SUMMARY
Current models of transcription termination factor recruitment to the RNA polymerase II (Pol II) transcription complex rely exclusively on the direct interaction between the termination factor and phosphorylated isoforms of the Pol II C-terminal domain (CTD). Here we report that the Pol II flap loop is needed for physical interaction of Pol II with the Pcf11/Clp1 subcomplex of Cleavage Factor IA (CF IA), which functions in both 3′ end processing and Pol II termination, and for proper termination of short RNAs in vitro and in vivo. Deletion of the flap loop reduces the in vivo interaction of Pol II with CF IA, but increases the association of Nrd1 during stages of the transcription cycle when the CTD is predominately Ser5-phosphorylated. We propose a model in which the flap loop coordinates a binding equilibrium between the competing termination factors Pcf11 and Nrd1 to Pol II during termination of short RNA synthesis.
INTRODUCTION
Pol II is responsible for transcribing all protein-coding RNAs (mRNAs) as well as some non-coding RNAs, such as small nucleolar RNAs (snoRNAs), in eukaryotes. The largest subunit of Pol II, Rpb1, contains a long, unstructured CTD composed of tandem hepta-peptide repeats. In a manner that is dependent upon the stage of transcription, the CTD is differentially phosphorylated at serines in positions 2, 5 and 7 and tyrosine at position 1 (Mayer et al., 2012; Heidemann, et al., 2013). These differential phosphorylation patterns help recruit and subsequently release specific classes of accessory factors to the Pol II transcription complex in ways that influence transcription initiation, elongation, termination and RNA processing (Hsin and Manley, 2012).
Evidence accumulated over the last decade points to the importance of specific protein-CTD interactions in choosing between two main termination pathways and facilitating proper Pol II termination. These two pathways include the poly(A)-dependent pathway (the Rat1 pathway) and the poly(A)-independent pathway (Sen1/Nrd1 pathway) (Kuehner et al., 2011; Mischo and Proudfoot, 2013). In the poly(A)-dependent pathway, Pcf11, a subunit of CF IA, and Rtt103 bind via a CTD-interacting domain (CID) to Pol II phosphorylated at position 2 of the heptad repeat (Ser2), a CTD marker indicative of the late elongation to termination transition on mRNA genes (Sadowski et al., 2003; Kim et al., 2004; Lunde et al., 2010). These termination factors in turn recruit the 5′-to-3′ exonuclease Rat1 (Luo et al., 2006). Pcf11- and Rtt103-mediated delivery of Rat1 to the site of RNA cleavage at the poly(A) site permits Rat1 to degrade the unprotected transcript and dismantle the transcription complex upon reaching Pol II. In addition to its role in poly(A)-dependent termination, CF IA cooperates with the Sen1 complex to promote termination of small mRNAs and short non-polyadenylated RNAs, such as snoRNAs (Kim et al., 2006; Steinmetz et al., 2006). Sen1, a putative RNA-DNA helicase (Kim et al., 1999; Porrua and Libri, 2013), associates with the RNA-binding termination factors Nrd1 and Nab3 to form the Sen1 complex (Vasiljeva et al., 2008; Nedea et al., 2008). Like Pcf11, Nrd1 contains a CID, but its CID shows greater specificity for Ser5-phosphorylated CTD, a marker of early elongation (Gudipati et al., 2008; Vasiljeva et al., 2008; Kubicek et al., 2012). The joint binding of Nrd1 to Pol II and of Nrd1 and Nab3 to specific RNA sequences may help recruit Sen1 to the transcription complex.
While it is clear that the CTD mediates timely recruitment of enzymatic termination activities to the transcription complex, evidence suggests that termination factor binding alone can influence Pol II release in ways that do not require enzymatic activities (Pearson and Moore, 2013; Zhang et al., 2005). A major unresolved issue is whether direct contact of termination factors with surface-exposed regions of Pol II outside of the CTD help release Pol II from the nucleic acid framework. Such regions may contribute to the larger network of interactions needed for effective and timely recruitment and positioning of termination enzymes, as well as for tuning Pol II to be more susceptible to these activities.
Insight into Pol II surface interactions critical for eukaryotic termination may be gleaned from a growing understanding of transcription termination in bacteria. For example, in E. coli, the transcription factor NusA binds to the β flap loop of RNA polymerase (RNAP), underneath which nascent RNA is extruded (Toulokhonov et al., 2001). Binding of NusA to both the flap domain and the exiting RNA produces an allosteric signal that is communicated directly to the polymerase active site and causes termination. In this report, we show that the evolutionarily conserved flap loop in the Pol II Rpb2 subunit contributes to proper Pol II termination on short genes. We find that the flap loop mediates interaction of CF IA with Pol II, thereby providing the first evidence that structural features of the Pol II body are important for termination factor binding.
