Abstract
Carbapenems are the last-resort antibiotics for the treatment of infections caused by multidrug-resistant Gram-negative bacilli. Klebsiella pneumoniae carbapenemase (KPC) hydrolyses β-lactam antibiotics including the carbapenems. KPCs have been detected in Enterobacteriaceae and Pseudomonas aeruginosa isolates worldwide associated with transposon Tn4401 commonly located in plasmids. Acinetobacter baumannii has become an important multidrug-resistant nosocomial pathogen capable of hydrolysing the carbapenem antibiotics. KPC-producing A. baumannii has so far only been reported in Puerto Rico. During a surveillance study, four KPC-producing A. baumannii with identical pulse type were identified in a single institution. The objectives of this study were to characterize the KPC genetic background and the allelic diversity of one of the isolates. Next-generation sequencing and multilocus sequence typing (MLST) were performed. Molecular characterization of the isolate demonstrated blaKPC in Tn4401b located in the bacterial chromosome within a 26.5 kb DNA fragment, which included a KQ-like element (18.9 kb) very similar to that described previously in a K. pneumoniae plasmid and a 7.6 kb DNA fragment with 98 % homology to a putative plasmid from Yersinia pestis strain PY-95. Our data suggested that the 26.5 kb DNA fragment harbouring blaKPC was integrated in the chromosome by a transposition event mediated by the transposase of ISEcp1 found in the KQ-like element. MLST showed a novel sequence type, ST250. To our knowledge, this is the first report of the identification of the genetic background of blaKPC in A. baumannii.
Introduction
Carbapenems are the antibiotics of choice for the treatment of serious infections caused by multidrug-resistant Gram-negative bacilli. Klebsiella pneumoniae carbapenemase (KPC) renders bacteria resistant to the β-lactam antibiotics, including the carbapenems. Most of these bacteria have acquired additional mechanism of resistance to other groups of antibiotics, leaving few or no available therapeutic options (Nordmann et al., 2009). KPCs have been detected in Enterobacteriaceae and Pseudomonas aeruginosa isolates worldwide (Nordmann et al., 2009). KPC global dissemination has been attributed to several factors, such as the propagation of epidemic strains like K. pneumoniae ST258, the difficulty in detecting KPC-producing organisms by susceptibility testing methods and its location in plasmids of different sizes harbouring transposon Tn4401 (Arnold et al., 2011; Cuzon et al., 2010; Mavroidi et al., 2012; Naas et al., 2008, 2012; Nordmann et al., 2009). In addition to plasmids, a chromosomal location of the KPC gene has been described in P. aeruginosa strains (Correa et al., 2012; Cuzon et al., 2011; Ge et al., 2011; Villegas et al., 2007). The structure of Tn4401 seems to be conserved except in the region located upstream of the blaKPC genes, which generate six different isoforms (a–f) (Bryant et al., 2013). Tn4401 isoform b is the most frequent structure encountered in the United States and shows a complete structure without deletions.
In 2009, our laboratory reported, for the first time, the presence of the KPC gene in Acinetobacter baumannii and to our knowledge KPC-producing A. baumannii has been reported only in Puerto Rico (Robledo et al., 2010). A. baumannii is an important pathogen associated with hospital outbreaks and life-threatening infections in vulnerable individuals (Adams et al., 2008). These infections are associated with high morbidity and mortality, and their treatment is hampered by the emergence of antibiotic-resistant isolates (Adams et al., 2008). The global epidemiology of multidrug-resistant A. baumannii was reviewed by Zarrilli et al. (2013), and they reported that it had a clonal population structure dominated by three international clonal lineages (I–III) and a few additional lineages that spread in single institutions and/or worldwide. The existence of KPC-producing Enterobacteriaceae and P. aeruginosa in different countries emphasizes the possibility of broader future dissemination of KPC-producing A. baumannii.
The objectives of this study were to characterize the localization and genetic background of the KPC gene in A. baumannii strain M3AC9-7 and to define its allelic diversity.
Methods
Bacterial strain.
