experiments (microarray, RNA-Seq, ChIP-Seq) to determine sets of targets controlled by a regulator (a transcription factor, chromatin remodeler, kinase, small molecule, etc.) and the logic of this regulation (activation or repression). IPA’s analysis method compares the overlap of each such set of targets with the subset of genes that are controlled by each regulator in an experimental gene list and reports the p-value of the overlap (Fisher’s exact test), If the overlap is significant, and the expression changes agree with those expected from the regulatory connection (genes activated by the regulator are activated in the analysis data set and repressed genes are repressed, for example), IPA analysis predicts whether the regulator associated with the gene targets is itself activated or inhibited. Analysis of overrepresented functional categories and upstream regulators in Ingenuity Pathway Analysis and heat map generation was performed as described (Bochkis et al., 2013).
ChIP-Seq analysis
Reads were aligned to the mouse genome (mm9; NCBI Build 37, Table S6) using BWA (Li and Durbin, 2009). Duplicate reads were removed. Reads (phred score > 20) that aligned uniquely were used for subsequent analysis. Data from two biological replicates were merged for each condition (Foxa2 young, Foxa2 old, Hdac3 young, and Hdac3 old). PeakSeq (Rozowsky et al., 2009) was used to identify bound peaks against input controls (Foxa2: FDR 10%, q-value =0.12; Hdac3: FDR 5%, q-value=0.2).
MNase-seq analysis
Reads were aligned using BWA (Li and Durbin, 2009) to mm9. Duplicate reads (replicate 1) and fragments (replicate 2) were removed from the analysis. Reads (replicate 1) and fragments (replicate 2) (phred score > 30) that aligned uniquely were used for subsequent analysis. Reads for each sample were down sampled to the same number (150 million). Nucleosome calls (~8.5 million nucleosome peaks) and changes in nucleosome occupancy were calculated by the DANPOS software (p-value < 1×10−7 in Poisson test, parameters: w=150, d=150, extend=148 for single-end, 74 for paired-end).
Regions annotation
Regions of changed nucleosome occupancy were associated to closest genes with the GREAT analysis tool (McLean et al., 2010). Overlap with histone modifications and CTCF binding (ENCODE data, 8 week mouse liver (Ding et al., 2012)) was computed using Galaxy genome analysis tools (Hillman-Jackson et al., 2012). Sequencing reads were visualized with the Integrative Genome Viewer (IGV) (Robinson et al., 2011). To compute correlations between the biological replicates, we trimmed reads from replicate one (single-end) to 25 bp and disregarded pairing information in reads from replicate two (paired-end) and used single reads (read 1) of 25 bp. Reads were aligned using BWA to Mus musculus genome build mm9 and duplicate reads were removed from the analysis. Each read alignment was extended to 150 bp (size of a nucleosome) and resulting data were used to compute genome coverage and Spearman correlation.
Motif analysis
MEME was used for de novo motif finding (default parameters: zero or one motif per sequence, min width=6, max width=50, max number of motifs=3) (Bailey et al., 2009). PscanChIP (Zambelli et al., 2013) was utilized to identify overrepresented positional weight matrices (PWMs) from Jaspar Version 5.0_Alpha (Vlieghe et al., 2006) and Transfac 7.0 Public (Matys et al., 2006) databases (parameters: assembly=mm9, background=mixed) among sequences corresponding to age-dependent change in nucleosome occupancy (150 bp nucleosome-wide regions with significant change in occupancy as calculated by DANPOS).
Statistical Analysis
Kolmogorov-Smirnov (K-S) test was performed to test the distributions of fold changes of targets differentially expressed in both mutant mice (Foxa2 (Bochkis et al., 2013), and Hdac3 (Knutson et al., 2008) and in aged wild type livers and fold changes of all genes differentially expressed in old vs. young livers. Chi-square test was used to assess whether a given chomatin mark is overrepresented in the set of age-dependent nucleosome occupancy changes (as compared to a background set).
Supplementary Material
Acknowledgments
We thank D. Thompson and R. Majovsky for critical reading of the manuscript, and L. Gaffney, R. Raychowdhury, H. Whitton, and I. Wortman for technical assistance. This work was supported by NHGRI CEGS P50 HG006193 and HHMI (AR). IMB was supported by National Diabetes and Digestive and Kidney Diseases Institute K01 award DK-101633.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Accession Numbers.
Genomic data from this study can be accessed at GEO under accession number GSE58006 (SubSeries GSE57809 for RNA-Seq, SubSeries GSE58005 for MNase-Seq, and SubSeries GSE60393 for ChIP-Seq).