RESULTS
The Rpb2 flap loop is conserved
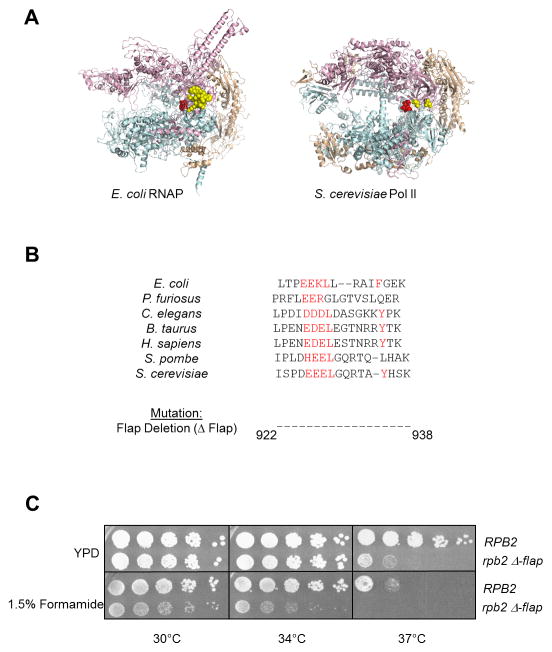
Previous studies have demonstrated that the flap loop of E. coli RNAP is important for regulation by elongation and termination factors (Toulokhonov et al., 2001; Nickels et al., 2005). This feature is conserved in eukaryotes, raising the possibility that it also contributes to proper Pol II termination (Kuehner et al., 2011). The placement of the Rpb2 flap loop in the overall yeast Pol ll quaternary structure is similar to that of E. coli RNAP (Fig. 1A). Because the yeast flap loop is too disordered to visualize in the crystal structure (Cramer et al., 2001), we have marked the first ordered flanking residues in space-filled rendering. These residues are positioned near the RNA exit channel in a similar orientation to that found in E. coli. Alignment of the yeast flap loop sequence to the same region of other species, including E. coli, reveals noticeable conservation, particularly the presence of a cluster of charged residues followed by leucine on one side of the loop and a hydrophobic residue, often tyrosine or phenylalanine, on the other side (Fig. 1B). Thus, the flap loop may serve similar functions across different species through evolutionarily conserved biological mechanisms.
Figure 1. Deletion of the Pol II flap loop in yeast causes phenotypic changes.
(A) Structures of Escherichia coli RNAP (left) and Saccharomyces cerevisiae Pol II (right) showing the β′/Rpb1 subunit in teal, β/Rpb2 subunit in pink, and all other subunits in wheat. The active site residues are shown in red. The E. coli flap loop is highlighted in yellow space-filled rendering, while only the amino-terminal and carboxy-terminal residues of the S. cerevisiae flap loop are shown due to the inherent disordered structure of the loop.
(B) Alignment of the flap loop primary structure across divergent species, with conserved residues highlighted in red. The region corresponding to the S. cerevisiae flap loop (amino acids 922–938 of Rpb2) was deleted to give the rpb2 Δ-flap mutant.
(C) Five-fold serial dilutions of Wt (RPB2) and mutant (rpb2 Δ-flap) Pol II strains are spotted onto YPD +/− 1.5% formamide and incubated at the indicated temperatures for 2–3 days.
To probe the function of the flap loop in yeast, we deleted residues 922–938 from S. cerevisiae Rpb2. Yeast expressing only mutant rpb2 were viable, but temperature-sensitive for growth at 37°C (Fig. 1C). This sensitivity is enhanced in the presence of 1.5% formamide, with slower growth at 30°C as well. Formamide sensitivity correlates with alterations in protein-protein interactions in vivo (Hampsey, 1997), suggesting that interactions with the flap loop may regulate Pol II activities.
The flap deletion reduces transcription termination in vitro
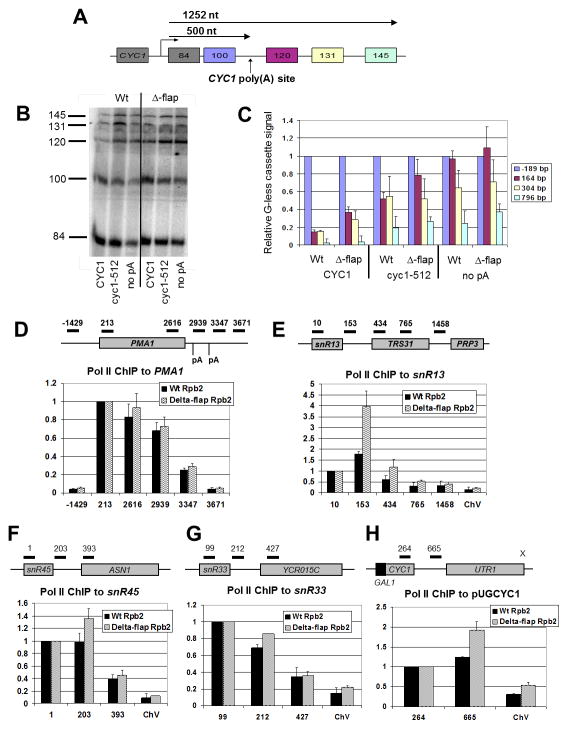
To explore the flap loop’s role in transcription, we performed an in vitro assay that couples mRNA 3′ end processing with transcription and can assess factor-dependent termination downstream of a poly(A) site (Mariconti et al., 2010). Body-labeled RNA was transcribed in Wt and Δ-flap extracts from three DNA templates whose two upstream G-less cassettes are separated from three downstream G-less cassettes by a complete CYC1 poly(A) site, a partial cyc1-512 poly(A) site, or no poly(A) site at all (Fig. 2A). After T1 RNase digestion, the different-sized G-less fragments were separated on a denaturing acrylamide gel (Fig. 2B). After normalizing the radioactivity in each downstream G-less cassette band to that of the cassette just upstream of the poly(A) site, the amount was plotted as a function of cassette positioning (Fig. 2C). With the CYC1 poly(A) site template, Wt Pol II demonstrates a ~15% increase in signal downstream of the poly(A) site, similar to that reported elsewhere (Mariconti et al., 2010). In contrast, Δ-flap Pol II shows 2-fold more signal from the two cassettes immediately downstream of the terminator. This increase in accumulation of RNA downstream of the poly(A) site could be due to a defect in Pol II termination and/or the ability of Rat1 to access and degrade the nascent RNA, which in turn is expected to lead to a termination defect, as has been shown in vivo and in vitro with exonuclease-defective Rat1 (Kim et al., 2004; Pearson and Moore, 2013). On templates without a poly(A) site or with the damaged poly(A) site, both Pol II species exhibit similar transcription through the downstream cassettes. This result shows that deletion of the flap loop does not affect the efficiency of elongation in vitro.