During a 6-month surveillance study in 2009, four multidrug-resistant KPC-producing A. baumannii strains, M3AC9-2, M3AC9-4, M3AC9-6 and MC3AC9-7, were identified in a single institution during a 30-day period (Robledo et al., 2010). These strains were isolated from four patients, three of them hospitalized in the intensive care unit with a diagnosis of ventilator-associated pneumonia and/or sepsis. In the fourth patient, the isolate was identified in a urine culture. These isolates were positive by PCR for KPC and OXA-51-like genes, but negative for other β-lactamase genes such as OXA-58, OXA-23, IMP, VIM, NDM-1, TEM, SHV and CTX-M. The KPC-3 gene variant was identified by DNA sequencing. The four isolates had identical antimicrobial susceptibility profiles. PFGE (data not shown) demonstrated identical pulse types in the four isolates, indicating a single clone. Therefore, the M3AC9-7 strain was selected for further characterization of the blaKPC genetic background and multilocus sequence typing (MLST).
Next-generation sequencing.
Whole-genome sequencing using an Illumina Miseq2×250 bp paired-end configuration and de novo assembly were performed commercially by Genewiz. ORFs were predicted using Glimmer 3.02 gene calling and automatic annotation was done by the NMPDR rast 4.0 server (http://www.nmpdr.org/FIG/wiki/view.cgi/Main/RAST; Overbeek et al., 2014). The predicted amino acid sequences were confirmed utilizing the online blast program (www.ncbi.nlm.nih.gov).
MLST.
MLST was performed using the Institut Pasteur MLST protocols and database (http://www.pasteur.fr/mlst). The amplicons obtained were sequenced commercially by GeneWiz, and the sequence alignments and analyses were performed utilizing the online blast program (www.ncbi.nlm.nih.gov). The eBURST v3 algorithm (eburst.mlst.net) was used to assess the genetic relationship of the sequence type obtained with those in the database with the most stringent definition of the groups by sharing alleles at six of seven loci.
Results and Discussion
Localization and genetic background of the KPC gene
Next-generation sequencing of the genome of the KPC-producing A. baumannii M3AC9-7 was performed to characterize the KPC genetic background. De novo assembly was conducted and the total length of the assembled genome was 4.06 Mbp with 3898 ORFs. The KPC gene was found inserted in the chromosomal DNA of A. baumannii, specifically in contig 36 (97 550 bp), which consists of 84 ORFs distributed as 70 proteins with known function and 14 hypothetical proteins. blaKPC-3 was identified in transposon Tn4401 isoform b (Figs 1 and 2). blaKPC-3 harbouring Tn4401b was found within a region very similar to the KQ (kpc and qnr) element identified in the 80 kb K. pneumoniae conjugative plasmid pLRM24, described by Rice et al. (2008). This previously described KQ element consists of the transposon Tn1331 backbone (GenBank accession number M55547.1) interrupted by Tn4401 at the transposase A (tnpA) gene and Tn5387 (GenBank accession number EU624315) inserted at the OXA-9 β-lactamase gene. Additional proteins in the KQ element include a resolvase (tnpR), two aminoglycoside resistance genes (aac(6′)-Ib and aadA1) and the β-lactamase TEM-1 gene (Sarno et al., 2002) from the Tn1331 backbone, and the insertion sequence ISEcp1 and the quinolone resistance gene qnrB19 from Tn5387 (Rice et al., 2008).
Fig. 1.