Author Contributions
IMB developed the project, performed experiments and data analysis, and wrote the manuscript. DP and JC analyzed the data. AR edited the manuscript and provided funding for the study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2013;49:67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Ye DZ, Kurinna S, Stratton SA, Barton MC, Kaestner KH. Genome-wide location analysis reveals distinct transcriptional circuitry by paralogous regulators Foxa1 and Foxa2. PLoS Genet. 2012;8:e1002770. doi: 10.1371/journal.pgen.1002770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Shin S, Kaestner KH. Bile acid-induced inflammatory signaling in mice lacking Foxa2 in the liver leads to activation of mTOR and age-onset obesity. Molecular Metabolism. 2013;2:447–456. doi: 10.1016/j.molmet.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celona B, Weiner A, Di Felice F, Mancuso FM, Cesarini E, Rossi RL, Gregory L, Baban D, Rossetti G, Grianti P, et al. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 2011;9:e1001086. doi: 10.1371/journal.pbio.1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013;23:341–351. doi: 10.1101/gr.142067.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Gao S, Scott RE. Senescence represses the nuclear localization of the serum response factor and differentiation regulates its nuclear localization with lineage specificity. 2001 doi: 10.1242/jcs.114.5.1011. [DOI] [PubMed] [Google Scholar]
- Ding W, Gao S, Scott RE. Senescence represses the nuclear localization of the serum response factor and differentiation regulates its nuclear localization with lineage specificity. 2012 doi: 10.1242/jcs.114.5.1011. [DOI] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler MB, Rudel LL, Brown JM. Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol. 2009;20:379–385. doi: 10.1097/MOL.0b013e32832fa5c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman ML. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol. 2009;20:121–126. doi: 10.1097/MOL.0b013e328328d0bb. doi:110.1097/MOL.1090b1013e328328d328320bb. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Cirillo LA. Chromatin opening and stable perturbation of core histone:DNA contacts by FoxO1. J Biol Chem. 2007;282:35583–35593. doi: 10.1074/jbc.M704735200. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A. 2010;107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman-Jackson J, Clements D, Blankenberg D, Taylor J, Nekrutenko A. Using Galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2012;Chapter 10(Unit 10):15. doi: 10.1002/0471250953.bi1005s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jin J, Iakova P, Jiang Y, Medrano EE, Timchenko NA. The reduction of SIRT1 in livers of old mice leads to impaired body homeostasis and to inhibition of liver proliferation. Hepatology. 2011;54:989–998. doi: 10.1002/hep.24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang GL, Iakova P, Shi X, Haefliger S, Finegold M, Timchenko NA. Epigenetic changes play critical role in age-associated dysfunctions of the liver. Aging Cell. 2010;9:895–910. doi: 10.1111/j.1474-9726.2010.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Wang GL, Timchenko L, Timchenko NA. GSK3beta and aging liver. Aging (Albany NY) 2009;1:582–585. doi: 10.18632/aging.100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Kim HJ, Kim HK, Lee JY, Kim DH, Jung KJ, Kim KW, Baik HS, Yoo MA, Yu BP, et al. The effect of age and calorie restriction on HIF-1-responsive genes in aged liver. Biogerontology. 2005;6:27–37. doi: 10.1007/s10522-004-7381-z. [DOI] [PubMed] [Google Scholar]
- Karakasilioti I, Kamileri I, Chatzinikolaou G, Kosteas T, Vergadi E, Robinson AR, Tsamardinos I, Rozgaja TA, Siakouli S, Tsatsanis C, et al. DNA Damage Triggers a Chronic Autoinflammatory Response, Leading to Fat Depletion in NER Progeria. Cell Metab. 2013;18:403–415. doi: 10.1016/j.cmet.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat Struct Mol Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Oldenburg AR, Delbarre E, Freberg CT, Duband-Goulet I, Eskeland R, Buendia B, Collas P. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013;23:1580–1589. doi: 10.1101/gr.159400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Panasyuk G, Espeillac C, Chauvin C, Pradelli LA, Horie Y, Suzuki A, Annicotte JS, Fajas L, Foretz M, Verdeguer F, et al. PPARgamma contributes to PKM2 and HK2 expression in fatty liver. Nature communications. 2012;3:672. doi: 10.1038/ncomms1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR research. 2010;2010 doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Muller DC, Metter EJ, Maggio M, Harman SM, Blackman MR, Andres R. Aging, androgens, and the metabolic syndrome in a longitudinal study of aging. J Clin Endocrinol Metab. 2007;92:3568–3572. doi: 10.1210/jc.2006-2764. [DOI] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DB. Mouse models of inherited lipodystrophy. Dis Model Mech. 2009;2:554–562. doi: 10.1242/dmm.002907. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Hoeijmakers JH, Garinis GA. Sealing the gap between nuclear DNA damage and longevity. Mol Cell Endocrinol. 2009;299:112–117. doi: 10.1016/j.mce.2008.10.031. [DOI] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- Sun K, Battle MA, Misra RP, Duncan SA. Hepatocyte expression of serum response factor is essential for liver function, hepatocyte proliferation and survival, and postnatal body growth in mice. Hepatology. 2009;49:1645–1654. doi: 10.1002/hep.22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Buhler M, et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R. Site-specific analysis of histone methylation and acetylation. Methods Mol Biol. 2004;287:99–120. doi: 10.1385/1-59259-828-5:099. [DOI] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45:279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. PscanChIP: Finding over-represented transcription factor-binding site motifs and their correlations in sequences from ChIP-Seq experiments. Nucleic Acids Res. 2013;41:W535–543. doi: 10.1093/nar/gkt448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.