Summary
Host cell factor-1 (HCF-1) is a metazoan transcriptional co-regulator essential for cell cycle progression and cell proliferation. Current models suggest a mechanism whereby HCF-1 functions as a direct co-regulator of E2F proteins, facilitating the expression of genes necessary for cell proliferation. In this report, we show that HCF-1 recruitment to numerous E2F-bound promoters is mediated by the concerted action of zinc finger transcription factors THAP11 and ZNF143, rather than E2F proteins directly. THAP11, ZNF143, and HCF-1 form a mutually dependent complex on chromatin, which is independent of E2F occupancy. Disruption of the THAP11/ZNF143/HCF-1 complex results in altered expression of cell cycle control genes and leads to reduced cell proliferation, cell cycle progression, and cell viability. These data establish a new model which suggests that a THAP11/ZNF143/HCF-1 complex is a critical component of the transcriptional regulatory network governing cell proliferation.
Introduction
Host cell factor-1 (HCF-1) is a ubiquitously expressed transcriptional co-regulator which has been identified in a variety of transcriptional regulatory complexes. HCF-1 is believed to function as a molecular scaffold, linking sequence-specific transcription factors with enzymes capable of altering the post-translational modifications of histones and other chromatin associated proteins (Ajuh et al., 2000; Liang et al., 2009; Vogel and Kristie, 2006, 2013; Wysocka et al., 2003). The biological significance of HCF-1 dependent gene expression is underscored by multiple studies demonstrating that HCF-1 function is critical for cell proliferation and cell cycle progression (Julien and Herr, 2003; Mangone et al., 2010; Reilly et al., 2002; Wysocka et al., 2001). Evidence suggesting that HCF-1 regulated transcription contributes to cell proliferation was initially provided by Wysocka et al., who showed that loss of HCF-1 chromatin association precedes growth arrest of temperature-sensitive tsBN67 hamster cells, which contain a single proline-to-serine missense mutation (HCF-1 P134S) previously known to disrupt HCF-1 association with the VP16 viral transactivator (Goto et al., 1997; Wysocka et al., 2001). Subsequent work from the same laboratory revealed that the S-phase defect in tsBN67 cells grown at the non-permissive temperature could be mitigated by inactivation of the retinoblastoma tumor suppressor protein (RB1), leading to the hypothesis that HCF-1 promotes cell proliferation by regulating E2F cell cycle control genes (Reilly et al., 2002; Tyagi et al., 2007). E2F family members E2F1, E2F4, and E2F3a have been shown to contain the tetrapeptide HCF-1 binding motif (HBM; [E/D]HxY) and physically interact with HCF-1 (Knez et al., 2006; Tyagi et al., 2007). E2F1 and E2F4 associate with HCF-1 in HeLa cells, and HCF-1 chromatin occupancy at E2F regulated genes has been suggested to occur by E2F-mediated recruitment of HCF-1 in a cell cycle-dependent manner (Tyagi et al., 2007).
Although the current model proposes that HCF-1 is a direct transcriptional co-regulator of E2F proteins, recent evidence suggests that other sequence-specific transcription factors may also play a role in HCF-1 recruitment at cell cycle and growth control genes. Yu et al. have demonstrated that a ternary complex composed of Yin Yang 1 (YY1), HCF-1, and deubiquitinase BRCA1 associated protein-1 (BAP1) regulates the expression of cell growth and proliferation genes (Yu et al., 2010). Additionally, work in our laboratory and by others has shown that the Thanatos associated protein (THAP) domain-containing family of atypical zinc finger transcription factors constitutes a large group of putative HCF-1-associated transcriptional regulators (Dejosez et al., 2008; Dejosez et al., 2010; Mazars et al., 2010; Parker et al., 2012). Mazars et al. have demonstrated that THAP1 recruits HCF-1 to the RRM1 promoter during endothelial cell proliferation, and both THAP1 and HCF-1 are necessary for RRM1 gene expression (Mazars et al., 2010). We have previously shown that THAP11 is an HCF-1-dependent transcriptional regulator and cell proliferation factor in human colon cancer cells (Parker et al., 2012). Interestingly, we found that THAP11 is recruited with HCF-1 at E2F target genes RBL1 and CDC25A, but the functional relevance of this observation remains unknown.
In this report, we provide evidence that HCF-1 recruitment to numerous E2F-bound promoters is mediated not by E2F factors directly, but instead by the joint occupancy of zinc finger transcription factors THAP11 and ZNF143. We further show that THAP11, ZNF143, and HCF-1 assemble into a complex on chromatin and that stable complex association with chromatin is mutually dependent on all three factors. The expression of cell proliferation and cell cycle control genes is found to be at least partially THAP11/ZNF143/HCF-1 complex-dependent, and disruption of this complex results in altered cell cycle progression and reduced cell proliferation. The present study changes our current view of the role of E2F proteins in HCF-1 recruitment and demonstrates a potentially new mechanism of THAP11/ZNF143-mediated HCF-1 recruitment to key cell cycle progression genes.
Results
HCF-1 Occupancy at E2F1-Bound Promoters Correlates with THAP11 and ZNF143 Binding
Our previous characterization of THAP11 function in human colon cancer cells unexpectedly revealed that THAP11 bound with HCF-1 at RBL1 and CDC25A promoters (Parker et al., 2012). HCF-1 recruitment to both RBL1 and CDC25A promoters has been previously shown to function in both an E2F- and cell cycle-dependent manner (Tyagi et al., 2007). Nonetheless, we speculated that THAP11 might also play a role in HCF-1 recruitment at these and, perhaps, other E2F target gene promoters. To explore this possibility, we analyzed ENCODE E2F1 chromatin occupancy datasets (Consortium, 2011) for the presence of a previously determined THAP11 binding motif (Dejosez et al., 2010). This analysis identified 1702 promoter-proximal candidate THAP11 binding sites located within 400 base pairs of E2F1-bound regions. Comparison of these regions with ENCODE transcription factor ChIP-seq data revealed that these putative THAP11 binding sites frequently lie within experimentally determined ZNF143-bound regions. Direct sequence comparison further showed that the THAP11 binding motif is highly similar to an extended ZNF143 binding motif (Myslinski et al., 2006; Wang et al., 2012), suggesting THAP11 and ZNF143 may occupy similar DNA sequences. To test if THAP11 or ZNF143 occupy chromatin with HCF-1 at these E2F1-bound promoters, we performed chromatin immunoprecipitation for each factor in HeLa cells and determined binding at 62 candidate THAP11/ZNF143, and E2F bound promoters, as well as two negative control promoters (HBB and CHRM1). We found that the chromatin occupancy of HCF-1, THAP11, and ZNF143 at these regions was highly correlated, with Pearson pairwise correlation coefficients exceeding 0.90 (Fig.1A and 1B), while E2F1 binding showed no correlation with either factor (r<0.19). ChIP-scanning assays performed to determine each factor's promoter-proximal positioning revealed that THAP11, HCF-1, and ZNF143 share a nearly identical binding profile that was distinct from that of E2F1 (Fig.1C). Moreover, the location of maximum ChIP signal observed for THAP11, HCF-1, and ZNF143 at CDC25A, CDC6, and MCM3 promoter-proximal regions corresponds with a conserved THAP11/extended ZNF143 binding motif (Fig.1D). Similar results demonstrating that HCF-1 binding is coincident with THAP11 and ZNF143 rather than E2F binding were also obtained in T98G glioblastoma cells, suggesting these findings are not cell type specific (Fig.S1). Together, these data suggest that at a subset of E2F target genes, THAP11 and/or ZNF143 may directly recruit HCF-1 to chromatin.
Figure 1. HCF-1 occupancy at E2F target genes correlates with THAP11 and ZNF143 binding.
