Introduction
Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy, accounting for 25% of all childhood cancers. In the United States, approximately 3000 children aged 1–19 years are diagnosed with ALL annually.1 Giant strides have been made in the management of childhood ALL over the past 50 years, which has resulted in improvement in cure rates from approximately 10% to approximately 90%.2, 3 The rational use of multi–agent systemic chemotherapy over a prolonged duration (2–3 years) and adequate central–nervous– system (CNS)-directed therapy as well as improved antibiotic and blood product support in the 1960s and 1970s were responsible for the early improvements in outcome. However, insights into the heterogenic biology of ALL and monitoring of minimal residual disease (MRD) have helped to refine therapy based on risk of relapse to maximize cure and minimize toxicities. For example, identification of the Philadelphia chromosome in a subset of patients with ALL has made it possible to incorporate ABL tyrosine kinase inhibitors into chemotherapy regimens. This targeted therapy approach has improved the cure rate of patients with Philadelphia chromosome– positive ALL from 35% to around 70% over the last 10 years, even without stem cell transplant.4
Leukemic cells have, and are being thoroughly investigated by methods ranging from karyotyping which identifies large chromosomal alterations, to whole genome sequencing, which identifies cryptic changes in the entire genome. ALL is particularly amenable to biologic studies because of the relative ease of obtaining sample, which in most cases is an enriched population of blasts. Moreover, because the majority of children with ALL are treated uniformly on large clinical trials, well–annotated clinical information is available to correlate with biologic findings. Extensive collaborative efforts among various study groups internationally have played a vital role in the remarkable progress made in not only improving therapeutic outcomes but also deciphering the complex biology of childhood ALL.5
In this review, we summarize various insights gained from biologic studies of childhood ALL, with a focus on recent studies. We also discuss genomic lesions and epigenetic regulatory mechanisms associated with leukemic transformation. Finally we highlight the importance of studying the biology of the host to understand additional heterogeneity in treatment response and toxicities.
B–ALL
Eighty–five percent of the cases of childhood ALL are of the B–lineage. To keep pace with the growing impact of biologic findings on treatment outcomes, in 2008 the World Health Organization revised the nomenclature from solitary precursor B–ALL to a classification based on 7 specific, recurring genetic lesions (e.g. B–ALL with ETV6–RUNX1, B–ALL with hyperdiploidy).6 Of note, the term B-ALL is not used for Burkitt leukemia/lymphoma which is a mature B-cell malignancy. As newer subtypes are identified, biology–centered classifications need to be continually reviewed and updated (Figure 1).
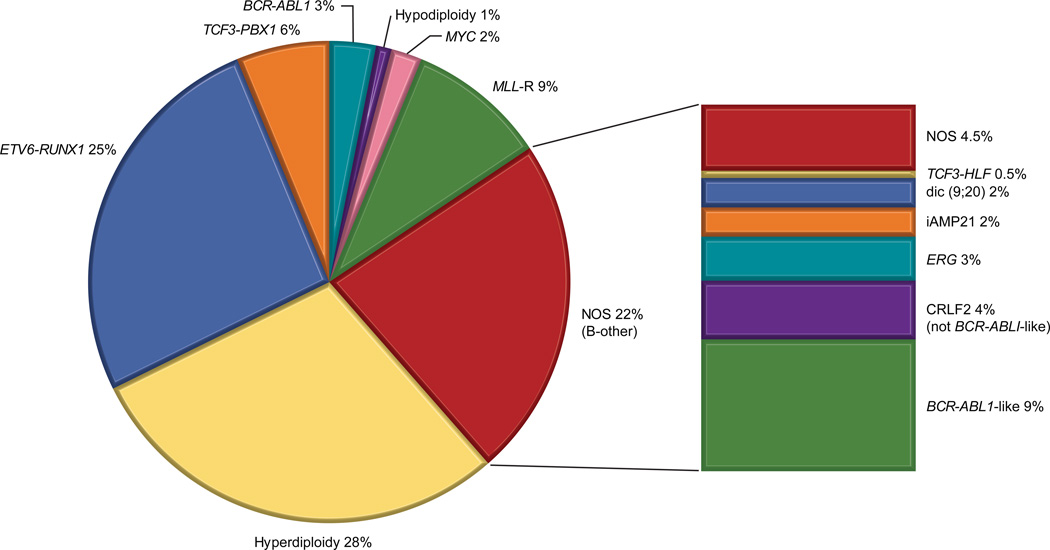
Figure 1. Distribution of molecular subtypes of childhood B-ALL.