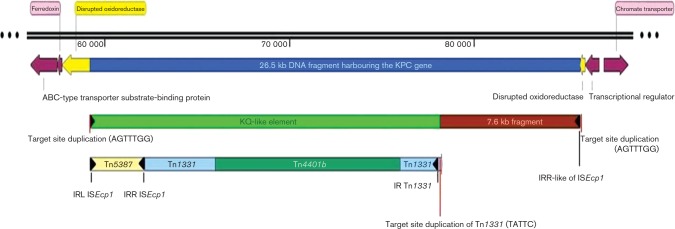
The 26.5 kb fragment harbouring the KPC gene in A. baumannii. The 26.5 kb DNA fragment (blue), which includes the 18.9 kb KQ-like element (bright green) and a 7.6 kb DNA fragment (brown) from Y. pestis PY-95 was inserted in the chromosome of A. baumannii. The KQ-like element consists of the truncated transposon Tn1331 backbone (light blue) interrupted by Tn5387 (light yellow) and Tn4401 (sea green). This 26.5 kb DNA fragment, flanked by ISEcp1 IRL and IRR-like sequences (black triangles), was inserted into an oxidoreductase gene (yellow arrow) generating a 7 bp duplication at the target sequence (vertical red line). Tn1331 was inserted in a truncated Y. pestis hypothetical protein (not shown) generating a 5 bp duplication at the target site (vertical red line) linking the KQ-like element with the 7.6 kb DNA fragment. Chromosomal proteins (plum arrows) flanking the 5′ and 3′ DNA region of the interrupted oxidoreductase gene were identified in the remaining sequence of contig 36. SnapGene 2.4.2 was used for map construction. ABC, ATP-binding cassette.
Fig. 2.
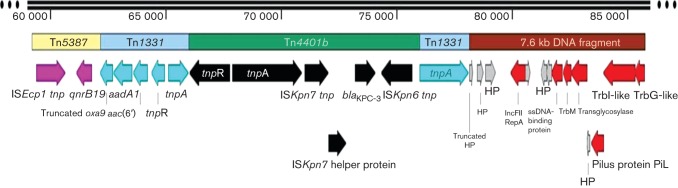
Protein predictions of the 26.5 kb fragment. The predicted proteins encoded in the KQ-like element and the 7.6 kb DNA fragment (brown) are shown. The KQ-like element contains 13 ORFs portrayed by coloured arrows based on predicted gene functions as follows: ISEcp1 tnp and qnrB19 of Tn5387 (pink); truncated oxa9, aadA1, aac(6'), tnpR and a disrupted tnpA of Tn1331 (turquoise); tnpR, tnpA, ISKpn7 tnp and helper protein, blaKPC-3, ISKpn6 tnp of Tn4401b (black). The 7.6 kb DNA fragment contains 13 ORFs including seven predicted plasmids (red arrows) and six hypothetical proteins (HPs; grey arrows) identified in Y. pestis PY-95, in the order: 3′ end of truncated HP, HP, HP, IncFII family plasmid replication initiator RepA, HP, HP, HP, ssDNA-binding protein, TrbM, transglycosylase, HP, conjugative pilus protein PilL, TrbI-like and TrbG-like. SnapGene 2.4.2 was used for map construction.
In the KPC-producing A. baumannii M3AC9-7, a 26.5 kb DNA fragment, which includes the KQ-like element (18.9 kb in the opposite orientation as described by Rice et al., 2008) and a 7.6 kb DNA fragment with homology to a putative plasmid from Y. pestis strain PY-95 (GenBank accession number NZ_AKTK01000456.1), was inserted into an oxidoreductase gene (yellow arrow in Fig. 1). This insertion generated a 7 bp duplication (AGTTTGG) at the target sequence, suggesting a transposition event. We speculate that the transposase of ISEcp1 present in Tn5387 mediated this transposition event as sequences with similarity to the ISEcp1 inverted repeats (IRs) described by Poirel et al. (2005) were found at both ends of the 26.5 kb DNA fragment (Fig. 1). The left IR (IRL) of ISEcp1 was detected at nt 59 217–59 238 and its 22 bp sequence (CCTAGATTCTACGTCAGTACTT) is identical to the previously described IR of ISEcp1 (nt 1–22 of GenBank accession number AJ242809). The right IR (IRR)-like of ISEcp1 was located at nt 85 742–85 755 and consists of a 14 bp sequence (CTTATTCTGCGGCG) in which the last 8 bp are identical to IRR4 found in the tnpA gene of IS903D described by Poirel et al. (2005). Other IRR sequences similar to those described by Poirel et al. (2005) and Tran et al. (2012) were found flanking the qnrB19 gene. Due to the ISEcp1-mediated transposition event, a truncated version of Tn1331 was observed, lacking the last 307 bp of blaOXA-9 and the entire blaTEM-1. Chromosomal proteins flanking the 5′ and 3′ DNA regions of the interrupted oxidoreductase gene described above were identified in the rest of contig 36 (plum arrows in Fig. 1).