(A) HCF-1, THAP11, ZNF143, and E2F1 chromatin occupancy determined by ChIP assay in HeLa cells at the indicated 62 E2F1 target genes containing putative THAP11/ZNF143 binding motifs. (B) Matrix of pairwise Pearson product-moment correlation coefficients from ChIP assays in panel A. (C) Distribution of THAP11, HCF-1, ZNF143, and E2F1 on the promoter proximal regions of E2F1 target genes determined by ChIP-scanning assays. (D) Schematic illustrating the positions of promoter proximal ChIP amplicons (black rectangles) analyzed in panel C. The approximate locations of putative THAP11/ZNF143 and known E2F1 binding sites are indicated. First exons of the indicated genes are denoted by blue rectangles. See also Figure S1.
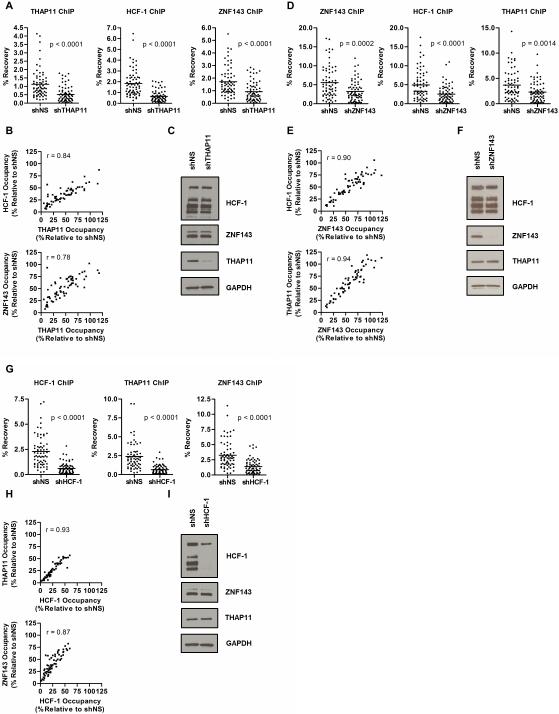
THAP11, ZNF143, and HCF-1 Chromatin Occupancy are Interdependent
To test the possibility that THAP11 and/or ZNF143 is necessary for HCF-1 recruitment, we next performed a series of ChIP assays examining occupancy of each factor at these 62 promoters following THAP11, ZNF143, or HCF-1 knockdown in HeLa cells. THAP11 knockdown resulted in a significant decrease of both HCF-1 and ZNF143 chromatin occupancy (Fig.2A) but depletion of THAP11 from chromatin was neither uniform nor complete. Pairwise scatterplots of HCF-1 or ZNF143 chromatin occupancy relative to THAP11 revealed good correlation between the extent of THAP11 depletion and corresponding loss of HCF-1 and ZNF143 binding (Fig.2B). Importantly, neither HCF-1 nor ZNF143 protein levels were changed in THAP11 knockdown cells (Fig.2C), suggesting that the decrease in HCF-1 and ZNF143 chromatin occupancy results from their altered recruitment to these promoters.
Figure 2. THAP11, HCF-1, and ZNF143 chromatin occupancy are interdependent.
THAP11, HCF-1, and ZNF143 chromatin occupancy was determined at each of the 62 genes shown in Fig.1A in THAP11, HCF-1 or ZNF143 knockdown HeLa cells. (A) ChIP in HeLa cells expressing either control (shNS) or THAP11 (shTHAP11) shRNA. (B) Scatterplots of HCF-1 (top panel) or ZNF143 (bottom panel) chromatin occupancy relative to THAP11 binding. (C) Immunoblots of THAP11, HCF-1, and ZNF143 in THAP11 knockdown HeLa cells. (D) ChIP in HeLa cells expressing either control or ZNF143 shRNA. (E) Scatterplots of HCF-1 (top panel) or THAP11 (bottom panel) chromatin occupancy relative to ZNF143 binding. (F) Immunoblots of THAP11, HCF-1, and ZNF143 in ZNF143 knockdown HeLa cells. (G) ChIP in HeLa cells expressing control or HCF-1 shRNA. (H) Scatterplots of THAP11 (top panel) or ZNF143 (bottom panel) chromatin occupancy relative to HCF-1. (I) Immunoblots of THAP11, HCF-1, and ZNF143 in HCF-1 knockdown HeLa cells. In panels A, B, D, E, G, and H each point represents one of 62 genes shown in Fig.1A. Values are expressed as percent recovery relative to input. In panels B, E, and H values are expressed as percent occupancy relative to control shRNA (shNS) expressing cells. Student's T-test p-values and Pearson product-moment correlation coefficient (r) values are indicated.
A similar set of experiments was performed to determine the role of ZNF143 in THAP11 and HCF-1 recruitment. As shown in Figure 2D, HeLa cells expressing ZNF143 shRNA show a significant reduction in average recruitment of ZNF143, THAP11, and HCF-1 at these 62 target genes. Similarly to what was observed for THAP11, the amount of ZNF143 that remained bound at these promoters was variable and ranged from unchanged to less than 25% remaining (Fig.2E), despite substantial depletion of ZNF143 protein (Fig.2F). Decreases in THAP11 and HCF-1 promoter occupancy upon ZNF143 knockdown closely paralleled that of ZNF143 (Fig.2E). Because THAP11 and HCF-1 total protein amounts were unchanged upon ZNF143 knockdown (Fig.2F), these results suggest that ZNF143 contributes to both THAP11 and HCF-1 recruitment to chromatin.
We previously demonstrated that THAP11 not only recruits but also requires HCF-1 for chromatin association (Parker et al., 2012). To determine if ZNF143 chromatin association has a similar dependence on HCF-1, we performed THAP11 and ZNF143 ChIP assays in HCF-1 knockdown cells. Confirming our previous observations, HCF-1 knockdown in HeLa cells resulted in a robust decrease in THAP11 promoter occupancy (Fig.2G). Unexpectedly, however, HCF-1 knockdown also resulted in a significant decrease in ZNF143 binding (Fig.2G). Unlike THAP11 and ZNF143 shRNAs, HCF-1 knockdown resulted in at least a 50% reduction of occupancy at most promoters examined (Fig.2H). Nevertheless, both THAP11 and ZNF143 relative promoter occupancy showed a strong linear relationship with remaining HCF-1 binding. Since neither THAP11 nor ZNF143 protein levels were changed (Fig.2I), these results likely suggest that HCF-1 is necessary for both THAP11 and ZNF143 binding at these promoters. Collectively, the results presented in Figure 2 indicate that THAP11 and ZNF143 are critical mediators of HCF-1 occupancy at these 62 E2F1-bound promoters and further demonstrate that THAP11, ZNF143, and HCF-1 chromatin binding are mutually dependent.
