Cholera reemerged in Latin America in 1991, after being absent for more than 100 years (Sepúlveda et al., 2006). Initially appearing in Peru in January, cholera reached Mexico in June, affecting 45 977 people in Mexico alone until 2002 (Sepúlveda et al., 2006). Vibrio cholerae, the causative agent of cholera, has more than 200 ‘O’ serogroups, but only O1 and O139 cause epidemic cholera; the remaining serogroups (non-O1/O139) are associated with sporadic diarrhoea. Serogroup O1 has two biotypes, namely classical (CL) and El Tor (ET). Historically, the CL biotype caused the fifth, sixth and presumably the earlier pandemics, while the ET biotype is responsible for the ongoing seventh pandemic that began in 1961. The major virulence factors for V. cholerae O1/O139 include the potent cholera toxin (CTX) and the toxin co-regulated pilus (TCP), which promotes bacterial intestinal colonization and serves as receptor for the lysogenic CTX prophage (CTXϕ). Naturally occurring V. cholerae populations can become toxigenic via acquisition of CTXϕ and those lacking CTXϕ are generally known as non-toxigenic. V. cholerae non-O1/O139 usually lack CTX and TCP; however, they possess virulence factors, namely ET-specific haemolysin, repeat in toxin (RTX), heat-stable enterotoxin (NAG-ST), cholix toxin and type three secretion system (T3SS) (Chatterjee et al., 2009; Dziejman et al., 2005). The T3SS is a virulence factor for several enteropathogens including Escherichia coli, Salmonella enterica, Shigella spp., Vibrio parahaemolyticus and V. cholerae non-O1/O139 (Dziejman et al., 2005; Coburn et al., 2007). Although the mechanism by which T3SS contributes to virulence and disease remains largely unknown, it has been shown to induce diarrhoea in animal models (Dziejman et al., 2005; Shin et al., 2011).
In the present study, 33 CTX− strains from a collection of 182 V. cholerae O1 isolates were analysed for virulence and related molecular traits. These isolates were obtained from clinical and environmental sources in Mexico between 1991 and 2008. PCR results revealed the presence of genes encoding ET-specific haemolysin and RTX, and the absence of NAG-ST (stn). Twenty-one CTX− strains had the tcpA gene encoding TCP. Despite belonging to biotype ET, most strains possessed TCP and all strains lacked the targeted ORFs of the Vibrio seventh pandemic island I (VSP-I; ORFs: VC0175, VC0178, VC0180, VC0183 and VC0185) and II (VSP-II; ORFs: VC0490, VC0493, VC0498, VC0502, VC0504, VC0511 and VC0516) including the ET marker VC2346. The non-toxigenic V. cholerae O1 existing in natural aquatic ecosystem can arise from toxigenic strains through excision of the CTXϕ (Alam et al., 2007). In Mexico, CTX− ET strains lacking the seventh pandemic ET markers VSP-I, VSP-II and VC2346 represented the typical genetic traits of the pre-seventh pandemic ET (Chun et al., 2009). Presumably, these CTX− strains were native to the ecosystem, or evolved from toxigenic ET via excisions of the gene islands such as CTXϕ, VSP-I and VSP-II. Nonetheless, T3SS+ non-toxigenic V. cholerae O1 strains lacking VSP-I and VSP-II in Mexico may have evolutionary implications for the bacterium.