The 7.6 kb DNA fragment present in the 26.5 kb fragment with 98 % homology to Y. pestis PY-95 mentioned above coded for the following plasmids proteins: TrbG-like, TrbI-like and TrbM conjugation proteins, conjugative pilus protein PilL, ssDNA-binding protein, transglycosylase, IncFII family plasmid replication initiator RepA, and six hypothetical proteins (red and grey arrows in Fig. 2). The order of the predicted proteins was identical to a whole-genome shotgun DNA sequence of Y. pestis, except for an 88 bp gap. The area of the gap occurred inside a hypothetical protein in which both organisms share the first 54 aa, but differ in the carboxylic end (11 aa for A. baumannii and 33 aa for Y. pestis). A hypothetical protein of Y. pestis was disrupted by the insertion of Tn1331 generating a 5 bp duplication (TATTC) target site (Fig. 1) described by Rice et al. (2008). This finding suggested that another transposition event linked the KQ-like element with this 7.6 kb DNA fragment from an unknown plasmid. Unfortunately, it is unknown if the 7.6 kb fragment was present in the 80 kb K. pneumoniae conjugative plasmid pLRM24 containing the KQ element as the complete DNA sequence of this plasmid was not available.
During the same 6-month surveillance study, KPC-producing K. pneumoniae, Escherichia coli and Pseudomonas aeruginosa were identified in the same institution as the KPC-producing A. baumannii isolates (Robledo et al., 2011). This suggested the possibility that the KQ-like element and the rest of the 7.6 kb DNA fragment were acquired horizontally by A. baumannii M3AC9-7 from an unknown plasmid and inserted into its chromosome by a transposition event. The host range of IncFII plasmid is limited to the family Enterobacteriaceae (Carattoli, 2009); therefore as an IncFII family plasmid replication initiator RepA was identified in the 7.6 kb fragment, we speculate that this fragment comes from an unknown Enterobacteriaceae plasmid. We hypothesize this IncFII plasmid entered A. baumannii, and due to its narrow host range could not replicate and be maintained stably in this organism. Therefore, to maintain the newly acquired antibiotic resistance genes, the 26.5 kb DNA fragment was integrated into the A. baumannii chromosome by a transposition event.
Allelic diversity by MLST
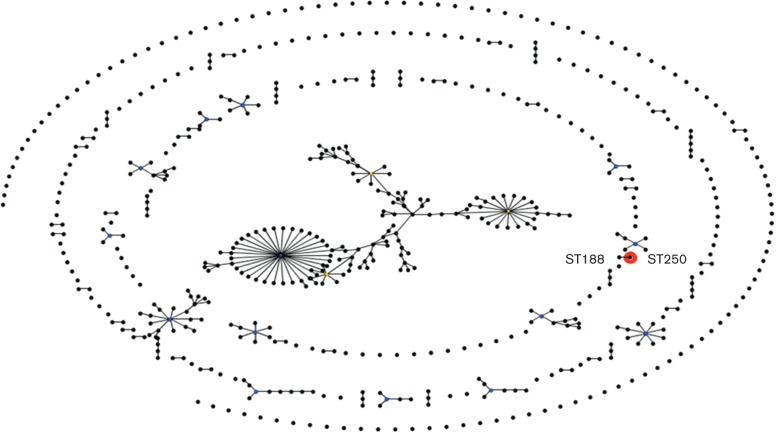
The Institut Pasteur MLST scheme assigned a novel sequence type, ST250, to the KPC-producing A. baumannii M3AC9-7. A new allele for the cpn60 locus was assigned, which differentiates our strain from the A. baumannii ST188 clone identified in Portugal (Manageiro et al., 2012), as shown by eBurst analysis (Fig. 3).