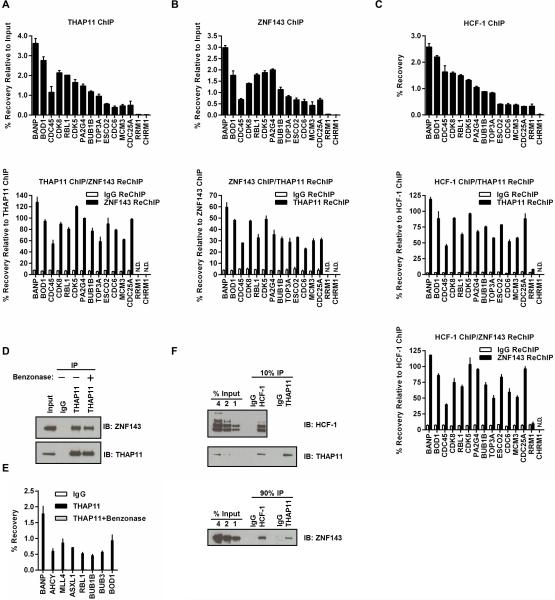
THAP11, ZNF143, and HCF-1 Form a Complex on Chromatin
The nearly identical promoter-proximal positioning (Fig.1C) and interdependent chromatin association (Fig.2) observed for THAP11, HCF-1 and ZNF143 could indicate formation of a complex between these factors. To explore this possibility further, a series of sequential chromatin immunoprecipitation experiments (ChIP-reChIP) was performed to determine if THAP11, HCF-1, and ZNF143 simultaneously co-occupy the promoters of E2F target genes involved in cell proliferation and cell cycle progression. Consistent with this hypothesis, we found that ZNF143 was robustly reChIP'ed at all THAP11 bound promoters examined (Fig.3A). Similar results were obtained when the order of antibodies was reversed (i.e. ZNF143 ChIP followed by THAP11 reChIP; Fig.3B) indicating that THAP11 and ZNF143 indeed occupy these promoters simultaneously. Sequential ChIP assays were next used to determine if THAP11 and ZNF143 co-occupy these promoters together with HCF-1. As shown in Figure 3C, HCF-1-bound promoters were effectively reChIP'ed by either anti-THAP11 or anti-ZNF143 antibodies, thus confirming that THAP11, ZNF143, and HCF-1 likely bind these promoters as a complex. The lack of reChIP signal observed for THAP11 and ZNF143 at RRM1 following HCF-1 ChIP (Fig.3C) is consistent with previous reports from our laboratory and others suggesting that HCF-1 recruitment to RRM1 is mediated not by THAP11 but instead by THAP1 (Mazars et al., 2010; Parker et al., 2012). This internal control not only confirms the fidelity of the sequential ChIP assay but also suggests that ZNF143 co-occupancy is not broadly applicable to all THAP protein/HCF-1 complexes.
Figure 3. THAP11, HCF-1, and ZNF143 simultaneously co-occupy chromatin.
(A) THAP11 ChIP (top panel) followed by ZNF143 or control IgG ReChIP (bottom panel). (B) ZNF143 ChIP (top panel) followed by THAP11 or control IgG ReChIP. (C) HCF-1 ChIP (top panel) followed by THAP11 ReChIP (middle panel) or ZNF143 ReChIP (bottom panel). (D) ZNF143 co-precipitation with THAP11 in formaldehyde-crosslinked SW620 cells is Benzonase-resistant. THAP11 ChIP-enriched material was treated with or without Benzonase. Digested nucleic acids and any associated proteins were removed by extensive washing, and remaining ZNF143 was detected by immunoblot. Input corresponds to 5% of starting material. (E) Confirmation of Benzonase digestion to remove THAP11 associated DNA was determined by qPCR using primers corresponding to THAP11 binding sites at indicated gene promoters. (F) ZNF143 associates with both THAP11 and HCF-1 in soluble nuclear extracts. HCF-1 and THAP11 were immunoprecipitated from SW620 nuclear extracts and co-precipitated ZNF143 was detected by immunoblot. In the top and middle panels, 10% of the immunoprecipitation was loaded for immunoblot. In the bottom panel, 90% of the immunoprecipitation was analyzed. In panels A-C and E values represent mean ± standard deviation of duplicate qPCR reactions from a single experiment performed at least three times with similar results. ChIP values are expressed as percent recovery relative to input. ReChIP values are expressed as percent recovery relative to the initial ChIP. qPCR amplicons below the limit of reliable detection are indicated as N.D. (not detected).
While these results strongly indicate the formation of a THAP11/ZNF143/HCF-1 complex, it remains possible that the observed THAP11 and ZNF143 chromatin co-occupancy may result from separate protein-DNA interactions that are indiscernible by conventional chromatin immunoprecipitation. To rule out this possibility, we performed THAP11 ChIP and subjected the protein-G bead-bound immunoprecipitated material to extensive Benzonase nuclease digestion followed by several stringent washes to remove proteins bound exclusively through protein-nucleic acid interactions. After formaldehyde crosslinks were reversed, ZNF143 recovery was monitored by western blot while the effectiveness of DNA digestion was determined by quantitative PCR of THAP11/ZNF143 bound promoters. As shown in Figure 3D, THAP11 ChIP recovered a significant fraction of chromatin-bound ZNF143 which was unaffected by Benzonase treatment despite a near complete digestion of THAP11 associated DNA (Fig.3E). Because the qPCR amplicons used to monitor Benzonase digestion span only 70-150bp and either flank or lie immediately adjacent to the presumptive THAP11/extended ZNF143 binding sites, the above results suggest that THAP11, HCF-1, and ZNF143 likely co-occupy these sites as a complex. To determine whether this complex is formed exclusively on chromatin, we performed co-immunoprecipitations from soluble nuclear extracts. As expected from previous studies, HCF-1 immunoprecipitation recovered a large amount of soluble THAP11 while from the same material a smaller fraction of ZNF143 was also recovered (Fig.3F). We find that under identical conditions, THAP11 immunoprecipitation recovered less albeit detectable ZNF143, indicating that THAP11 can interact with both HCF-1 and ZNF143 in soluble nuclear extracts as well as on chromatin.
E2F Binding is Dispensable for HCF-1 Recruitment to Chromatin
The finding that THAP11 and ZNF143 are necessary for HCF-1 recruitment to E2F target genes was somewhat surprising since previous reports suggested that E2F proteins (E2F1, E2F3a, and E2F4) directly recruit HCF-1 to chromatin (Tyagi et al., 2007; Tyagi and Herr, 2009). To independently examine the role of E2F binding activity in HCF-1 recruitment, we performed HCF-1 ChIP in E2F1 knockdown HeLa cells. We find that under conditions where E2F1 protein and chromatin occupancy are significantly depleted, HCF-1 recruitment at the same loci is mostly unchanged (Fig.4A and 4B). E2F4 knockdown similarly failed to disrupt HCF-1 promoter occupancy at prototypical E2F target genes RBL1, CDC25A, CDC6, and MCM3 (Fig.S2A). Since remaining E2F members may compensate for loss of individual E2F proteins (Kong et al., 2007), we next used knockdown of the E2F obligate heterodimeric partner DP1 to determine if E2F functional redundancy explains why HCF-1 promoter occupancy is insensitive to knockdown of individual E2Fs. We find that, while depletion of DP1 is sufficient to simultaneously reduce both E2F1 and E2F3 binding at these promoters, HCF-1 binding is largely unchanged (Fig.4C and 4D), suggesting direct E2F-mediated recruitment may not be the primary mechanism of HCF-1 occupancy at these E2F-bound promoters after all. To confirm this finding, we generated T98G cells containing a chromosomally integrated wildtype or E2F binding site mutant RBL1 promoter construct (Fig.4E) and monitored HCF-1 occupancy at these ectopic loci by ChIP. The specific E2F binding site mutations employed here have been previously shown to eliminate E2F binding at the endogenous Rbl1 promoter in both mouse embryonic stem cells and fibroblasts (Burkhart et al., 2010). Likewise, we find that E2F recruitment, as determined by DP1 ChIP, is reduced to near-background levels at the mutant RBL1 promoter (Fig.4F, compare DP1 ChIP at promoter and luciferase). HCF-1 occupancy, however, was unperturbed by the E2F binding site mutations (Fig.4F), further reinforcing the idea that HCF-1 promoter occupancy is independent from that of E2F proteins.
Figure 4. E2F binding is dispensable for HCF-1 recruitment to chromatin.
(A) E2F1 and HCF-1 ChIP in HeLa cells expressing either control (shNS) or E2F1 (shE2F1) shRNA. Each point represents one of 62 genes shown in Fig.1A. Student's T-test p-values are indicated. (B) Immunoblots from control and E2F1 knockdown cells shown in panel A. (C) HCF-1, DP1, E2F1, and E2F3 ChIP assays in HeLa cells expressing either control or DP1 shRNA. (D) Immunoblots from control and DP1 knockdown cells shown in panel C. (E) Schematic of the RBL1 promoter construct stably integrated in T98G cells. Numbers indicate position relative to the annotated RBL1 transcription start site. The putative THAP11/ZNF143 binding site and known E2F binding sites are indicated by an oval and circles, respectively. The relative position of the qPCR primers used to detect occupancy at the promoter or luciferase coding sequence are indicated by closed and open arrowheads, respectively. (F) HCF-1 and DP1 binding in T98G cells at the chromosomally integrated RBL1 wildtype (WT) and E2F binding site mutant promoters as determined by ChIP. (G) THAP11, HCF-1, and DP1 occupancy at the indicated E2F binding site in T98G cells as determined by ChIP assays. (H) E2F1 and HCF-1 ChIP in HeLa cells expressing either control (shNS) or E2F1 shRNAs. Binding was determined at E2F-bound promoters shown to be either THAP11/ZNF143-bound (positive) or unbound (negative). In panels C, F, G and H values represent mean ± standard deviation of duplicate qPCR reactions from a single experiment performed at least three times with similar results. See also Figure S2.