Figure 2. Δ-flap Pol II accumulates in the intergenic region downstream of short genes.
(A) Tandem G-less cassette transcription template. The transcription start site, the position and lengths of the G-less cassettes, the position of the inserted CYC1 poly(A) element, and the distance in nucleotides (nt) to the poly(A) site and the end of the last cassette are indicated.
(B) Radio-labeled G-less cassette transcription fragments synthesized in Wt and Δ-flap extracts are shown resolved on a 6% polyacrylamide/7M urea gel. The three transcription templates contain the complete CYC1 poly(A) signal (CYC1), a mutated poly(A) signal in which the positioning and efficiency elements have been removed (cyc1-512), or no poly(A) elements (no pA). Lengths, in bases, of the G-less cassettes produced upon T1 RNase digestion of transcript are indicated.
(C) Quantification of transcription products in (B). Bars are shaded to correspond to the G-less cassettes shown in (A). G-less cassettes downstream of the CYC1 poly(A) signal are normalized to the 100 nt G-less cassette. Error bars represent the standard deviation from the average values generated from three independent experiments.
(D–H) Relative Pol II occupancy in Wt (black) and Δ-flap (wavy lines) Pol II strains at indicated PMA1, snR13, snR45, snR33 and CYC1 (pUGCYC1) positions, respectively. Schematics of the qPCR primer sets used to measure occupancy are included. The promoter of the convergent UTR1 gene on pUGCYC1 has been inactivated (indicated by an X). Pol II ChIP signals were obtained with the 4H8 antibody, which recognizes both phosphorylated and unphosphorylated Ser residues on the CTD. The signal at position −1429 is taken as background signal at PMA1, and the signal at ChV intergenic region is taken as background signal at the snoRNA and CYC1 genes. Error bars represent the standard error and the standard deviation, respectively, calculated from either two or three independent biological replicates from cells shifted to 37°C for 30 min., with each qPCR experiment performed in duplicate.
Flap loop removal alters Pol II occupancy downstream of short genes, but not a long mRNA gene
To obtain an in vivo snap-shot of Pol II distribution at a classic mRNA locus, we measured Pol II Chromatin Immunoprecipitation (ChIP) signals on PMA1. Wt and mutant Pol II exhibit similar occupancies downstream of the poly(A) sites (Fig. 2D), indicating no termination defect on this mRNA gene. While the in vitro termination reporter contains a poly(A) sequence within 0.5 kb of the promoter (Fig. 2A), the PMA1 poly(A) sequences are roughly 3 kb from the promoter (Fig. 2D). Since termination pathways tend to be chosen based upon the distance of the terminator signal from the transcription start site (Steinmetz et al., 2006; Gudipati et al., 2008), termination on the in vitro reporter (a “short” gene, <1 kb) may involve a pathway different from that used on PMA1 (a “long” gene, >1 kb). To assess the occupancy of Δ-flap Pol II on shorter genes, we measured Pol II ChIP signals on the snoRNA gene snR13 and its associated 3′ sequences (Fig. 2E). Compared to Wt, there was a striking two-fold increase in Δ-flap Pol II occupancy just downstream of the snR13 coding sequence (Fig. 2E, positions +153 and +434). Similarly, Δ-flap Pol II demonstrates greater occupancy than Wt downstream of the snoRNA gene snR45, though to a lesser extent than what is observed on snR13 (Fig. 2F, position +203). At the snR33 locus, the increase in Pol II occupancy in the intergenic region is even less pronounced (Fig. 2G). This difference may be related to the observation that snR13 and snR45 sequences contain more Nrd1-binding sites than snR33 and may be more dependent on Nrd1 for proper termination (Carroll et al., 2004; Kim et al., 2006). Furthermore, PAR-CLIP data have ranked snR13 and snR45 #6 and #10, respectively, in the list of the top 100 Nrd1 cross-linked sites, while snR33 was not on this list (Creamer et al., 2011). This ranking is consistent with our data.
We also examined Pol II occupancy profiles on a GAL-inducible, plasmid-borne copy of CYC1. In a sen1 mutant background, Pol II density shifts downstream of this and of several other “short” mRNA genes (Steinmetz at al., 2006). We also detect an increase in Δ-flap Pol II occupancy in the CYC1 intergenic region (Fig. 2H, position +665), which is consistent with increased transcriptional readthrough or delayed kinetics of termination at the CYC1 p(A) site in the Δ-flap mutant. Taken together, these results suggest that the flap loop contributes to proper in vivo termination at “short” genes.