Fig. 3.
Population snapshot of A. baumannii. The entire A. baumannii MLST database from the Institut Pasteur is displayed as a single eBURST diagram. Results show the KPC-producing A. baumannii M3AC9-7, ST250 (coloured red), as distinct but genetically related (sharing alleles at six of seven loci) to the KPC-negative A. baumannii ST188 (connected by a line) identified in Portugal. The sequence types that are subgroup founders (ancestral genotype) are coloured yellow and the primary founder of the group is coloured blue.
Conclusions
To our knowledge, this is the first report of the identification of the genetic background of blaKPC in A. baumannii, which was chromosomally located due to an ISEcp1-mediated transposition event. This finding is not surprising as ISEcp1 has been implicated previously in the mobilization of blaCTX-M from chromosome to plasmid (Rice et al., 2008) and vice versa (Hudson et al., 2014). ISEcp1 has also been associated with the mobilization of other β-lactamases such as blaCMY and qnrB19 resistance genes in different Enterobacteriaceae (D’Andrea et al., 2011; Tran et al., 2012). Moreover, Potron et al. (2011) reported the mobilization of blaCTX-M-5 from a narrow-range plasmid to the chromosome of A. baumannii by an ISEcp1-mediated transposition event similar to what we observed in this study.
The identification of a novel local sequence type carrying the KPC gene could explain why this organism has not yet disseminated to other geographical areas. However, amongst other factors, the ease of travel between Puerto Rico, the United States and other geographical areas may help spread this unique sequence type. The results further illustrate A. baumannii’s genetic plasticity and its ability to acquire antibiotic resistance genes.
Acknowledgements
This work was supported by MBRS/RISE (R25GM061838), RCMI/NIH (G12-MD 007600), Associate Deanship for Biomedical Sciences Graduate Program School of Medicine, Medical Sciences Campus, University of Puerto Rico and Merck Sharp & Dohme Inc. We thank the platform for Genotyping of Pathogens and Public Health (Institut Pasteur, Paris, France) for coding MLST alleles and profiles available at Institut Pasteur MLST Databases (http://www.pasteur.fr/mlst). We also thank Drs Wieslaw J. Kozek and Carlos H. Ramirez Ronda for reviewing the manuscript. This research project constitutes a partial fulfilment of the doctoral thesis dissertation of T. M.
Abbreviations:
- IR
inverted repeat
- IRL
left inverted repeat
- IRR
right inverted repeat
- KPC
Klebsiella pneumoniae carbapenemase
- MLST
multilocus sequence typing
References
- Adams M. D., Goglin K., Molyneaux N., Hujer K. M., Lavender H., Jamison J. J., MacDonald I. J., Martin K. M., Russo T. & other authors (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190, 8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold R. S., Thom K. A., Sharma S., Phillips M., Kristie Johnson J., Morgan D. J. (2011). Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J 104, 40–45. 10.1097/SMJ.0b013e3181fd7d5a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant K. A., Van Schooneveld T. C., Thapa I., Bastola D., Williams L. O., Safranek T. J., Hinrichs S. H., Rupp M. E., Fey P. D. (2013). KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57, 37–41. 10.1128/AAC.01062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53, 2227–2238. 10.1128/AAC.01707-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A., Montealegre M. C., Mojica M. F., Maya J. J., Rojas L. J., De La Cadena E. P., Ruiz S. J., Recalde M., Rosso F. & other authors (2012). First report of a Pseudomonas aeruginosa isolate coharboring KPC and VIM carbapenemases. Antimicrob Agents Chemother 56, 5422–5423. 10.1128/AAC.00695-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Naas T., Truong H., Villegas M.-V., Wisell K. T., Carmeli Y., Gales A. C., Navon-Venezia S., Quinn J. P., Nordmann P. (2010). Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg Infect Dis 16, 1349–1356. 10.3201/eid1609.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon G., Naas T., Villegas M. V., Correa A., Quinn J. P., Nordmann P. (2011). Wide dissemination of Pseudomonas aeruginosa producing β-lactamase blaKPC-2 gene in Colombia. Antimicrob Agents Chemother 55, 5350–5353. 10.1128/AAC.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea M. M., Literacka E., Zioga A., Giani T., Baraniak A., Fiett J., Sadowy E., Tassios P. T., Rossolini G. M. & other authors (2011). Evolution and spread of a multidrug-resistant Proteus mirabilis clone with chromosomal AmpC-type cephalosporinases in Europe. Antimicrob Agents Chemother 55, 2735–2742. 10.1128/AAC.01736-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C., Wei Z., Jiang Y., Shen P., Yu Y., Li L. (2011). Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J Antimicrob Chemother 66, 1184–1186. 10.1093/jac/dkr060 [DOI] [PubMed] [Google Scholar]