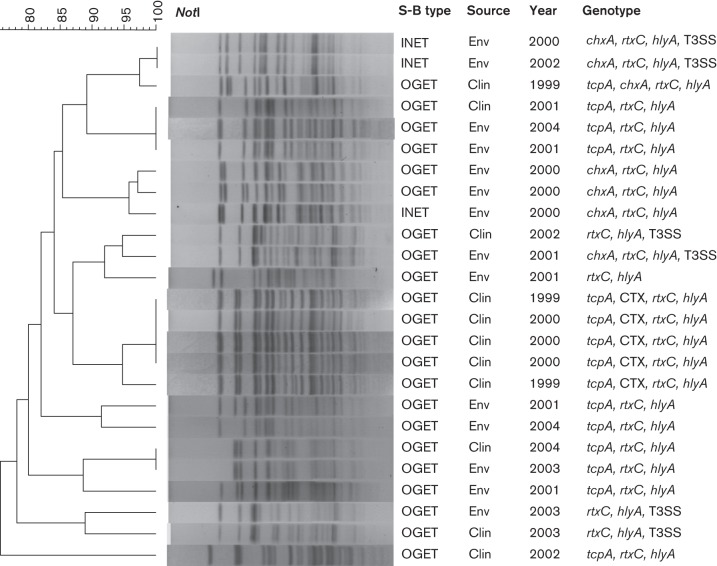
Eight of the Mexican CTX− ET strains possessed the gene encoding the cholix toxin (chxA), and six harboured T3SS specific genes (structural: vcsJ2, vspD, vcsVUQ2 and vcsRTCNS2; regulatory: vttRA and vttRB; effector: vopF). Notably, all T3SS+ strains lacked TCP. Year-wise data revealed the presence of a T3SS gene cluster from ET strains isolated from the environmental samples in 2000, 2001, 2002 and 2003, and finally from human samples in 2002 and 2003. DNA sequence analysis of vttRA (630 bp), vttRB (432 bp) and vopF (1590 bp) (GenBank accession nos KJ641467–72) revealed the deduced amino acid sequences share >98 % homology with respective genes of AM-19226 (GenBank accession no. DQ124262), a T3SS+ V. cholerae O39 clinical isolate capable of colonizing infant mice and developing diarrhoea in rabbits despite lacking TCP and CTX (Dziejman et al., 2005; Shin et al., 2011). T3SS+ V. cholerae non-O1/O139 has been reported from wide-ranging ecosystems (Octavia et al., 2013). PFGE of NotI-digested genomic DNA revealed high genetic divergence among the non-toxigenic V. cholerae O1 stains including the T3SS+ ET, which differed from the toxigenic isolates (Fig. 1). Molecular fingerprinting data suggest that the T3SS+ ET strains do not belong to a specific clone, and likely arose independently in the aquatic ecosystem of Mexico (Alam et al., 2010).
Fig. 1.
Dendrogram of PFGE patterns showing the genomic fingerprinting of V. cholerae O1 CTX− ET isolates including those possessing T3SS gene clusters in Mexico. The dendrogram was prepared by Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) clustering methods by using PFGE images of NotI-digested genomic DNA. The scale bar at the top (left) indicates the correlation coefficient (%). No clustering of the PFGE patterns revealed divergence among the V. cholerae CTX− ET possessing T3SS gene clusters, suggesting that they are not clonal. INET, Inaba ET; OGET, Ogawa ET; env, environment; clin, clinical.
The consistent isolation of CTX− V. cholerae O1 strains was unique between 1999 and 2004, a time when V. cholerae O1 isolated from human and environmental samples were predominantly CTX− in Mexico (data not shown). Notably, CTX− ET strains were T3SS+ in Mexico since 2000, following CTX− ET was first confirmed from the environment in 1999; the CTX−, T3SS+ ET strains were found in human samples from 2002. This is unique and may have epidemiological significance as T3SS is present in non-O1/O139 and rarely in CTX− V. cholerae O1 (Octavia et al., 2013). The mechanism explaining how CTX−, TCP− ET strains acquired the island encoding the T3SS is unclear, although the naturally occurring V. cholerae population can become competent in the presence of chitin and can horizontally acquire genes and gene clusters such as those encoding the T3SS (Morita et al., 2013). Recently, toxigenic V. cholerae O1 altered ET, ET and CL biotype strains have been reported from human and surface water sources in Mexico between 1991 and 1997 (Alam et al., 2010). Toxigenic V. cholerae O1 lacks genetic elements such as T3SS; thus, it is plausible that non-toxigenic O1 strains acquired T3SS horizontally rather than the assumption that T3SS+ non-O1/O139 strains switched serologically and converted to O1 by acquiring O1 biosynthesis genes. Previously, T3SS and CTXϕ have been shown to be present in the same V. cholerae non-O1/O139 host, although the number of isolates was relatively few (Octavia et al., 2013). Nevertheless, it remains a possibility that the non-toxigenic T3SS+ strains could become CTX+ via acquisition of TCP, a receptor for CTXϕ (Rahman et al., 2008). The clinical relevance of the naturally occurring V. cholerae O1 carrying T3SS in Mexico can pose health threat considering that such CTX− V. cholerae O1 can cause disease and also serve as a progenitor to become toxigenic by laterally acquiring the CTX prophage.