While the above results demonstrate that E2F binding is dispensable for HCF-1 recruitment at promoters also co-occupied by THAP11 and ZNF143, E2Fs might still play an important role in HCF-1 recruitment, especially at genes that lack THAP11 and ZNF143 occupancy. To test this hypothesis, we first determined HCF-1 occupancy on promoters of E2F target genes without discernible THAP11/ZNF143 binding motifs near their known E2F binding sites. In both T98G (Fig.4G, left panel) and HeLa cells (data not shown), we find that HCF-1 recruitment to THAP11/ZNF143-free MCM4 and PCNA promoters is markedly lower than HCF-1 binding at the E2F/THAP11/ZNF143 co-occupied RBL1 promoter, despite comparable levels of E2F occupancy. Similar results were found when HCF-1 recruitment at the CDC25A promoter, bound by THAP11, ZNF143, and E2F, was compared to HCF-1 binding at E2F-only promoters E2F1, CCNA2, CCNE1, and CDK1 (Fig.4G, right panel). In both instances, total E2F binding, as determined by DP1 ChIP, was similar between THAP11/ZNF143 bound and unbound promoters, suggesting that E2F binding alone may be insufficient to mediate substantial HCF-1 recruitment.
The small amount of HCF-1 bound at these promoters precluded a more detailed investigation of whether HCF-1 recruitment to THAP11/ZNF143-free promoters is dependent on E2F1. To circumvent this issue, we used available ChIP-seq datasets to identify a candidate set of promoters that were enriched for HCF-1 and E2F1 but devoid of THAP11 and ZNF143. Fourteen of these promoters were chosen at random and validated by ChIP-qPCR to have virtually no detectable THAP11 and ZNF143 binding (Fig.S2B). Knockdown of E2F1 in HeLa cells resulted in substantial depletion of E2F1 across all chosen THAP11/ZNF143-free promoters, yet no change in HCF-1 occupancy was observed (Fig.4H). These findings strongly suggest that HCF-1 recruitment at E2F target genes is independent of E2F binding regardless of THAP11 and ZNF143 co-occupancy.
To extend the above observations to a genome-wide scale, we analyzed previously published and publicly available ChIP-seq data. While no single dataset covered all of the proteins of interest, we chose datasets that were generated in the same cell line (HeLa-S3). E2F1 ChIP-seq has been performed by the ENCODE consortium (Consortium, 2011), while ChIP-seq data for THAP11, ZNF143, and HCF-1 was recently published by Michaud et al. (2013). In order to compare these data, we used identical parameters to first align raw sequencing reads from all datasets to the human hg19 reference genome and then call peaks using HOMER ChIP-seq analysis software (see Methods for details).
Proteins that are found on chromatin in a single complex and are responsible for each other's occupancy should have significant spatial overlap, as observed by ChIP-scanning in Figure 1C. To extend this observation to a genome-wide scale for ZNF143, THAP11, and E2F1 in reference to HCF-1 binding, we analyzed the distribution of HCF-1 ChIP-seq peaks relative to the three transcription factors. For co-localized peaks (no more than 500 bases apart), E2F1 is found in the vicinity of more HCF-1 peaks (2019) than either ZNF143 (1300) or THAP11 (471, Fig.S3A). However, at the genomic loci where these proteins are co-localized, HCF-1 appears to bind very closely to THAP11 and ZNF143 binding sites (90% of peaks within ~100 bases), but has a much broader distribution relative to E2F1 (90% of peaks within ~350 bases, Fig.5A). Since E2F1 and THAP11/ZNF143/HCF-1 data were generated by independent laboratories, we wondered whether the difference in the HCF-1 relative distribution could be attributed to variations between experiments. To this end, we performed the same analysis on an additional ZNF143 dataset published by ENCODE (Fig.S3B). While we observed an increased range of HCF-1 peak distances to ENCODE ZNF143 (90% of peaks within 200 bases), this distribution was still significantly narrower than that of ENCODE E2F1 peaks, suggesting that the observed difference in relative HCF-1 distribution is unlikely to be an experimental artifact.
Figure 5. THAP11 and ZNF143, but not E2F1, genome-wide chromatin occupancy correlates with HCF-1 binding.

(A) For E2F1, THAP11, or ZNF143 ChIP-seq peaks within 500 bases of HCF-1 peaks, the absolute distances between peak centers are plotted as box-and-whisker plots. Whiskers indicate bottom 10% and top 90% of the population. (B, C, D) At co-localized regions, ChIP-seq tags for each protein were counted in the 1000bp region surrounding the HCF-1 binding sites. The total number of peak pairs (n) and Pearson product-moment correlation coefficient (r) are indicated. See also Figure S3.
Additionally, we analyzed the relationship of ChIP-seq tag densities (corresponding to amount of chromatin occupancy) between HCF-1 and the other proteins. Within a chromatin-bound complex, it can be expected that the chromatin occupancy of one protein should correlate with the occupancy of proteins in the same complex. We have observed this phenomenon for THAP11, ZNF143, and HCF-1 but not E2F1 (Fig.1A, Fig.2B, E, H and Fig.4A). Similarly, when ChIP-seq tag densities are compared between these four proteins on a genome-wide scale, we observe a much better pair-wise correlation between HCF-1 and THAP11 (Fig.5B) or ZNF143 (Fig.5C) than we do between HCF-1 and E2F1 (Fig.5D). Relatively good correlation (r = 0.67) is also observed between HCF-1 and ZNF143 peaks derived from the ENCODE dataset (Fig.S3C).
We considered the possibility that the dominant role of THAP11/ZNF143 in HCF-1 recruitment may obscure the effect that E2F1 has on HCF-1 occupancy at some of these loci. This could potentially explain the poor correlation observed for E2F1 and HCF-1 chromatin occupancy when THAP11/ZNF143 co-occupied loci are included in the analysis. To overcome this possible limitation, we focused on loci where E2F1 ChIP-seq peaks were directly overlapping with HCF-1 (by at least 50% of the peak width) and excluded E2F1-HCF-1 loci co-occupied by THAP11 or ZNF143 (Fig.S3D). Such filtering should increase the likelihood of identifying E2F1-dependent HCF-1 recruitment to chromatin. Nevertheless, this restrictive subset of E2F1 peaks still showed no increase in correlation with chromatin occupancy of HCF-1. The genome-wide data analysis strongly supports our gene-specific observations (Fig.4G, H) and suggests that THAP11 and ZNF143 but not E2F1 play a dominant role in HCF-1 recruitment to chromatin.