The flap loop affects recruitment of termination factors to snoRNA genes
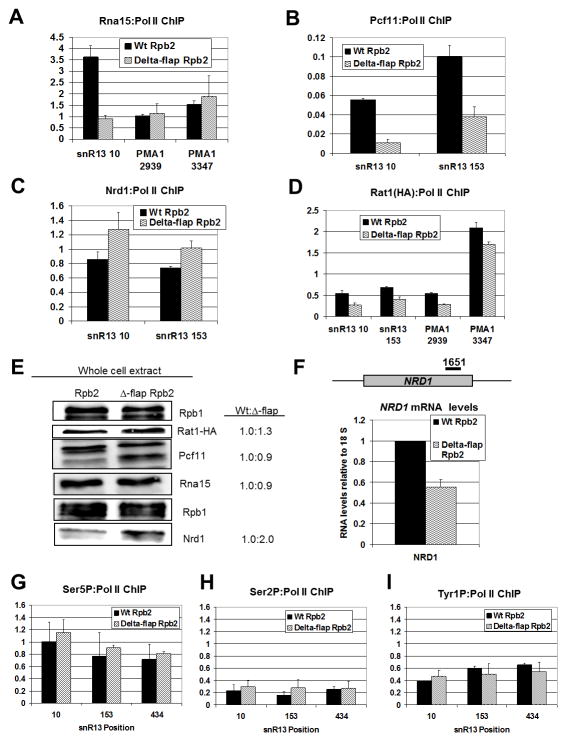
Since Δ-flap Pol II impacts both poly(A)-dependent and poly(A)-independent termination on short transcripts, we examined CF IA recruitment, as CF IA assists in termination at both classes of genes (Kim et al., 2006; Sadowski et al., 2003). We found that co-localization of the CF IA Rna15 subunit with Δ-flap Pol II at the PMA1 3′ UTR is similar to that of Wt Pol II (Fig. 3A). In contrast, Rna15 recruitment to snR13 is markedly reduced in the Δ-flap background (Fig. 3A). In support of this result, recruitment of Pcf11, another CF IA subunit, to Δ-flap Pol II is also strongly reduced (Fig. 3B). These results suggest that the flap loop deletion significantly alters the in vivo recruitment of CF IA to a snoRNA gene and not to a long mRNA gene.
Figure 3. Flap loop removal alters Pol II occupancy and termination factor recruitment on snoRNA genes.
(A–D) ChIP signals of Rna15, Pcf11, Nrd1 and Rat1-HA normalized to Pol II occupancy are displayed, respectively, as a percentage of input for Wt (black) and Δ-flap (wavy lines) strains at the indicated PMA1 and snR13 positions. ChIP for Rna15, Pcf11 and Nrd1 was performed with antibodies against these proteins. An HA antibody was used for Rat1 ChIP. Error bars represent the standard error calculated from two independent biological replicates from cells shifted to 37°C for 30 min., with each qPCR experiment performed in duplicate.
(E) Western blot of Rpb1, Rat1, Pcf11, Nrd1 and Rna15 in Wt and Δ-flap extracts is shown with quantified Wt-to-mutant ratios of normalized protein abundance (right). The relative level of protein in the Wt strain is set to 1.0 and normalization is to Rpb1. Normalization to beta-actin gives similar values (data not shown).
(F) Full-length NRD1 transcript levels in Wt (black) and D-flap (wavy lines) strains as measured by RT-qPCR. The position of the qPCR primer set is shown. All qPCR signals were normalized to the ribosomal 18S qPCR signal, and the relative amount of NRD1 RNA in the Wt strain was set to 1. Error bars represent the standard deviation calculated from three independent biological replicates from cells shifted to 37°C for 30 min., with each qPCR experiment performed in duplicate.
(G–I) ChIP signals of Ser5P, Ser2P and Tyr1P normalized to total Pol II occupancy are displayed, respectively, presented as a percentage of input for Wt (black) and Δ-flap (wavy lines) strains at the indicated snR13 positions. ChIP for Ser5P, Ser2P and Tyr1P was performed with 3E8, H5 and 3D12 antibodies, respectively. Error bars represent the standard error calculated from two independent biological replicates from cells shifted to 37°C for 30 min., with each qPCR experiment performed in duplicate.
Since Nrd1 is recruited to both mRNA and snoRNA genes (Nedea et al., 2003; Steinmetz et al., 2001) and the observation here that the Δ-flap snoRNA termination deficiency seems to correlate with the extent of Nrd1 binding sites, we examined the recruitment of Nrd1 (Fig. 3C). Unlike CF IA (Figs. 3A–B), Nrd1 occupancy at snR13 is somewhat enhanced in the mutant when compared to Wt (Fig. 3C). These results suggest that the flap loop deletion correlates with increased recruitment of Nrd1 to snoRNA genes.
We also investigated recruitment of the Rat1 termination factor and found that it is slightly reduced at the snR13 gene and the 3′ UTR of PMA1 in the Δ-flap strain when compared to the Wt (Fig. 3D). Reduction of Rat1 recruitment in the mutant may be related to the reduced recruitment of Pcf11, as Pcf11 recruits Rat1 to the transcription complex (Luo et al., 2006). Whether this reduction in Rat1 recruitment is related to the increase in Δ-flap Pol II occupancy in the intergenic region of snR13 remains unclear.