The pie chart on the left depicts molecular subtypes that were identified prior to 2004, incorporated in the 2008 WHO classification and are currently used for risk stratification. Subtype was unknown in 22% of patients (termed B-other). Since then, various novel molecular subtypes have been characterized shown in the bar graph on the right. Data has been modified from Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350: 1535–1548; and from Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: Where are we going and how do we get there? Blood. 2012;120: 1165–1174.
NOS: not otherwise specified
Conventional chromosomal alterations
ALL commonly arises from a series of genetic alterations, and in the majority of ALL subtypes, the interplay of these alterations. For the last 3 decades, several conventional cytogenetic studies of genetic aberrations that include chromosomal translocations and alterations in chromosome number have provided information on the pathogenesis of ALL. Common translocations in children with B–ALL include t(12;21) [ETV6–RUNX1](25%), t(1;19) [TCF3–PBX1](5%), t(9;11) [BCR–ABL1](3%) and translocations involving the MLL gene with various partner fusion genes (5%). Gains in whole chromosomes, or high–hyperdiploidy (>50 chromosomes) accounts for 25% of childhood ALL, whereas hypodiploidy (< 44 chromosomes) accounts for approximately 1% of cases. Several of these genetic changes have prognostic and therapeutic implications and are important in risk stratification schemes.7 The overall survival (OS) of patients with ETV6–RUNX1 or high–hyperdiploid ALL is more than 93%;8, 9 therefore these patients are treated on less intensive regimens, provided that they have an adequate early response to remission induction therapy (as currently assessed by the measurement of MRD). Contemporary therapy has abolished the previously unfavorable prognostic impact of TCF3– PBX1(t[1;19]). However, because bone marrow and CNS relapse could be competitive events, patients with TCF3–PBX1 may need more intensive intrathecal therapy to reduce the risk of CNS relapse with improved systemic therapy.10 Similarly, the addition of tyrosine kinase inhibitors has dramatically improved the bone marrow control of patients with BCR–ABL1–positive ALL4 and attention should be paid to optimal intrathecal therapy in this genetic subtype of ALL. Hypodiploidy continues to be a high–risk feature,11 necessitating further understanding of oncogenic mechanisms and the rational use of targeted therapy (e.g. for RAS pathway inhibition).12 The frequency and prognostic impact of MLL rearrangements differ by age. Approximately 80% of infants younger than 1 year harbor MLL rearrangements and their overall outcome is poor (5–year survival 50%) despite receiving very intensive therapy.13
Fusion gene products that result from chromosomal translocations provide the lymphoid progenitor or stem cell with leukemogenic potential, such as constitutional activation of tyrosine kinases (e.g. ABL1) or disruption of genes that regulate normal lymphoid development (e.g. ETV6, PAX5). Additional genetic hits are often required for the ultimate development of leukemia and include loss of the tumor suppressor gene CDKN2A and deletion of the non– translocated ETV6 allele in ETV6–RUNX1–positive ALL.14 High–resolution genomic studies are revealing the entire range of co–operating genetic changes and pathogenic mechanisms.12, 15–20
Submicroscopic alterations
The advent of genome–wide profiling of RNA and DNA and next–generation sequencing (NGS) technologies has greatly increased our ability to identify and catalog submicroscopic genetic alterations and sequence mutations in ALL, which in turn help define new molecular subtypes. Some novel genomic lesions have prognostic and therapeutic significance and may be used to refine risk stratification schemes in the near future; for example, IKZF1 deletion predicts poor prognosis in children with B–ALL.18 In addition, the recognition of specific molecular lesions and critical oncogenic pathways paves the way for developing novel targeted approaches to therapy (e.g. inhibition of the ABL tyrosine kinase or JAK–STAT pathways). Single nucleotide polymorphism (SNP) array analyses revealed that gross genomic instability is not present in most children with ALL.17 A mean of 6.4 genomic lesions were present per case, with wide variability within the genetic subtypes of ALL. Structural alterations in genes encoding transcriptional regulators of B–lymphoid development and differentiation occur in more than 40% of patients with B–ALL. PAX5 is the most common target; others targets include IKZF1and EBF1. A number of additional lesions have also been identified in lymphoid signaling, transcription factors and tumor suppressors. Several of these alterations cooperate in leukemogenesis. For example, the deletion of IKZF1 accelerates the onset of ALL in murine models of BCR–ABL1–ALL.20 The assortment and accumulation of “driver” and “passenger” mutations and the sequence of events in leukemia development and progression continue to be investigated.