Acknowledgements
This research was supported in part by the NIAID grant no. 1RO1A13912901, and NIID, Japan. The icddr,b is thankful to the governments of Australia, Bangladesh, Canada, Sweden and the UK for providing core/unrestricted support. We acknowledge colleagues from icddr,b, Universidad Nacional Autónoma de México and Centro de Investigación Científica y de Educación Superior de Ensenada, Baja California, Mexico for their kind support.
Abbreviations:
- CL
classical
- CTX
cholera toxin
- ET
El Tor
- NAG-ST
heat-stable enterotoxin
- RTX
repeat in toxin
- TCP
toxin co-regulated pilus
- T3SS
type three secretion; system
- VSP
Vibrio seventh pandemic island
References
- Alam M., Sultana M., Nair G. B., Siddique A. K., Hasan N. A., Sack R. B., Sack D. A., Ahmed K. U., Sadique A. & other authors (2007). Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. Proc Natl Acad Sci U S A 104, 17801–17806. 10.1073/pnas.0705599104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M., Nusrin S., Islam A., Bhuiyan N. A., Rahim N., Delgado G., Morales R., Mendez J. L., Navarro A. & other authors (2010). Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol 48, 3666–3674. 10.1128/JCM.00866-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Ghosh K., Raychoudhuri A., Chowdhury G., Bhattacharya M. K., Mukhopadhyay A. K., Ramamurthy T., Bhattacharya S. K., Klose K. E., Nandy R. K. (2009). Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol 47, 1087–1095. 10.1128/JCM.02026-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Grim C. J., Hasan N. A., Lee J. H., Choi S. Y., Haley B. J., Taviani E., Jeon Y. S., Kim D. W. & other authors (2009). Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106, 15442–15447. 10.1073/pnas.0907787106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Sekirov I., Finlay B. B. (2007). Type III secretion systems and disease. Clin Microbiol Rev 20, 535–549. 10.1128/CMR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M., Serruto D., Tam V. C., Sturtevant D., Diraphat P., Faruque S. M., Rahman M. H., Heidelberg J. F., Decker J. & other authors (2005). Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A 102, 3465–3470. 10.1073/pnas.0409918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Yamamoto S., Hiyoshi H., Kodama T., Okura M., Arakawa E., Alam M., Ohnishi M., Izumiya H., Watanabe H. (2013). Horizontal gene transfer of a genetic island encoding a type III secretion system distributed in Vibrio cholerae. Microbiol Immunol 57, 334–339. 10.1111/1348-0421.12039 [DOI] [PubMed] [Google Scholar]
- Octavia S., Salim A., Kurniawan J., Lam C., Leung Q., Ahsan S., Reeves P. R., Nair G. B., Lan R. (2013). Population structure and evolution of non-O1/non-O139 Vibrio cholerae by multilocus sequence typing. PLoS ONE 8, e65342. 10.1371/journal.pone.0065342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. H., Biswas K., Hossain M. A., Sack R. B., Mekalanos J. J., Faruque S. M. (2008). Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol 27, 347–355. 10.1089/dna.2008.0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda J., Valdespino J. L., García-García L. (2006). Cholera in Mexico: the paradoxical benefits of the last pandemic. Int J Infect Dis 10, 4–13. 10.1016/j.ijid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- Shin O. S., Tam V. C., Suzuki M., Ritchie J. M., Bronson R. T., Waldor M. K., Mekalanos J. J. (2011). Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio 2, e00106–e00111. 10.1128/mBio.00106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]