We performed Western blots for the above factors in crude whole cell extract and found that altered recruitment of CF IA to mutant Pol II is not due changes in the level of protein, as the amount of Rna15 and Pcf11 in Wt and mutant extracts is similar (Fig. 3E). We sometimes observe Pcf11 species of varying mobilities, but these seem independent of the RPB2 genotype. Interestingly, the mutant extract exhibits twice the level of Nrd1 when compared to Wt extract (Fig. 3E). Increased production of Nrd1 protein suggests that attenuation on the autoregulated NRD1 gene is altered in the mutant strain (Arigo et al., 2006) and is consistent with deficient Nrd1-dependent termination in the Δ-flap mutant (Fig. 2). We were surprised, however, that the level of NRD1 mRNA is reduced by half in the Pol II mutant (Fig. 3F). While previous work has demonstrated a positive correlation between levels of full-length NRD1 mRNA and levels of Nrd1 protein (Arigo et al., 2006), in our experience, protein levels do not always correlate with mRNA levels (Graber et al., 2013). A recent report has also documented read-through of a truncated NRD1 attenuator in a CUP1 reporter by a sen1 mutant without concomitant increase in NRD1 mRNA levels (Chen et al., 2014). It is possible that the observed discrepancy between NRD1 mRNA and Nrd1 protein levels (Figs. 3E and 3F) may point to additional mechanisms that control Nrd1 expression.
The Pol II flap loop does not affect CTD phosphorylation patterns
In light of evidence that CF IA and Nrd1 recruitment to Δ-flap Pol II is altered, we wished to determine whether alterations in CTD modifications were responsible for this effect. We assessed the levels of phosphorylation at Ser5 and Ser2 of the CTD heptad repeat, normalized to total Pol II levels, and found that the phosphorylation of Ser5 and Ser2 (Figs. 3G–H) across the snR13 locus is comparable in both Wt and mutant. Since Tyr1 phosphorylation has been shown to impair termination factor recruitment to Pol II (Mayer et al., 2012), we examined Tyr1 phosphorylation and found that these levels are similar between Wt and mutant Pol II (Fig. 3I). These results suggest that the flap loop does not affect Nrd1 and CF IA interactions by modulating CTD phosphorylation.
CF IA interacts with the Pol II flap loop
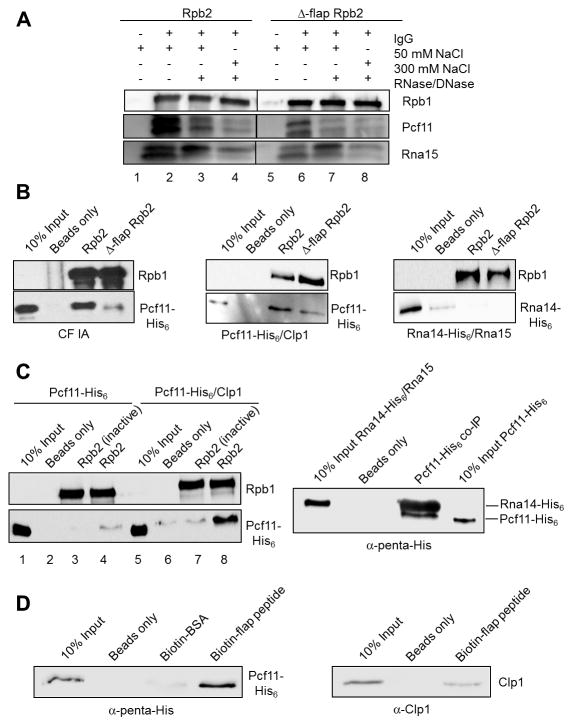
To study the association of CF IA with Pol II (Figs. 3A–B), we immunoprecipitated TAP-tagged Wt and Δ-flap Pol II from extract and found that Δ-flap Pol II pulls down much less endogenous Pcf11 and Rna15 from extract than does Wt Pol II (Fig. 4A, compare lanes 2 and 6). Nuclease treatment, in conjunction with a higher salt wash, reduces the amount of Pcf11 and Rna15 immunoprecipitated with Pol II, suggesting that the association of CF IA subunits with Pol II is stabilized by multi-partite interactions (Fig. 4A, compare lanes 2, 4, 6, and 8). The importance of RNA in mediating Pcf11-Pol II interactions has been underscored previously (Zhang et al., 2005). We asked next if recombinant CF IA, consisting of stable subcomplexes of Pcf11/Clp1 and Rna14/Rna15 (Gordon et al., 2011), associate with Pol II in vitro. We extensively washed Wt and Δ-flap Pol II immunoprecipitated from transcription extract, added CF IA, and found that Δ-flap Pol II pulls down much less CF IA compared to Wt (Fig. 4B, left panels). Similarly, much less Pcf11/Clp1 heterodimer is pulled down with Δ-flap Pol II compared to Wt (Fig. 4B, middle panels). The Rna14/Rna15 heterodimer does not associate with either form of Pol II (Fig. 4B, right panels). Taken together, these results indicate that the interaction between CF IA and the Pol II flap loop is mediated by Pcf11/Clp1 and not Rna14/Rna15.
Figure 4. CF IA makes direct contact with the Pol II flap loop.
(A) Western blot of CF IA subunits Pcf11 and Rna15 co-immunoprecipitated with Wt and Δ-flap Pol II from extract using the TAP tag on Rpb2. As indicated, precipitates were treated with 50 or 300 mM NaCl washes or with an RNase/DNase mixture before elution. The samples were loaded onto the same gel, and the intervening lanes were removed as demarked by the line between the Wt and the mutant fractions.
(B) Western blots of recombinant CF IA (left panels), recombinant Pcf11/Clp1 heterodimer (middle panels) and recombinant Rna14/Rna15 heterodimer (right panels) co-immunoprecipitated with TAP-purified Wt and Δ-flap Pol II and detected with His6 antibody.