High-risk subtypes of B–ALL
BCR–ABL–like ALL
IKZF1 deletions are a hallmark of BCR–ABL1–positive ALL, but these deletions also occur in a subset of patients with poor-response, high-risk ALL without any known chromosomal rearrangement.18 Using genome-wide analyses, two groups of investigators independently identified a subgroup of B-ALL which has a gene expression profile similar to that of BCR–ABL1–positive ALL including a high frequency of IKZF1 alterations but lacks the BCR–ABL1 fusion protein; they termed this genetic subtype as BCR–ABL1–like or Philadelphia chromosome-like ALL.18, 21 This subtype comprises 10% of the cases of B–ALL in children and 25% of the cases of ALL in adolescents and young adults.22 NGS techniques and downstream functional experiments show that BCR–ABL1–like ALL is characterized by genetic changes which result in constitutive activation of cytokine receptor and/or tyrosine kinase signaling.19, 22 The spectrum of genetic alterations is extremely diverse; however, several rearrangements involve tyrosine kinases such as ABL and PDGFR which respond to imatinib and dasatinib in vitro and in vivo.22, 23 Even though risk-directed therapy including intensive chemotherapy with or without transplant based on MRD level during remission induction therapy can abolish the poor prognosis of this group of patients, it is important to look for genetic lesions responsive to ABL tyrosine kinase inhibitor so that some patients can be spared from transplantation.24 Several other rearrangements target JAK and EPOR, which are sensitive to JAK inhibitors in preclinical models.25 In addition, re–arrangements involving the cytokine receptor gene CRLF2 have been identified in 50% of patients with BCR–ABL1–like ALL, with frequent co–existing JAK mutations, also potentially sensitive to JAK inhibition.19, 25 In view of the therapeutic implications for this high–risk subset of patients, array and sequencing based methodologies are being developed for rapid classification of patients with BCR–ABL1–like ALL and identification of targetable lesions; the incorporation of tyrosine kinase inhibitors in frontline therapy is also planned.26
ALL with intrachromosomal amplification of chromosome 21 (iAMP 21)
iAMP21 ALL was originally discovered by the observation of multiple copies of the RUNX1 gene during routine screening for ETV6–RUNX1 by fluorescent in situ hybridization. This particular ALL subtype is characterized by the instability of chromosome 21.27 The incidence of iAMP21 is approximately 2%, and the median age of patients is 9–11 years. Intensification of chemotherapy has abolished the poor prognosis once associated with this ALL subtype.28
Down syndrome ALL
Patients with Down syndrome are at an approximately 20–fold increased risk of developing ALL, though the precise role of the extra chromosome 21 in leukemogenesis is unknown.29 These patients have low frequencies of T–ALL and common ALL translocations such as ETV6–RUNX1. Patients with Down syndrome ALL have inferior outcome due to increased risk of relapse and high rate of treatment–related mortality.30 High–resolution SNP profiling has identified a submicroscopic deletion of the pseudoautosomal regions of chromosomes X and Y which leads to the P2RY8–CRLF2 fusion in approximately 50% of patients with Down syndrome–ALL.31 These fusions and other CRLF2 alterations were associated with JAK mutations. Together, these lesions activate the JAK–STAT pathway and promote cytokine–independent growth. Therefore, the inhibition of JAK tyrosine kinase is a potentially useful therapeutic strategy in patients with Down syndrome–ALL.