HCF-1 recruitment at several of the genes identified here has been shown to occur in a cell cycle-dependent manner (Tyagi et al., 2007). To determine if THAP11 and ZNF143 contribute to the cell cycle-dependent recruitment of HCF-1 previously attributed to E2F proteins, we examined the occupancy of HCF-1, THAP11, ZNF143, E2F1, and E2F4 by ChIP at RBL1, CDC25A, CDC6, and MCM3 promoters in synchronized HeLa cells. HeLa cells were enriched at prometaphase by sequential thymidine and nocodazole treatments. Following release from nocodazole block, cells were collected every one to two hours. Progression through mitosis into G1 and S-phases was monitored by immunoblotting for histone H3 serine 10 phosphorylation (H3S10P) and cyclin E1 (Fig.6A). THAP11, ZNF143, and HCF-1 associate with E2F target promoters in early G1 (at approximately 2 hours after nocodazole release) and coincided with maximal E2F4 recruitment (Fig.6B). Since E2F4 knockdown in asynchronously growing HeLa cells failed to reduce HCF-1 occupancy at these promoters (Fig.S2A), we suggest that recruitment of HCF-1 in early G1 is likely to be mediated by THAP11 and ZNF143. Additionally, while E2F4 binding steadily decreases as cells progress through G1, THAP11, ZNF143, and HCF-1 remained promoter bound with maximum occupancy achieved prior to peak E2F1 binding (Fig.6B). The remarkable similarity in the cell cycle-dependent binding profiles of THAP11, HCF-1, and ZNF143, in conjunction with our ChIP data in knockdown cells, strongly suggests that THAP11 and ZNF143 are the primary determinants of cell cycle-dependent HCF-1 recruitment at these promoters.
Figure 6. HCF-1 cell cycle-dependent promoter recruitment correlates with THAP11 and ZNF143 occupancy.
(A) Immunoblots from thymidine-nocodazole synchronized HeLa cells upon nocodazole release. (B) ChIP assays from synchronized HeLa cells in panel A. ChIP signal is expressed as a percent of the maximum signal observed throughout the time course. Values represent mean ± standard deviation of duplicate PCR reactions from a single experiment performed at least three times with similar results.
THAP11/ZNF143/HCF-1 Complex Regulates Expression of Cell Cycle Genes and is Necessary for Cell Proliferation
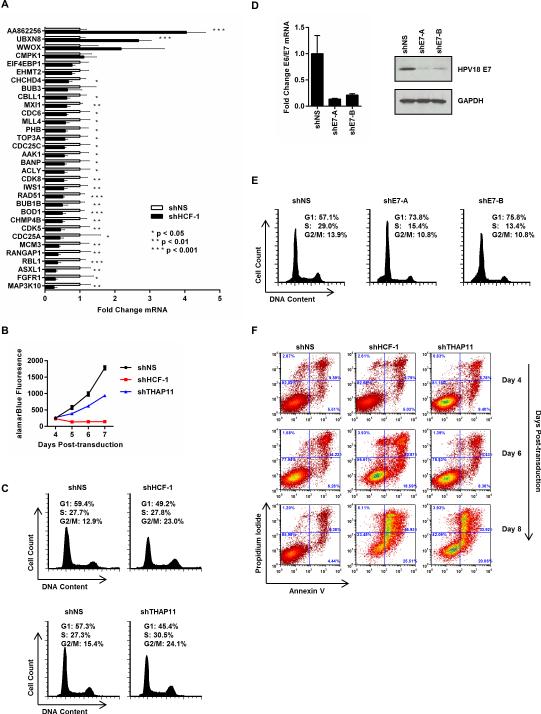
To determine if the promoters occupied by THAP11, ZNF143, and HCF-1 are also regulated by this complex, we examined the mRNA expression of THAP11/ZNF143/HCF-1 target genes in HCF-1 knockdown HeLa cells. HCF-1 knockdown was used to deplete the THAP11/ZNF143/HCF-1 complex from target gene promoters because HCF-1 knockdown results in a greater decrease in complex occupancy than knockdown of either THAP11 or ZNF143 alone (Fig.2). HeLa cells expressing HCF-1 shRNA were collected 4 days post-transduction and used to determine the mRNA expression of 32 genes that showed the greatest change in complex occupancy following HCF-1 knockdown (Fig.2G). As shown in Figure 7A, HCF-1 knockdown resulted in a significant change in the expression of 27 out of the 32 genes. Notable among the mRNAs downregulated by HCF-1 knockdown are key genes involved in cell proliferation (MAP3K10, FGFR1, ACLY), G1/S-phase progression (CDC25A, CDC6, MCM3, RBL1), and mitosis (BUB1B, BOD1, RANGAP1). Similar results were obtained when the THAP11/ZNF143/HCF-1 complex was disrupted by simultaneous siRNA-mediated knockdown of THAP11 and ZNF143 (Fig.S4A and S4B), suggesting that these genes are indeed functional targets of a THAP11/ZNF143/HCF-1 complex.
Figure 7. THAP11/ZNF143/HCF-1-dependent gene expression contributes to cell proliferation and cell cycle progression.
(A) Quantitative RT-PCR-determined mRNA expression changes at THAP11/HCF-1/ZNF143 co-occupied E2F-bound genes in HeLa cells expressing either control or HCF-1 shRNA. Values represent the mean ± standard deviation of four independent experiments. Student's T-test p-values are indicated. (B) HeLa cells expressing the indicated shRNA were plated in 96-well plates (5000 cells/well) four days post-transduction and viable cells detected using alamarBlue every 24 hours for 4 days. Values represent the mean ± standard deviation from two independent experiments. (C) Cell cycle analysis of HeLa cells collected four days post-transduction with HCF-1, THAP11 or control (shNS) shRNA. (D, left panel) Quantitative RT-PCR determined HPV18 E6/E7 transcript levels in HeLa cells collected four days post-transduction with control or two independent HPV18 E7 shRNAs. Values represent the mean ± standard deviation of two independent experiments. (D, right panel) E7 immunoblot in HeLa cells expressing the indicated shRNA. (E) Cell cycle analysis of E6/E7 knockdown HeLa cells in panel D. (F) HeLa cells expressing the indicated shRNA were collected at 4, 6, and, 8 days post-transduction and analyzed for Annexin V and propidium iodide staining by flow cytometry. See also Figure S4.
Consistent with HCF-1 dependent expression of cell proliferation and cell cycle progression genes, we found that HCF-1 depletion resulted in an almost complete reduction in cell proliferation (Fig.7B). Similar to our previous results in colon cancer cells (Parker et al., 2012), we found that THAP11 knockdown also decreases HeLa cell proliferation, albeit less than HCF-1 knockdown (Fig.7B). Cell cycle analysis revealed that HCF-1 knockdown results in an almost two-fold increase in G2/M-phase HeLa cells (Fig.7C, top panels), which is consistent with previous reports suggesting a role for HCF-1 during cytokinesis (Julien and Herr, 2003, 2004; Reilly et al., 2002). Importantly, we found that THAP11 knockdown recapitulates the G2/M cell cycle defect observed in HCF-1 knockdown HeLa cells (Fig.7C, bottom panels) suggesting this phenotype may arise from perturbation of the THAP11/ZNF143/HCF-1 complex. Surprisingly, no accumulation of G1-phase HeLa cells was observed after HCF-1 knockdown despite previous reports indicating otherwise (Julien and Herr, 2003, 2004; Tyagi et al., 2007). However, HCF-1 knockdown did result in significant increases in G1-phase cells in both T98G glioblastoma and SW620 colon carcinoma cells (Fig.S4C), suggesting that the cell cycle defects resulting from HCF-1 depletion are context dependent. The lack of G1/S arrest observed in HCF-1 knockdown HeLa cells may stem from the expression of human papillomavirus (HPV) E6 and E7 oncoproteins in these HPV18-positive cells. High-risk HPV E6 targets p53 for proteasome mediated degradation while E7 binds to and inactivates hypophosphorylated RB1 (Dyson et al., 1992; Dyson et al., 1989; Munger et al., 1989; Scheffner et al., 1990). The E7 mediated disruption of repressive RB1/E2F complexes results in aberrant E2F target gene expression including genes critical for G1/S-phase progression (reviewed in Moody and Laimins, 2010). Consistent with numerous previous studies demonstrating a role for E6 and E7 expression in HeLa cell proliferation (DeFilippis et al., 2003; Goodwin and DiMaio, 2000; Gu et al., 2006; Hall and Alexander, 2003; Johung et al., 2007; Psyrri et al., 2004; Tang et al., 2006), we find that knockdown of HPV18 E6/E7 mRNA (Fig. 7D) alone is sufficient to result in a significant G1/S arrest (Fig. 7E). This result suggests that role THAP11/ZNF143/HCF-1 in HeLa cell G1/S progression is likely masked by the expression of HPV18 E6 and E7 in these cells.