(C) Western blots of recombinant Pcf11 and recombinant Pcf11/Clp1 heterodimer co-immunoprecipitated with TAP-purified Wt Pol II (left panels) and of recombinant Pcf11 co-immunoprecipitated with recombinant Rna14/Rna15 heterodimer (right panel). In the experiment shown in the left panels, ”Rbp2 (inactive)” indicates Pol II immunoprecipitated from transcriptionally inactive extract. In the experiment shown in the right panel, a rabbit polyclonal anti-Rna15 antibody was used to immunoprecipitate Rna14/Rna15 pre-incubated with Pcf11 and Rna14 and Pcf11 detected by antibody against the His6 tag on both proteins.
(D) Western blots of recombinant Pcf11/Clp1 heterodimer detected with an anti-penta-His antibody (left panel) and recombinant Clp1 detected with a rabbit polyclonal anti-Clp1 antibody (right panel) co-immunoprecipitated with biotinylated flap loop peptide.
All co-immunoprecipitation experiments were performed two-four times each.
In an attempt to study the direct interaction between Pcf11 and Pol II, we found that Pcf11 alone is inefficiently pulled down with even Wt Pol II (Fig. 4C, left panel, lane 4). To confirm that recombinant Pcf11 is physically functional, we performed co-immunoprecipitation experiments with recombinant Rna14/Rna15 heterodimer. We found that Pcf11 does indeed associate stably with the Rna14/Rna15 heterodimer (Fig. 4C, right panel), indicating that misfolding cannot account for why Pcf11 alone fails to interact stably with Wt Pol II. However, the amount of Pcf11 interacting with Pol II was greatly increased if the Pcf11/Clp1 heterodimer was used in pulldowns (Fig. 4C, left panel, lane 8). These results indicate that Clp1 is required for a stronger interaction of Pcf11 with complete Pol II. We also performed co-immunoprecipitation experiments with Pol II from transcriptionally inactive extracts, made using a step after cell lysis that efficiently releases proteins from chromatin. Pol II from the transcriptionally inactive extracts was not capable of pulling down Pcf11 alone or even the Pcf11/Clp1 heterodimer (Fig. 4C, lanes 3 and 7).
Finally, we explored a possible direct interaction between the Pcf11/Clp1 heterodimer and the flap loop by attempting to co-immunoprecipitate Pcf11/Clp1 with biotinylated flap loop peptide. To our surprise, we found that the flap loop peptide is sufficient to pull down Pcf11/Clp1 (Fig. 4D, left panel). Biotinylated BSA does not pull down Pcf11/Clp1 (Fig. 4D, left panel), confirming the specificity of the flap loop interaction. There is a less efficient interaction when precipitations are done with recombinant Clp1 only (Fig. 4D, right panel), in agreement with the finding from Fig. 4C using the entire Pol II that the strongest association with the flap loop requires a complex of Pcf11 and Clp1. Together, these results support the conclusion that Pcf11/Clp1 interacts directly with the Pol II flap loop.
DISCUSSION
Here we show that the conserved, surface-exposed flap loop of yeast Pol II facilitates proper in vitro and in vivo termination on short genes, but not on a longer gene. The altered termination is not a result of aberrant transcription elongation, and is consistent with the observation that flap loop deletion in human Pol II does not disrupt transcription initiation or elongation (Palangat et al., 2011). We also demonstrate that deletion of the flap loop alters the recruitment of CF IA and Nrd1 to a snoRNA gene in vivo. Additionally, this deletion drastically reduces the physical association of the Pcf11/Clp1 heterodimer with Pol II in vitro. While previous work has shown that the body of Pol II is important in mediating proper termination (Nag et al., 2007), to our knowledge this work is the first demonstration of the requirement of a specific Pol II surface-exposed region outside of the CTD for coordinating and stabilizing the binding of an essential termination factor.
Pcf11 functions in proper termination of both long mRNA gene transcription, characterized by Ser2-phosphorylation marks, and short mRNA/non-coding RNA gene transcription, characterized by Ser5-phosphorylation marks (Sadowski et al., 2003; Kim et al., 2006). The ability to operate in both termination pathways may be due to the apparent flexibility that Pcf11 exhibits in binding various CTD phosphoisoforms, with only slight preference for Ser2P (Lunde et al., 2011; Noble et al., 2005). In this property, Pcf11 is unlike other CID-containing termination factors, such as Nrd1 and Rtt103, which display strong, distinct preferences for specific phospho-patterns. It has been suggested that the CTD-binding by Pcf11 is stabilized by additional features, such as cooperative binding with Rtt103 and other CF IA subunits, some of which are associated with the nascent transcript at the 3′ ends of mRNA genes (Luo et al., 2006; Lunde et al., 2011).