T–ALL
T–ALL accounts for 10%–15% of the cases of childhood ALL. The outcome of children with T–ALL, which has been historically poor, has improved gradually with the use of intensified therapy, including dexamethasone, asparaginase, and high–dose methotrexate.3, 8 However, children who relapse have a dismal outcome even with hematopoietic stem cell transplantation.32 Therefore, it is critical to identify aberrant molecular pathways and targets for therapeutic intervention for T–ALL. Genetic lesions in T–ALL are diverse and complex and a multitude of alterations contribute in the pathogenesis of various subtypes of T–ALL.33, 34 Chromosomal translocations are present in approximately 50% of patients with T–ALL cases, but unlike B–ALL, their prognostic impact is not well defined and they are not used for risk stratification. Some translocations result in the juxtaposition of oncogenes to T–cell receptor (TCR) genes, leading to overexpression of the oncogene in T–cell progenitor cells (e.g. TLX1– TCRδ), whereas others result in the fusion of 2 transcription factor oncogenes (e.g. STIL–TAL1). In addition, rearrangements of the MLL gene occur in 5%–10% of patients with T–ALL. Gene expression profiling studies have identified 4 major subtypes of T–ALL on the basis of the predominant oncogenic pathway activation (TLX1, LYL1, TAL/LMO2 and TLX3).35
NOTCH activation in T–ALL
Constitutive activation of NOTCH signaling, primarily via somatic mutations, is seen in more than 50% of patients with T–ALL; a finding that is not restricted to specific subtypes of T– ALL.36 In general, the presence of NOTCH1 mutations indicates a favorable prognosis. NOTCH1 is a transmembrane receptor crucial for T–cell development, lineage commitment, cell growth, and survival. Activation of NOTCH1 and the presence of co–operating lesions, such as deletion of the tumor suppressor CDKN2A (found in 70% of patients with T–ALL) can lead to leukemic transformation. In addition, mutations in FBXW7, which encodes an ubiquitin protein ligase (found in 8%–10% of patients with T–ALL), attenuate the degradation of activated NOTCH1, further enhancing its downstream signaling.37 Thus, the inhibition of NOTCH1, either by small molecule inhibitors of gamma secretase (that impede the release of activated NOTCH1) or by anti–NOTCH1 antibodies is being actively pursued as a therapeutic strategy for T–ALL.38,39
Early T–cell precursor ALL
Early T–cell precursors (ETPs) are a subset of immature thymocytes that retain stem-cell–like features and can differentiate into multiple lineages, including lymphoid and myeloid lineage. Complementary studies of flow cytometry, gene expression and DNA copy number, showed that the genetic profile of approximately 12% of patients with T–ALL is similar to that of these immature thymocytes.40 A whole–genome study showed that ETP–ALL has frequent mutations of genes involved in hematopoietic development, cytokine receptor and RAS signaling, and chromatin modification.41 The incidence of activating NOTCH1 mutations is low in ETP–ALL which also lacks a unifying chromosomal abnormality. In general, the outcome of patients with ETP–ALL is poor, but myeloid–directed and epigenetic therapies may be beneficial for these patients.41 A recent, small study suggested that patients with ETP-ALL have an intermediate outcome when treated with intensive chemotherapy that includes pegylated asparaginase and dexamethasone (5-year event-free survival of 76.7%);42 a finding that requires confirmation.
Epigenetics in ALL
In recent years, the importance of epigenetic regulatory mechanisms in normal and malignant hematopoiesis has become increasingly evident. Alterations in the methylation of DNA promoters and the modification of histone can significantly perturb transcriptional regulation and modify gene expression. Different subtypes of ALL are characterized by distinct DNA methylation signatures, which in turn correlate with gene expression profiles.15 Several genes related to lymphoid development that are targets of somatic mutations in ALL are also inactivated by aberrant methylation, suggesting that multiple mechanism of silencing of critical genes may contribute to leukemic transformation. Mutations in histone writers, erasers and readers are more frequent in T–ALL than in most other pediatric cancers.43 Also, several studies show that various MLL fusion proteins characteristically modulate chromatin structure through histone modifications; thus MLL–rearranged leukemia is considered an epigenetic malignancy.13, 44 Epigenetic can also influence chemoresistance in ALL as manifested by increased global promoter methylation at relapse.16 Therapy with demethylating agents led to re–expression of hypermethylated genes and restored chemosensitivity in an experimental model.45 The interplay among the altered epigenetic landscape and structural changes in the genome, and the development of epigenetic therapies such as histone deacetylase inhibitors and demethylating agents provides exciting opportunities for therapeutic interventions.