Because cell death is a frequent consequence of mitotic delay (Neumann et al., 2010), we next measured apoptosis in HeLa cells undergoing prolonged HCF-1 or THAP11 knockdown. At four days post-transduction, HCF-1 and THAP11 shRNA-expressing HeLa cells showed no change in viability: Annexin V positive, propidium iodide (PI) positive, or Annexin V/PI double-positive cells were comparable in HCF-1, THAP11 and control knockdown cells (Fig.7F, top panels). HCF-1 and THAP11 knockdown HeLa cells at six days post-transduction had a significant increase in both Annexin V positive and Annexin V/PI double-positive cells (Fig.7F, middle panels). Two days later, only 23% of HCF-1 knockdown cells and 42% of THAP11 knockdown cells were viable (Annexin V and PI negative) compared to 86% viability for control knockdown cells (Fig.7F bottom panels). The larger decrease in cell viability observed in HCF-1 versus THAP11 knockdown (Fig.7F) likely explains the proliferation difference observed in Figure 7B. Collectively, this data suggests that a transcriptional regulatory complex consisting of HCF-1, THAP11, and ZNF143 contributes to the expression of genes critical for cell proliferation and cell cycle progression.
Discussion
HCF-1 is a well-studied transcriptional co-regulator with important functions in cell proliferation and cell cycle progression. In this work, we demonstrate that a complex comprised of HCF-1 and zinc-finger transcription factors THAP11 and ZNF143 is the predominant mechanism for HCF-1 recruitment at many E2F-regulated promoters. Furthermore, we demonstrate that THAP11/ZNF143/HCF-1 complex-dependent expression of cell cycle control genes is necessary for cell proliferation, cell cycle progression, and maintenance of cell viability. This work significantly alters the current model of HCF-1-directed transcription and also suggests that the THAP11/ZNF143/HCF-1 complex is a novel component of the transcriptional regulatory network controlling cell proliferation.
The discovery that HCF-1 is recruited to E2F-target genes offers an appealing explanation for its role in cell cycle regulation. The current model of HCF-1 recruitment at these promoters is based on a previous investigation (Tyagi et al., 2007) that suggests HCF-1 is a direct cofactor for E2F proteins; evidenced by the finding that HCF-1 physically interacts with E2Fs and occupies the promoters of several E2F-regulated cell cycle control genes. We now make several key observations that alter this model of E2F-dependent HCF-1 recruitment and suggest a previously unrecognized role for THAP11 and ZNF143 in HCF-1 recruitment to, and regulation of, E2F-target genes. First, we find that the amount of HCF-1 bound at E2F1-occupied promoters correlates with THAP11 and ZNF143 rather than E2F1. Additionally, both the cell cycle-dependent recruitment and spatial distribution of promoter bound HCF-1 are highly coincident with THAP11 and ZNF143 but not E2F1 occupancy, suggesting that THAP11 and ZNF143 are likely the key determinants of HCF-1 recruitment to these loci. Strongly supporting this hypothesis is the finding that knockdown of either THAP11 or ZNF143 is sufficient to significantly reduce HCF-1 recruitment at E2F-bound promoters. The inability to release HCF-1 from these promoters following E2F1, E2F4, or DP1 knockdown is a key feature of this work and, to our knowledge, is the first attempt to directly ascertain the role of E2F proteins in HCF-1 recruitment at cell cycle control genes. While these findings suggest that HCF-1 may not be a direct E2F-recruited co-regulator, multiple studies have nonetheless affirmed that E2F-dependent recruitment of chromatin modifying activities such as CBP/p300, KAT2A/GCN5, KAT2B/PCAF, and KAT5/TIP60 histone acetyltransferase complexes is essential for expression of cell cycle control genes (Brehm et al., 1998; Litovchick et al., 2007; Louie et al., 2004; Magnaghi-Jaulin et al., 1998; Taubert et al., 2004). Accordingly, we suggest that distinct THAP11/ZNF143/HCF-1 and E2F-recruited transcriptional regulatory complexes may cooperate to control the expression of genes necessary for cell proliferation.
The importance of THAP11 in HCF-1 recruitment to E2F-regulated promoters is further strengthened by the similarity of cell physiological response to the knockdown of these factors. Under both THAP11 and HCF-1 knockdown conditions, we observe a substantial G2/M arrest in HeLa cells, which likely leads to the subsequent apoptosis observed in both cases. Consistent with these results, HCF-1 depletion has been shown to result in upregulated SETD8/KMT5A/ PR-Set7 leading to aberrant mitotic histone H4K20 monomethylation and corresponding defects in mitosis and cytokinesis (Julien and Herr, 2004). In addition to this mechanism, the data reported here suggest that HCF-1 may also contribute to G2/M-phase progression by directly regulating genes involved in mitotic spindle formation. The THAP11/ZNF143/HCF-1 complex occupies the promoters of mitotic spindle assembly checkpoint genes BUB1B and BUB3 (Lara-Gonzalez et al., 2012), the Ran GTPase activating protein RANGAP1 (Joseph et al., 2004), the cohesion acetyltransferase ESCO2 (Whelan et al., 2012), and BOD1, a recently identified kinetochore protein necessary for chromosome bi-orientation (Porter et al., 2007; Porter et al., 2013). Depletion of HCF-1, or THAP11 and ZNF143, was sufficient to reduce BUB1B, BOD1, and RANGAP1 expression. ESCO2 expression has been previously shown to involve ZNF143 (Nishihara et al., 2010). Unlike previous reports (Julien and Herr, 2003, 2004; Tyagi et al., 2007), we did not observe an increase in G1-phase HeLa cell population upon HCF-1 knockdown. However, based on numerous published studies and our present observation, we speculate that this discrepancy may manifest as the result of RB1 inactivation by HPV18 E7 oncoprotein expressed in these cells, which results in deregulated E2F-target gene expression and G1 to S-phase transition (Moody and Laimins, 2010). Consistent with this possibility, we find that RB-intact T98G glioblastoma and SW620 colon carcinoma cells do arrest in G1 following HCF-1 knockdown. Furthermore, we have previously shown that THAP11 is a cell proliferation factor in SW620 as well as other colon cancer cells (Parker et al., 2012) and HCF-1 depletion results in significant proliferation defects in these cells as well (J.B.P, unpublished observations).
Another novel feature of this work is the observation that THAP11, ZNF143, and HCF-1 binding at promoters containing THAP11/extended ZNF143 binding motifs is mutually dependent on all three factors. The extended ZNF143 binding motif, also referred to as Staf binding sequence 2 (SBS2), differs from the canonical ZNF143 binding motif (SBS1) by the addition of a 5’ ACTACA sub-motif adjacent to the core TCCCA ZNF143 recognition sequence (Myslinski et al., 2006). While shown to be dispensable for ZNF143 binding in vitro, the ACTACA sub-motif is nonetheless functional, since mutation of this sequence significantly blunts ESCO2 promoter-driven luciferase activity (Nishihara et al., 2010). In this context, we speculate that the ACTACA sub-motif may be a sequence specific binding site for THAP11. Structure-function analyses have revealed that THAP proteins bind specific DNA sequences through bipartite recognition of adjacent DNA major and minor grooves (Campagne et al., 2010; Sabogal et al., 2010). However, THAP11 has been suggested to be a relatively poor DNA binding protein due to its uniquely shortened DNA minor groove binding loop 4 region (Campagne et al., 2010; Gervais et al., 2013; Sabogal et al., 2010). Because of this, we hypothesize that HCF-1 may serve as a bridge between THAP11 and chromatin-bound ZNF143, thereby enabling stable association of the THAP11-HCF-1 complex to promoters. Consistent with this hypothesis are our results and the recent observation that HCF-1 physically associates with ZNF143 and occupies extended, rather than canonical, ZNF143 binding sites (Michaud et al., 2013; Ngondo-Mbongo et al., 2013). This model would explain HCF-1- and ZNF143- dependent THAP11 recruitment and, since HCF-1 does not bind DNA directly, also HCF-1's requirement for THAP11 and ZNF143. Less clear, however, is why ZNF143 binding is sensitive to THAP11 and HCF-1 depletion.