Our experiments demonstrate that CF IA recruitment to Δ-flap Pol II is reduced only on a short gene where Ser5P patterns are dominant and suggest that the flap loop contributes to the recruitment of CF IA and specifically the stable binding of Pcf11 under conditions of heavy Ser5 phosphorylation. As noted above, the absence of the flap loop may not significantly affect Pcf11 binding when Ser2P is dominant because additional protein-protein and protein-RNA interaction may secure the Pcf11-Pol II interaction. This hypothesis is supported by evidence that Δ-flap Pol II cannot efficiently pull-down recombinant Pcf11/Clp1 heterodimer, perhaps because other mitigating protein and RNA binding determinants are not present. Unlike the cooperative recruitment patterns of Rtt103 and Pcf11 to Ser2P-CTD, Nrd1 actively competes with Pcf11 when the CTD is Ser5-phosphorylated (Singh et al., 2009; Honorine et al., 2010). Therefore, in the absence of the flap loop and under conditions of Ser5P, Pcf11 does not have any assists in maintaining interactions with Pol II. Our data showing enhanced Nrd1 recruitment to a snoRNA gene in the Δ-flap strain suggests that the unstable binding of Pcf11 to the CTD allows the competitor Nrd1 to associate freely. Such an unchallenged association between Nrd1 and Δflap Pol II may be enhanced in part by the increase in Nrd1 expression in the Δ-flap strain. It has been proposed that persistent association of Nrd1 with Pol II results in defective or delayed termination events (Lenstra et al., 2013; Singh et al., 2009), which would be consistent with the impaired termination in the Δ-flap strain. It will be interesting in future studies to determine if termination in the Δ-flap mutant is not absent, but simply delayed because Pcf11-mediated reorganizations are less efficient. Additionally, recent work in transcriptional pausing in E. coli provides evidence that the interaction between the RNAP flap-tip and nascent RNA in the exit channel is coordinated energetically with the active site through conformational changes in the RNAP clamp-domain (Hein et al., 2014). Therefore, perturbations in Pol II flap loop interactions may be communicated to the active site, possibly resulting in delayed kinetics of termination.
Previous work has identified five evolutionarily conserved surface-exposed domains on Pol II beyond the CTD that might mediate interactions with transcription factors (Garcia-Lopez and Navarro, 2011). These domains include the jaw, funnel, dock and foot domains of Rpb1 and the wall domain of Rpb2, where the flap loop is located. Of particular note with respect to our work, the CTD is an important binding determinant for the RNA capping enzyme (CE) (McCracken et al., 1997; Cho et al., 1997). However, recent work demonstrated that the foot domain is also critical for the interaction between CE and Pol II (Suh et al., 2010). Suh et al. proposed that factors involved in other Pol II-associated processes may recognize surfaces of Pol II outside of the CTD in ways that fine-tune Pol II activity at precise stages of the transcription cycle.
Interestingly, the CTD can be relocated to other Pol II subunits and still support viability, provided the CTD remains on the same face of Pol II (Suh et al., 2013). This observation suggests that the CTD must be spatially coordinated with other Pol II domains located at defined positions on the surface of Pol II and supports the hypothesis that a dual CTD-Pol II body interface supports termination factor recruitment. Additional evidence for such a model comes from in vitro studies showing that pausing of Pol II just downstream of a mammalian poly(A) site, which is thought to be the first step leading to Pol II release, does not require the CTD but instead, interaction of the Cleavage/Polyadenylation Specificity Factor (CPSF) with an undefined region on the body of Pol II (Nag et al., 2007). Nag et al. proposed that recognition of RNA signal sequences and recruitment of the CstF factor then led to CTD interaction, RNA cleavage, and release of Pol II from the DNA. Other evidence suggests that binding to Pol II may not always be coordinated with the CTD. For example, mutations identified in Rpb3 and Rpb11, located at the back end of Pol II, result in transcriptional read-through on both mRNA and snoRNA genes (Steinmetz et al., 2006). Therefore, additional Pol II regions outside of the potential CTD spatial range could affect termination factor recruitment.
EXPERIMENTAL PROCEDURES
Yeast strains
Construction of Wt Pol II (yBC25), Δ-flap Pol II (yEP6) and Rat-HA tagged (yEP12 and yEP13) strains is described in the Supplemental Experimental Procedures. All strains were grown in YPAD (YPD rich medium supplemented with adenine) at 30°C, unless otherwise indicated.
In vitro transcription reactions
To generate extract for in vitro transcription, yeast (1L) were grown to an OD600 of 2.0–5.0 and processed as detailed in the Supplemental Experimental Procedures. Transcription reactions were performed as described previously (Mariconti et al., 2010), except that 100 μg extract and 0.5 μg plasmid DNA were used.
Chromatin Immunopreciptation (ChIP), quantitative PCR (qPCR) and RT-qPCR
Strains (50 ml) were grown to OD600 0.6–0.8, shifted to 37°C for 30 minutes andfixed with formaldehyde. Sheared chromatin was immunoprecipitated and analyzed by qPCR using standard techniques as detailed in the Supplemental Experimental Procedures. Total RNA was extracted from mid-log cells shifted to 37°C for 30 minutes using the standard hot phenol method. cDNA was synthesized and analyzed by qPCR using standard techniques as detailed in the Supplemental Experimental Procedures. Sequences of ChIP-qPCR and RT-qPCR primers are listed in Table S1 in the Supplemental Experimental Data section. PMA1 primer sets have been published elsewhere (Johnson et al., 2009).
In vitro co-immunoprecipitation experiments
Pol II was immunoprecipitated by incubation of 500 μg transcription extract with IgG-coupled sepharose beads (GE). Purified recombinant CF IA, Pcf11/Clp1, Pcf11, or Rna14/Rna15 (1–3 μg) were incubated with washed, immunoprecipitated Pol II. CF IA, Pcf11/Clp1 and Rna14/Rna15 were kind gifts of the Bohm laboratory (Gordon et al., 2011), and Pcf11 was purified via nickel affinity chromatography. Immobilized biotinylated flap peptide (the sequence specified in Fig. 1B with the addition of N- and C-terminal flanking residues: Rpb2 amino acids 915–943) was incubated with recombinant Pcf11/Clp1, Pcf11, or Clp1. For the pull-downs in crude extract, 50 ml cultures were grown to an OD600 of 0.6–0.8, shifted to 37°C for 30 minutes, and extracts prepared as detailed in the Supplemental Experimental Procedures. Extract (1.5 mg) was incubated with IgG-coupled sepharose beads. Additional details can be found in the Supplemental Experimental Procedures.