Biology of relapsed ALL
Studies of matched diagnosis–relapse samples have shed light on the clonal evolution leading to relapse, pathways associated with chemoresistance, and potential targets for therapy. In one study, 86% of the patients at relapse had outgrowth of a minor subclone present at diagnosis, which has genetic alterations both similar to and different from the major clone at diagnosis.46 In some patients, relapse may have genetic alterations either identical to, or entirely different from those seen at diagnosis. The latter scenario likely represents a second malignancy. Preclinical studies and clinical experience show that leukemic blasts are more resistant to various chemotherapeutic agents at relapse than at initial diagnosis.47, 48 Mechanisms of resistance may include selection of a pre–existing resistant subclone or the acquisition of additional genomic lesions under the selective pressure of chemotherapy. In a recent study, gain–of–function mutations in NT5C2 were identified in leukemic blasts of approximately 20% of patients at relapse.49, 50 NT5C2 encodes a 5’nucleotidase enzyme that catalyzes the inactivation of nucleoside analogs such as mercaptopurine and thioguanine. As mercaptopurine and methotrexate are the mainstay of the maintenance therapy for ALL, acquisition of the NT5C2 mutation can lead to emergence of drug–resistant clones and early relapse. Other genomic lesions at relapse include mutations in CREBBP (which mediates glucocorticoid response and histone acetylation)51 and focal deletions in the mismatch repair gene MSH652 and the glucocorticoid receptor NR3C1.53
Biology of the host
ALL susceptibility
Besides constitutional trisomy 21 (Down syndrome) and rare DNA damage repair defects (e.g. ataxia–telangiectasia and Bloom syndrome), little is known about the genetic predisposition to ALL.54 The frequency of hematologic malignancies is 4% in patients with common cancer predisposition syndromes such as Li–Fraumeni syndrome (caused by inherited mutations in TP53), approximately half of which are ALL.55 However, a study of hypodiploid ALL with whole genome and exome sequencing revealed that 91% of patients with low–hypodiploid ALL (32–39 chromosomes) harbored somatic TP53 alterations.12 In 43% of these patients, TP53 mutations were also present in non–tumor DNA, indicating a previously unrecognized link of Li–Fraumeni syndrome to low-hypodiploid ALL and implications for genetic counseling. Recently, whole exome sequencing identified a new familial leukemia syndrome in kindreds harboring a novel germline variant in PAX5 on chromosome 9.56 PAX5 is a lymphoid transcription factor that plays a crucial role in B–cell development and is a common target for submicroscopic deletion in B–ALL blasts. Leukemic samples from family members with ALL showed 9p deletion with loss of heterozygosity and retention of the mutant PAX5 allele, leading to significantly reduced transcriptional activity and perhaps leukemic transformation.
In the search for common, inherited ALL susceptibility variants, several large genome–wide association studies (GWAS) have identified polymorphisms in ARID5B, IKZF1, CEBPE, CDKN2A and PIP4K2A–BMI1 were over–represented in patients with ALL compared with non– ALL controls.57–59 There were significant differences in the prevalence of these variants among patients of different ancestries, possibly contributing to racial differences in the incidence of ALL. Each of these variants account for a modest increase in the risk of developing ALL (odds ratio 1.5–2), but they independently and cumulatively contribute to genetic susceptibility to ALL.58 CDKN2A and IKZF1 are also targeted by somatic alterations in ALL,17 suggesting that both inherited and somatic genetic variations cooperate in the pathogenesis of ALL. Interestingly, the risk of developing various subtypes of ALL is also influenced by genetic inheritance. An intronic SNP in TP63 (a member of the TP53 family of transcription factors) conferred susceptibility to ETV6–RUNX1–positive ALL60 and SNPs in GATA3 conferred susceptibility to BCR–ABL1–like ALL and its underlying somatic lesions (IKZF1 deletions, CRLF2 rearrangements and JAK mutations).61
Toxicity
An individual’s genetic make–up can influence drug transport and metabolism and subsequently efficacy and toxicity. Thiopurine S–methyl transferase (TPMT) enzymatic activity is deficient in approximately 10% of individuals with polymorphisms in the TPMT gene. The reduced activity of TPMT leads to excessive cellular accumulation of active thiopurine metabolites and thereby excessive hematopoietic toxicity. In one study, the cumulative incidences of mercaptopurine–related myelosuppression were 100%, 35%, and 7% for patients with homozygous, heterozygous, and wild–type genotypes respectively.62 Tailoring the dose of mercaptopurine on the basis of TPMT genotype results in equivalent systemic exposure, tolerance, and efficacy, and is an excellent example of widely used pharmacogenetic–guided therapy in clinical care. Germline variants associated with steroid–induced osteonecrosis,63 vincristine–induced peripheral neuropathy,64 anthracycline–induced cardiotoxicity65 and asparaginase allergy66 have also been identified by candidate gene and genome–wide studies. Validation of these associations is warranted to determine their clinical relevance.