- Hudson C. M., Bent Z. W., Meagher R. J., Williams K. P. (2014). Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS ONE 9, e99209. 10.1371/journal.pone.0099209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manageiro V., Jones-Dias D., Ferreira E., Louro D., Caniça M., Antimicrobial Resistance Surveillance Program in Portugal (2012). Genetic diversity and clonal evolution of carbapenem-resistant Acinetobacter baumannii isolates from Portugal and the dissemination of ST118. Int J Antimicrob Agents 40, 398–403. 10.1016/j.ijantimicag.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Mavroidi A., Miriagou V., Malli E., Stefos A., Dalekos G. N., Tzouvelekis L. S., Petinaki E. (2012). Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int J Antimicrob Agents 39, 247–250. 10.1016/j.ijantimicag.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Villegas M. V., Lartigue M. F., Quinn J. P., Nordmann P. (2008). Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob Agents Chemother 52, 1257–1263. 10.1128/AAC.01451-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Truong H. V., Nordmann P. (2012). Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56, 4753–4759. 10.1128/AAC.00334-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Cuzon G., Naas T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9, 228–236. 10.1016/S1473-3099(09)70054-4 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., Edwards R. A., Gerdes S., Parrello B. & other authors (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Lartigue M. F., Decousser J. W., Nordmann P. (2005). ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49, 447–450. 10.1128/AAC.49.1.447-450.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Munoz-Price L. S., Nordmann P., Cleary T., Poirel L. (2011). Genetic features of CTX-M-15-producing Acinetobacter baumannii from Haiti. Antimicrob Agents Chemother 55, 5946–5948. 10.1128/AAC.05124-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. B., Carias L. L., Hutton R. A., Rudin S. D., Endimiani A., Bonomo R. A. (2008). The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Antimicrob Agents Chemother 52, 3427–3429. 10.1128/AAC.00493-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo I. E., Aquino E. E., Santé M. I., Santana J. L., Otero D. M., León C. F., Vázquez G. J. (2010). Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob Agents Chemother 54, 1354–1357. 10.1128/AAC.00899-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo I. E., Aquino E. E., Vázquez G. J. (2011). Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother 55, 2968–2970. 10.1128/AAC.01633-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno R., McGillivary G., Sherratt D. J., Actis L. A., Tolmasky M. E. (2002). Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother 46, 3422–3427. 10.1128/AAC.46.11.3422-3427.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T., Andres P., Petroni A., Soler-Bistué A., Albornoz E., Zorreguieta A., Reyes-Lamothe R., Sherratt D. J., Corso A., Tolmasky M. E. (2012). Small plasmids harboring qnrB19: a model for plasmid evolution mediated by site-specific recombination at oriT and Xer sites. Antimicrob Agents Chemother 56, 1821–1827. 10.1128/AAC.06036-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas M. V., Lolans K., Correa A., Kattan J. N., Lopez J. A., Quinn J. P., Colombian Nosocomial Resistance Study Group (2007). First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 51, 1553–1555. 10.1128/AAC.01405-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrilli R., Pournaras S., Giannouli M., Tsakris A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41, 11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]