Supplementary Material
ChIP-qPCR and RT-qPCR primer sequences
Acknowledgments
We are grateful to Domenico Libri, Odil Porrua, Stephen Buratowski, Steven Hanes, David Brow, Craig Kaplan, Patrick Cramer and members of the Bohm laboratory for helpful discussion and to Mike Hampsey, Bernhard Dichtl, Nick Proudfoot and Benoit Coulombe for sharing reagents. This work was supported by NIH grant GM68887 and NSF grant MCB-1244043 to C. Moore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arigo J, Carroll K, Ames J, Corden J. Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Carroll K, Pradhan D, Granek J, Clarke N, Corden J. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol. 2004;24:6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Muller U, Sundling KE, Brow D. Saccharomyces cerevisiae Sen1 as a model for the study of mutations in human Senataxin that elicit cerebellar ataxia. Genetics. 2014 doi: 10.1534/genetics.114.167585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Takagi T, Moore C, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science. 2001;292:1863–1875. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Creamer T, Darby M, Jamonnak N, Schaughency P, Hao H, Wheelan S, Corden JL. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 2011;7:e1002329. doi: 10.1371/journal.pgen.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez M, Navarro F. RNA polymerase II conserved protein domains as platforms for protein-protein interactions. Transcription. 2011;2:193–197. doi: 10.4161/trns.2.4.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Shikov S, Kuehner J, Liriano M, Lee E, Stafford W, Poulsen M, Harrison C, Moore C, Bohm A. Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry. 2011;50:10203–10214. doi: 10.1021/bi200964p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber J, Nazeer F, Yeh P, Kuehner J, Borikar S, Hoskinson D, Moore C. DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Res. 2013;23:1690–1703. doi: 10.1101/gr.144964.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati R, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Hein P, Kolb K, Windgassen T, Bellecourt M, Darst S, Mooney R, Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorine R, Mosrin-Huaman C, Hervouet-Coste N, Libri D, Rahmouni Nuclear mRNA quality control in yeast is mediated by Nrd1 co-transcriptional recruitment, as revealed by the targeting of Rho-induced aberrant transcripts. Nucleic Acids Res. 2010;39:2809–2820. doi: 10.1093/nar/gkq1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J, Manley J. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Cubberley G, Bentley D. Cotranscriptional Recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Choe J, Seo Y. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan N, Vasiljeva L, Rando OJ, Nedea E, Greenblatt J, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, Stefl R. Serine phosphorylation and proline isomerization in RNAPII CTD control recruitment of Nrd1. Genes Dev. 2012;26:1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner J, Pearson E, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Tudek A, Clauder S, Xu Z, Pachis S, van Leenen D, Kemmeren P, Steinmetz L, Libri D, Holstege F. The Role of Ctk1 kinase in termination of small non-coding RNAs. PLoS One. 2013;8:e80495. doi: 10.1371/journal.pone.0080495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde B, Reichow S, Kim M, Suh H, Leeper T, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson A, Bentley D. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Loll B, Schlinkmann K, Wengi A, Meinhart A, Dichtl B. Coupled RNA polymerase II transcription and 3′ end formation in yeast whole-cell extracts. RNA. 2010;16:2205–2217. doi: 10.1261/rna.2172510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley D. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo H, Proudfoot N. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochem et Biophys Acta. 2013;1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Narsinh K, Martinson H. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat Struct Mol Biol. 2007;14:662–669. doi: 10.1038/nsmb1253. [DOI] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- Nedea E, Nalbant D, Xia D, Theoharis NT, Suter B, Richardson CJ, Tatchell K, Kislinger T, Greenblatt J, Nagy P. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA gene. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Nickels B, Garrity S, Mekler, Minakhin L, Severinov K, Ebright R, Hochschild A. The interaction between sigma70 and the beta-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc Natl Acad Sci USA. 2005;102:4488–4493. doi: 10.1073/pnas.0409850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C, Hollingworth D, Martin S, Ennis-Adeniran V, Smerdon S, Kelly G, Taylor I, Ramos A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat Struct Mol Biol. 2005;12:144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- Palangat M, Grass J, Langelier M, Coulombe B, Landick R. The RPB2 flap loop of human RNA polymerase II is dispensable for transcription initiation and elongation. Mol Cell Biol. 2011;31:3312–3325. doi: 10.1128/MCB.05318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson E, Moore C. Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. J Biol Chem. 2013;288:19750–19759. doi: 10.1074/jbc.M112.434985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol. 2013;20:884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W. Independent function of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Ma Z, Gemmill T, Wu X, DeFiglio H, Rossettini A, Rabeler C, Beane O, Morse R, Palumbo M, Hanes S. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol Cell. 2009;36:255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E, Conrad N, Brow D, Corden J. RNA-binding protein Nrd1 directs poly(A)-independent 3′–end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz E, Ng S, Cloute J, Brow D. cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol. 2006;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E, Warren C, Kuehner J, Panbehi B, Ansari A, Brow D. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suh M, Meyer P, Gu M, Ye P, Zhang M, Kaplan C, Lima C, Fu J. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J Biol Chem. 2010;285:34027–34038. doi: 10.1074/jbc.M110.145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Hazelbaker DZ, Soares L, Buratowski S. The C-terminal domain of Rpb1 functions on other RNA polymerase II subunits. Mol Cell. 2013;51:850–858. doi: 10.1016/j.molcel.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour D. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ChIP-qPCR and RT-qPCR primer sequences