Two recent genome-wide investigations have revealed that THAP11, HCF-1, and ZNF143 have at least partially overlapping cistromes (Michaud et al., 2013; Ngondo-Mbongo et al., 2013). When interpreted in the context of our findings, we suggest that these reports may be indicative of a more broadly relevant THAP11/ZNF143/HCF-1 transcriptional regulatory complex. We note, however, that this interpretation contrasts with the model reported by Ngondo-Mbongo et al. (2013) who suggested that THAP11 and ZNF143 occupancy is mutually exclusive. Their conclusion is based in large part on the inability of recombinant THAP11 and ZNF143 DNA-binding domains to simultaneously occupy an extended ZNF143 oligonucleotide probe in electrophoretic mobility shift assays (EMSA). Because this assay utilizes truncated proteins and is devoid of HCF-1, we suggest that this result likely reflects a limitation of EMSA rather than a misinterpretation of our sequential ChIP results. Nonetheless, we anticipate that additional experimentation will be required to clarify these conflicting results.
In summary, we have identified transcription factors THAP11 and ZNF143 as key determinants of HCF-1 recruitment to, and regulation of, E2F target genes. These findings significantly change our understanding of the mechanism of HCF-1-mediated gene expression and cell proliferation. Furthermore, these results suggest that the THAP11/ZNF143/HCF-1 complex is an important component of the transcriptional regulatory network controlling cell cycle progression. Future work will be required to elucidate the mechanism of THAP11/ZNF143/HCF-1 directed gene expression and to determine the interplay between this complex and E2F-associated transcriptional regulatory complexes on cell cycle control genes.
Experimental Procedures
Cell Culture and Cell Synchronization
293T/17, HeLa, SW620 and T98G cells were purchased from ATCC. 293T/17, HeLa, and SW620 cells were maintained in Dulbecco's modified Eagle medium (high glucose) supplemented with 10% fetal bovine serum. T98G were maintained in minimal essential medium supplemented with 10% fetal bovine serum. All cells were grown at 37°C in a humidified cell culture incubator containing 5% CO2.
HeLa cells were enriched at prometaphase using thymidine-nocodazole block. Cells (~30% confluence) were cultured in medium containing 2mM thymidine for 18 hours, after which the thymidine-DMEM medium was removed, and culture dishes were rinsed. Cells were then cultured in DMEM medium containing 100ng/mL nocodazole (Sigma) for 13 hours. HeLa cells were subsequently harvested for the G2/M population (0 hour) or transferred to normal medium and harvested at indicated time points for flow cytometry, immunoblots, or ChIP assays.
RBL1 promoter construct stable transfection
T98G cells containing chromosomally integrated wildtype or E2F binding site mutant RBL1 promoter constructs were created using phiC31 integrase as previously described (Hillman et al., 2012). Briefly, the luciferase reporter plasmid pGL4.21 (Promega) was modified to contain an attB phiC31 attachment sequence, creating pGL4.21-attB. An RBL1 promoter fragment (-287 to +42) or its tandem E2F binding site mutant (Burkhart et al. 2010) were cloned into pGL4.21-attB. T98G cells were co-transfected with a codon-optimized phiC31 expression vector (Raymond et al., 2007) (Addgene plasmid #13795) and either pGL4.21-attB RBL1-WT or pGL4.21-attB RBL1-E2F mutant plasmids using FugeneHD (Promega) transfection reagent. Pools of stably transfected cells were selected with 1μg/ml puromycin. Full details are available in the Supplemental Material.
Retroviral shRNA transduction and siRNA transfection
Non-silencing control, THAP11, and HCF-1 pSuper.Retro.Puro retroviral shRNA expression vectors have been described previously (Parker et al., 2012). ZNF143, E2F1, E2F4, DP1 and human papillomavirus type 18 (HPV18) E7 shRNAs were cloned into pSuper.Retro.Puro using the sequences listed in the Supplemental Material. VSV-G pseudotyped retrovirus was produced and used to spin-infect cells as previously described (Parker et al., 2012). Two days post-transduction, cells were split into media containing 2μg/ml puromycin and selected for at least two days or as indicated.
For knockdown using siRNA, HeLa cells were transfected with 30nM non-silencing control siRNA (Thermo Scientific) or 15nM each of THAP11 and ZNF143 siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Transfected HeLa cells were harvested for RNA and protein isolation after 72 hour incubation. THAP11 and ZNF143 siRNA oligonucleotides corresponding to the shRNA targeting sequences were synthesized by Thermo Scientific.
Immunoblots and Quantitative RT-PCR
Whole cell extracts were prepared from cells expressing shRNA four days post-transduction and immunoblotted as previously described (Parker et al., 2012). Antibodies and working dilutions used for immunoblotting are provided in the Supplemental Material.
Total RNA was prepared from shRNA expressing cells four days post-transduction or from siRNA transfected cells 3 days post-transfection and analyzed by quantitative RT-PCR as previously described (Parker et al., 2012).
Immunoprecipitation, Chromatin Immunoprecipitation (ChIP), Sequential ChIP, and ChIP-Western
Nuclear extract preparation and co-immunoprecipitation were performed as described previously (Parker et al., 2012) with minor modifications. Full details are provided in the Supplemental Material. Immunoprecipitation was performed using the following antibodies: THAP11 (R&D Systems #MAB5727, 2μg), HCF-1 (Bethyl Laboratories #A301-399A, 1μg).
ChIP and sequential ChIP (ChIP-reChIP) assays were performed as described (Parker et al., 2012). Antibodies used for ChIP are as follows: THAP11 (R&D Systems #MAB5727, 2μg), HCF-1 (Bethyl Laboratories #A301-399A, 1μg), ZNF143 (Proteintech Group #16618-1-AP, 2μg), E2F1 (Millipore #05-379, 2μg), E2F3 (Millipore #05-551, 2μg), E2F4 (Santa Cruz Biotechnology #A-20x, 2μg), DP1 (Santa Cruz Biotechnology #sc-610x, 2μg).
ChIP-Western assays were performed identically as conventional ChIP except that following the final RIPA wash (50mM HEPES-KOH pH 7.6, 500mM LiCl, 1mM EDTA, 0.7% Na-Deoxycholate, 1% IGEPAL CA-630), beads were washed twice with 10mM Tris-HCl pH 7.6, 50mM NaCl, 2mM MgCl2, 0.1% Triton X-100 and resuspended in 100μl of the same buffer. Where indicated, 1μl (250U) of Benzonase nuclease (Sigma) was added and samples were incubated for one hour at 25°C in a Thermomixer (Eppendorf) at 900rpm. The beads then washed three times with ice-cold RIPA and once with 10mM Tris pH 7.6, 1mM EDTA, 50mM NaCl. Twenty percent of the bead-bound immunoprecipitated material was reserved for qPCR and the remaining 80% was eluted in 2×Laemelli sample buffer containing 5% β-mercaptoethanol at 100°C for 5min. Eluted protein was separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted as described above.
Flow Cytometry
Cell cycle analysis was determined by flow cytometry using propidium iodide (PI) stained cells as described previously (Loomis et al., 2009), with minor modifications.
Apoptosis was determined by flow cytometry using Annexin V/PI staining reagents (Invitrogen) according to manufacturer's instructions. Full details of flow cytometry procedures are provided in the Supplemental Procedure.
In Silico Analyses
To determine potential THAP11/HCF-1 binding sites co-localized with E2F1, E2F1-bound loci determined by ENCODE consortium (Consortium, 2011) were scanned using MAST software v4.8.1 (Bailey et al., 2009; Bailey and Gribskov, 1998) and a previously determined THAP11 DNA motif (Dejosez et al., 2010).