Response
Inherited genetic variation can contribute to inter–patient variability in ALL treatment response, by influencing host disposition of anti–leukemic agents, interactions between tumor– microenvironment and ALL, and tumor biology itself. At a genome–wide level, a study of more than 400,000 host germline polymorphisms in 487 children with ALL, identified 102 SNPs (representing 71 unique genomic loci) that were significantly associated with MRD at the end of remission induction therapy67. Twenty percent of these SNP genotypes associated with high MRD were related to decreased exposure to methotrexate and etoposide, either because of increased clearance of these agents or decreased intracellular accumulation of active methotrexate polyglutamates. These findings underscore the effect of host genetic makeup on the response to multiple agents. A subsequent GWAS of 2,535 children with ALL identified 134 SNPs consistently associated with outcome, most of which remained prognostic even after adjusting for known risk factors (ex. MRD, molecular subtype). In particular, risk variants in PDE4B predisposed patients to relapse, plausibly via affecting methotrexate pharmacodynamics and tumor sensitivity to steroids.68
The outcome of Hispanic children with ALL is historically inferior to that of patients of European descent. This racial disparity may in part be due to increased frequency of germline variants associated with Native American ancestry in Hispanic patients which negatively influence MRD and relapse.69 Patients with Hispanic ethnicity are also predisposed to developing high–risk subtypes of ALL (e.g. GATA3 germline polymorphism in BCR–ABL1–like ALL).61 Importantly, the addition of an extra block of delayed intensification therapy for Hispanic children seemed to mitigate the ancestry–related difference in outcome.69
Summary
Modern–day management of childhood ALL exemplifies the successful integration of biology into therapeutic decision making. In addition to the prognostic impact of conventional chromosomal translocations and aneuploidy, functional studies of key genetic alterations have contributed to our understanding of ALL pathogenesis. With the advent of high–throughput genomics and NGS technologies, knowledge of specific molecular lesions and critical pathways of leukemogenesis has exponentially increased. The incorporation of targeted therapy is expected to improve outcome for high–risk patients, particularly patients with BCR–ABL1–positive ALL and those with the novel BCR–ABL1–like ALL subtype. In addition to genomic lesions, alterations in the epigenome modulate gene expression and contribute significantly to leukemic transformation and resistance to therapy. Therefore, epigenetic therapy is another strategy being actively pursued in the clinic. A deeper understanding of the effect of inherited genetic variations can provide the opportunity to modify therapy to decrease toxicity without compromising efficacy. Furthermore, strong associations have been identified between inherited genetic variants and ALL susceptibility. Many of these polymorphisms are in genes that are targets of somatic lesions in ALL, highlighting plausible interactions between the biology of the disease and the host (Figure 2).
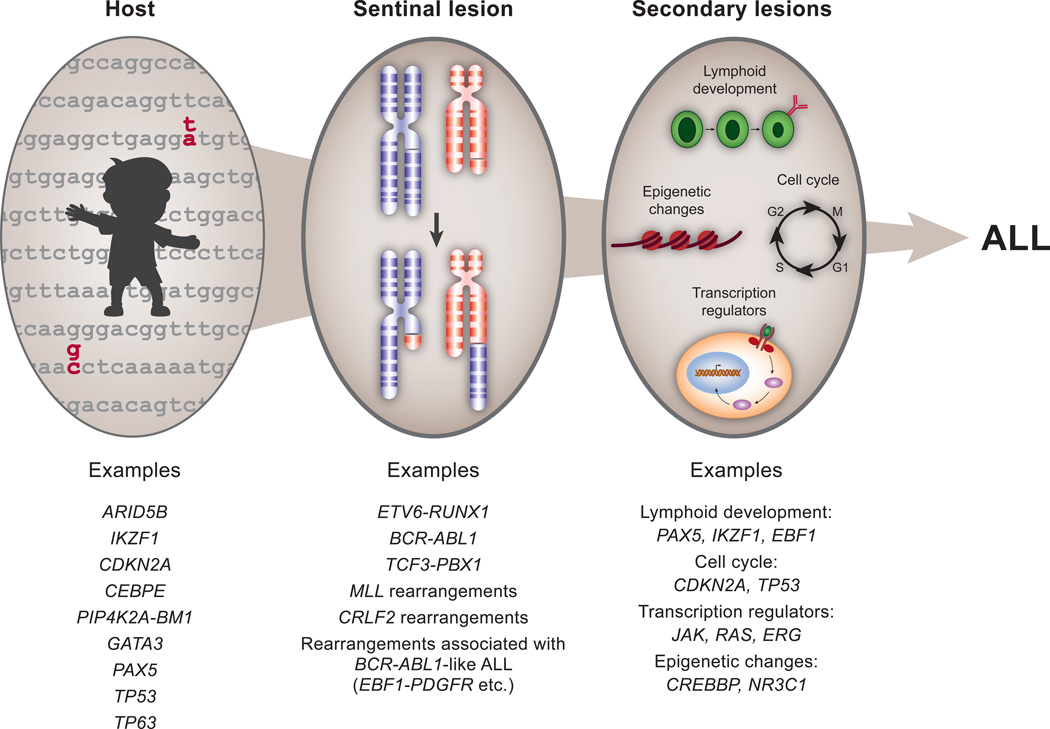
Figure 2. Pathogenesis of ALL.
Several inherited variants associated with susceptibility to ALL have been identified by genome– wide association studies and studies of familial ALL. Within hematopoietic cells, in addition to a sentinel event such as a chromosomal translocation, a multitude of secondary genetic events contribute to leukemic transformation.
Key points.
Childhood ALL is a heterogeneous disease with multiple distinct biologic subtypes.
High-throughput genomic profiling and next–generation sequencing technologies have identified submicroscopic genomic lesions and sequence mutations that define novel subtypes of ALL.
The discovery of various oncogenic pathways and candidate genes has led to the development of biologically based targeted therapy.
Host germline polymorphisms influence susceptibility to ALL, chemotherapy-related toxicities and response to therapy.
Acknowledgements
The authors thank Klo Spelshouse (Department of Biomedical Communications, St Jude Children’s Research Hospital) for assistance with illustrations and Vani Shanker (Department of Scientific Editing, St Jude Children’s Research Hospital) for assistance with editing the manuscript.
Funding: National Institutes of Health grant P30-CA021765 and the American Lebanese Syrian Associated Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Deepa Bhojwani, Email: deepa.bhojwani@stjude.org.
Jun J. Yang, Email: jun.yang@stjude.org.
Ching-Hon Pui, Email: ching-hon.pui@stjude.org.
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Evans WE. A 50-year journey to cure childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:185–196. doi: 10.1053/j.seminhematol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunger SP, Lu X, Devidas M, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. J Clin Oncol. 2012;30:1663–1669. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz KR, Carroll A, Heerema NA, et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children's Oncology Group study AALL0031. Leukemia. 2014;28:1467–1471. doi: 10.1038/leu.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pui CH, Yang JJ, Hunger SP, et al. Childhoof Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol. 2014 doi: 10.1200/JCO.2014.59.1636. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 7.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children's Cancer Group (CCG) Blood. 2007;109:926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360:2730–2741. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moorman AV, Chilton L, Wilkinson J, Ensor HM, Bown N, Proctor SJ. A population-based cytogenetic study of adults with acute lymphoblastic leukemia. Blood. 2010;115:206–214. doi: 10.1182/blood-2009-07-232124. [DOI] [PubMed] [Google Scholar]
- 10.Jeha S, Pei D, Raimondi SC, et al. Increased risk for CNS relapse in pre-B cell leukemia with the t(1;19)/TCF3-PBX1. Leukemia. 2009;23:1406–1409. doi: 10.1038/leu.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachman JB, Heerema NA, Sather H, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110:1112–1115. doi: 10.1182/blood-2006-07-038299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmfeldt L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pieters R. Infant acute lymphoblastic leukemia: Lessons learned and future directions. Curr Hematol Malig Rep. 2009;4:167–174. doi: 10.1007/s11899-009-0023-4. [DOI] [PubMed] [Google Scholar]
- 14.Kim DH, Moldwin RL, Vignon C, et al. TEL-AML1 translocations with TEL and CDKN2 inactivation in acute lymphoblastic leukemia cell lines. Blood. 1996;88:785–794. [PubMed] [Google Scholar]
- 15.Figueroa ME, Chen SC, Andersson AK, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. J Clin Invest. 2013;123:3099–3111. doi: 10.1172/JCI66203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan LE, Meyer JA, Yang J, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 18.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virely C, Moulin S, Cobaleda C, et al. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010;24:1200–1204. doi: 10.1038/leu.2010.63. [DOI] [PubMed] [Google Scholar]
- 21.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase activating lesions in Ph-like acute lymphoblastic leukemia of BCR-ABL1-like acute lymphoblastic leukemia. N Engl J Med. 2014 doi: 10.1056/NEJMoa1403088. (in press): abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weston BW, Hayden MA, Roberts KG, et al. Tyrosine kinase inhibitor therapy induces remission in a patient with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol. 2013;31:e413–e416. doi: 10.1200/JCO.2012.47.6770. [DOI] [PubMed] [Google Scholar]
- 24.Roberts KG, Pei D, Campana D, et al. Outcomes of Children With BCR-ABL1-Like Acute Lymphoblastic Leukemia Treated With Risk-Directed Therapy Based on the Levels of Minimal Residual Disease. J Clin Oncol. 2014 doi: 10.1200/JCO.2014.55.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maude SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey RC, Kang H, Roberts AW, et al. Development and validation of a highly sensitive and specific gene expression classifier to prospectively screen and identify B-precursor acute lymphoblastic leukema patients with a Philadelphia Chromosome-like ("Ph-like" or BCR-ABL1-like") signature for therapeutic targeting and clinical intervention. Blood. 2013;122:826. [Google Scholar]