ChIP-seq data was retrieved from the NCBI Sequence Read Archive and mapped to the human hg19 genome using bowtie (v1.0.0, http://bowtiebio.sourceforge.net). Peaks were called using HOMER (v4.3, http://homer.salk.edu). Co-localized peaks were defined as those with centers no more than 500bp apart. To determine chromatin occupancy correlation, tag densities surrounding co-localized peaks were compared. E2F1-HCF-1 peaks devoid of THAP11/ZNF143 were defined as E2F1 peaks directly overlapping HCF-1 with no THAP11 or ZNF143 peaks called within 500bp. Full details are available in the Supplemental Material.
Statistics
Student's T-test and Pearson product-moment correlation coefficients were calculated using GraphPad Prism version 5.
Supplementary Material
Acknowledgements
We thank Drs. Ira Schulman, Curt Horvath, Hiroaki Kiyokawa, and Sui Huang for suggestions and comments on the manuscript. Flow cytometry work was performed at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University Flow Cytometry Core Facility, which is supported by Cancer Center Support Grant NCI CA060553. This work was supported by NCI grant R01 CA133755 (D.C.). J.B.P. is supported by NCI Institutional NRSA Training Program in Signal Transduction and Cancer grant T32 CA070085.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
J.B.P and D.C. conceived the research plan. J.B.P, H.Y., and D.C. designed experiments and analyzed data. J.B.P, H.Y, and A.V. performed research and A.V performed in silico analyses. J.B.P and D.C. wrote the manuscript with contributions from H.Y. and A.V. All authors reviewed and edited the manuscript.
The authors declare no conflict of interest.
References
- Ajuh PM, Browne GJ, Hawkes NA, Cohen PT, Roberts SG, Lamond AI. Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res. 2000;28:678–686. doi: 10.1093/nar/28.3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Wirt SE, Zmoos AF, Kareta MS, Sage J. Tandem E2F binding sites in the promoter of the p107 cell cycle regulator control p107 expression and its cellular functions. PLoS Genet. 2010;6:e1001003. doi: 10.1371/journal.pgen.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne S, Saurel O, Gervais V, Milon A. Structural determinants of specific DNA-recognition by the THAP zinc finger. Nucleic Acids Res. 2010;38:3466–3476. doi: 10.1093/nar/gkq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis RA, Goodwin EC, Wu L, DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J Virol. 2003;77:1551–1563. doi: 10.1128/JVI.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, Barton MC, Gunaratne PH, Young RA, Zwaka TP. Ronin/HCF-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 2010;24:1479–1484. doi: 10.1101/gad.1935210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Gervais V, Campagne S, Durand J, Muller I, Milon A. NMR studies of a new family of DNA binding proteins: the THAP proteins. J Biomol NMR. 2013;56:3–15. doi: 10.1007/s10858-012-9699-1. [DOI] [PubMed] [Google Scholar]
- Goodwin EC, DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc Natl Acad Sci U S A. 2000;97:12513–12518. doi: 10.1073/pnas.97.23.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- Gu W, Putral L, Hengst K, Minto K, Saunders NA, Leggatt G, McMillan NA. Inhibition of cervical cancer cell growth in vitro and in vivo with lentiviral-vector delivered short hairpin RNA targeting human papillomavirus E6 and E7 oncogenes. Cancer Gene Ther. 2006;13:1023–1032. doi: 10.1038/sj.cgt.7700971. [DOI] [PubMed] [Google Scholar]
- Hall AH, Alexander KA. RNA interference of human papillomavirus type 18 E6 and E7 induces senescence in HeLa cells. J Virol. 2003;77:6066–6069. doi: 10.1128/JVI.77.10.6066-6069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman RT, Calos MP. Site-specific integration with bacteriophage PhiC31 integrase. Cold Spring Harb Protoc. 20122012 doi: 10.1101/pdb.prot069211. [DOI] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–725. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Knez J, Piluso D, Bilan P, Capone JP. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol Cell Biochem. 2006;288:79–90. doi: 10.1007/s11010-006-9122-x. [DOI] [PubMed] [Google Scholar]
- Kong LJ, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Mangone M, Myers MP, Herr W. Role of the HCF-1 basic region in sustaining cell proliferation. PLoS One. 2010;5:e9020. doi: 10.1371/journal.pone.0009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazars R, Gonzalez-de-Peredo A, Cayrol C, Lavigne AC, Vogel JL, Ortega N, Lacroix C, Gautier V, Huet G, Ray A, et al. The thap-zinc finger protein thap1 associates with coactivator HCF-1 and O-GLcNAc transferase: A link between DYT6 and DYT3 dystonias. J Biol Chem. 2010;285:13364–13371. doi: 10.1074/jbc.M109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud J, Praz V, James Faresse N, Jnbaptiste CK, Tyagi S, Schutz F, Herr W. HCFC1 is a common component of active human CpG-island promoters and coincides with ZNF143, THAP11, YY1, and GABP transcription factor occupancy. Genome Res. 2013;23:907–916. doi: 10.1101/gr.150078.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myslinski E, Gerard MA, Krol A, Carbon P. A genome scale location analysis of human Staf/ZNF143-binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J Biol Chem. 2006;281:39953–39962. doi: 10.1074/jbc.M608507200. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngondo-Mbongo RP, Myslinski E, Aster JC, Carbon P. Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11. Nucleic Acids Res. 2013;41:4000–4014. doi: 10.1093/nar/gkt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara M, Yamada M, Nozaki M, Nakahira K, Yanagihara I. Transcriptional regulation of the human establishment of cohesion 1 homolog 2 gene. Biochem Biophys Res Commun. 2010;393:111–117. doi: 10.1016/j.bbrc.2010.01.094. [DOI] [PubMed] [Google Scholar]
- Parker JB, Palchaudhuri S, Yin H, Wei J, Chakravarti D. A Transcriptional Regulatory Role of the THAP11-HCF-1 Complex in Colon Cancer Cell Function. Mol Cell Biol. 2012;32:1654–1670. doi: 10.1128/MCB.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter IM, McClelland SE, Khoudoli GA, Hunter CJ, Andersen JS, McAinsh AD, Blow JJ, Swedlow JR. Bod1, a novel kinetochore protein required for chromosome biorientation. The Journal of Cell Biology. 2007;179:187–197. doi: 10.1083/jcb.200704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter IM, Schleicher K, Porter M, Swedlow JR. Bod1 regulates protein phosphatase 2A at mitotic kinetochores. Nat Commun. 2013;4:2677. doi: 10.1038/ncomms3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psyrri A, DeFilippis RA, Edwards AP, Yates KE, Manuelidis L, DiMaio D. Role of the retinoblastoma pathway in senescence triggered by repression of the human papillomavirus E7 protein in cervical carcinoma cells. Cancer Res. 2004;64:3079–3086. doi: 10.1158/0008-5472.can-03-3739. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly PT, Wysocka J, Herr W. Inactivation of the retinoblastoma protein family can bypass the HCF-1 defect in tsBN67 cell proliferation and cytokinesis. Mol Cell Biol. 2002;22:6767–6778. doi: 10.1128/MCB.22.19.6767-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC. THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat Struct Mol Biol. 2010;17:117–123. doi: 10.1038/nsmb.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Tang S, Tao M, McCoy JP, Jr., Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Tyagi S, Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 2009;28:3185–3195. doi: 10.1038/emboj.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors. Proc Natl Acad Sci U S A. 2006;103:6817–6822. doi: 10.1073/pnas.0602109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JL, Kristie TM. The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection. Viruses. 2013;5:1272–1291. doi: 10.3390/v5051272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan G, Kreidl E, Wutz G, Egner A, Peters JM, Eichele G. Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J. 2012;31:71–82. doi: 10.1038/emboj.2011.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ, 3rd, Drobetsky E, Milot E, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.