- 27.Rand V, Parker H, Russell LJ, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011;117:6848–6855. doi: 10.1182/blood-2011-01-329961. [DOI] [PubMed] [Google Scholar]
- 28.Moorman AV, Robinson H, Schwab C, et al. Risk-directed treatment intensification significantly reduces the risk of relapse among children and adolescents with acute lymphoblastic leukemia and intrachromosomal amplification of chromosome 21: a comparison of the MRC ALL97/99 and UKALL2003 trials. J Clin Oncol. 2013;31:3389–3396. doi: 10.1200/JCO.2013.48.9377. [DOI] [PubMed] [Google Scholar]
- 29.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 30.Buitenkamp TD, Izraeli S, Zimmermann M, et al. Acute lymphoblastic leukemia in children with Down syndrome: a retrospective analysis from the Ponte di Legno study group. Blood. 2014;123:70–77. doi: 10.1182/blood-2013-06-509463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen K, Devidas M, Cheng SC, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children's Oncology Group study. Leukemia. 2008;22:2142–2150. doi: 10.1038/leu.2008.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 34.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 36.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 37.Thompson BJ, Buonamici S, Sulis ML, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Real PJ, Tosello V, Palomero T, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 40.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patrick K, Wade R, Goulden N, et al. Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014;166:421–424. doi: 10.1111/bjh.12882. [DOI] [PubMed] [Google Scholar]
- 43.Huether R, Dong L, Chen X, et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun. 2014;5:3630. doi: 10.1038/ncomms4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 45.Bhatla T, Wang J, Morrison DJ, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood. 2012;119:5201–5210. doi: 10.1182/blood-2012-01-401687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klumper E, Pieters R, Veerman AJ, et al. In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood. 1995;86:3861–3868. [PubMed] [Google Scholar]
- 48.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14:e205–e217. doi: 10.1016/S1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 49.Meyer JA, Wang J, Hogan LE, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzoneva G, Perez-Garcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang JJ, Bhojwani D, Yang W, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuster L, Grausenburger R, Fuka G, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–2667. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- 54.Seif AE. Pediatric leukemia predisposition syndromes: clues to understanding leukemogenesis. Cancer Genet. 2011;204:227–244. doi: 10.1016/j.cancergen.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 55.Kleihues P, Schauble B, zur Hausen A, Esteve J, Ohgaki H. Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol. 1997;150:1–13. [PMC free article] [PubMed] [Google Scholar]
- 56.Shah S, Schrader KA, Waanders E, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trevino LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu H, Yang W, Perez-Andreu V, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105:733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellinghaus E, Stanulla M, Richter G, et al. Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia. 2012;26:902–909. doi: 10.1038/leu.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Andreu V, Roberts KG, Harvey RC, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45:1494–1498. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Relling MV, Hancock ML, Rivera GK, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst. 1999;91:2001–2008. doi: 10.1093/jnci/91.23.2001. [DOI] [PubMed] [Google Scholar]
- 63.French D, Hamilton LH, Mattano LA, Jr, et al. A PAI-1 (SERPINE1) polymorphism predicts osteonecrosis in children with acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2008;111:4496–4499. doi: 10.1182/blood-2007-11-123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diouf B, Crews K, Lew G, et al. Genome-wide association analysis identify susceptibility loci for vincristine-induced peripheral neuropathy in children with acute lymphoblastic leukemia. Blood. 2013;122:618. ASH annual meeting abstract. [Google Scholar]
- 65.Wang X, Liu W, Sun CL, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children's oncology group. J Clin Oncol. 2014;32:647–653. doi: 10.1200/JCO.2013.50.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez CA, Smith C, Yang W, et al. HLA-DRB1*07:01 is associated with a higher risk of asparaginase allergies. Blood. 2014 doi: 10.1182/blood-2014-03-563742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. Jama. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang JJ, Cheng C, Devidas M, et al. Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood. 2012;120:4197–4204. doi: 10.1182/blood-2012-07-440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang JJ, Cheng C, Devidas M, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43:237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 71.Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120:1165–1174. doi: 10.1182/blood-2012-05-378943. [DOI] [PMC free article] [PubMed] [Google Scholar]