Abstract
Oxidative stress is implicated in the development of non-alcoholic steatohepatitis (NASH). The Nrf2-antioxidant response element pathway protects cells from oxidative stress. Studies have shown that global Nrf2 deficiency hastens the progression of NASH. The purpose of this study was to determine whether long-term hepatocyte-specific activation of Nrf2 mitigates NASH progression. Transgenic mice expressing a constitutively active Nrf2 construct in hepatocytes (AlbCre+/caNrf2+) and littermate controls were generated. These mice were fed standard or methionine-choline-deficient (MCD) diet, a diet used to induce NASH development in rodents. After 28 days of MCD dietary feeding, mice developed significant increases in steatosis, inflammation, oxidative stress, and HSC activation compared with those mice on standard diet. AlbCre+/caNrf2+ animals had significantly decreased serum transaminases and reduced steatosis when compared with the AlbCre+/caNrf2− animals. This significant reduction in steatosis was associated with increased expression of genes involved in triglyceride export (MTTP) and β-oxidation (CPT2). However, there were no differences in the increased oxidative stress, inflammation, and HSC activation from MCD diet administration between the AlbCre+/caNrf2− and AlbCre+/caNrf2+ animals. We conclude that hepatocyte-specific activation of Nrf2-mediated gene expression decreased hepatocellular damage and steatosis in a dietary model of NASH. However, hepatocyte-specific induction of Nrf2-mediated gene expression alone is insufficient to mitigate inflammation, oxidative stress, and HSC activation in this nutritional NASH model.
Keywords: liver, Nrf2, antioxidants, steatosis, NASH, transgenic mice
Obesity is one of the major health epidemics facing the world today. More than one-third of the U.S. adult population is considered obese with body mass index greater than 30 kg/m2 (Flegal et al., 2012). In the liver, obesity leads to non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH). The prevalence of NAFLD has been estimated to be between 15% and 50%, and approximately 30% of patients with NAFLD will eventually progress into NASH (Adams and Lindor, 2007; Farrell and Larter, 2006).
Although NAFLD and NASH have been recognized as two of the most common liver disorders, their pathogenesis is complex and remains to be elucidated. The current thought regarding the development of NAFLD and NASH involves the “two-hit” hypothesis. With caloric excess, there is increased adiposity, insulin resistance, and development of hepatic steatosis, or NAFLD. This eventually leads to increased lipotoxic metabolites and increased β-oxidation, which results in increased oxidative stress and lipid peroxidation. These processes lead to a second hit, resulting in inflammation and the development of NASH (Rolo et al., 2012). There is currently no standard therapy to decrease the progression of NAFLD to NASH and cirrhosis.
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a basic leucine zipper transcription factor with a Cap “n” Collar structure that is key to the cellular antioxidative response. Nrf2 is normally bound in the cytoplasm to the Keap1 protein via the Neh2 domain on the N-terminal of Nrf2. Keap1 acts as a negative regulator of Nrf2 by targeting Nrf2 toward ubiquitination and degradation (Zhang and Hannink, 2003). The presence of oxidative stress leads to modification of Keap1’s cysteine thiols, resulting in the weakening of the Nrf2-Keap1 interaction, which allows newly synthesized Nrf2 to be translocated into the nucleus (Wakabayashi et al., 2004). Once in the nucleus, Nrf2 binds the antioxidant response element (ARE), and initiates the transcription and translation of several antioxidative and other cytoprotective genes (Lee et al., 2003).
Previous experiments have demonstrated that the global absence of Nrf2 leads to exacerbated development of NASH in a dietary-induced murine model of NASH (Chowdhry et al., 2010; Sugimoto et al., 2010; Tanaka et al., 2008). In addition, globally enhanced expression of Nrf2 leads to a reduction in the onset of fatty liver disease in mice fed a methionine and choline deficient (MCD) diet (Zhang et al., 2010). However, less is known regarding the effect of cell-specific Nrf2 overexpression in NASH development. Given that hepatocyte are primarily responsible for lipid metabolism and detoxification, we wanted to determine whether induction of the Nrf2-ARE pathway selectively in hepatocytes would mitigate NASH development similarly to that seen in the other published studies that utilized mice with global Nrf2 overexpression.
To achieve this aim, we used transgenic mice expressing a constitutively active Nrf2 (caNrf2) mutant that lacks the N-terminal Neh2 domain under control of a ubiquitously active β-actin promoter and a CMV enhancer (Schafer et al., 2012). Due to the lack of the Keap1-binding domain, caNrf2 is stabilized and continuously translocates to the nucleus, resulting in overexpression of Nrf2 target genes. Upon mating these mice with transgenic mice expressing Cre under the control of the hepatocyte-specific albumin promoter (AlbCre), we obtained mice with hepatocyte-specific overactivation of the Nrf2-ARE pathway (Kohler et al., 2014).
The MCD diet was used to establish NASH in our murine model. The MCD diet is a well-established experimental model of NASH development in rodents. It rapidly generates the clinical histologic features of NASH, including steatosis, ballooning degeneration, focal inflammation, hepatic necrosis, and fibrosis (Rinella et al., 2008). The pathological features of NASH may be seen in as short as 2–5 days of diet (Zhang et al., 2010). The mechanism for which MCD is thought to lead to NASH development is partly due to choline deficiency. This leads to decreased generation of phosphatidylcholine, which is required for generation of very low-density lipoprotein (VLDL) for triglyceride export from the liver. Methionine deficiency also leads to decreased generation of S-adenosylmethionine and glutathione, which results in increased oxidative stress and inflammation, and simulates the second hit of the “two-hit” hypothesis (Caballero et al., 2010; Rinella et al., 2008).
The objectives of this study were to determine whether long-term, cell-specific activation of the Nrf2-ARE pathway in hepatocytes could decelerate the progression of NASH. More specifically, we assessed whether this hepatocellular expression of caNrf2 leads to a reduction in steatosis, oxidative stress, inflammation, and hepatic stellate cell activation in mice subjected to nutritionally induced NASH.
MATERIALS AND METHODS
Animals
Mice expressing a constitutively active Nrf2 (caNrf2) under the control of a CMV promoter and β-actin enhancer (Schafer et al., 2012) in hepatocytes postnatally have previously been described (Kohler et al., 2014). To avoid constitutive expression of the transgene in all cells, the caNrf2 cDNA is preceded by a floxed transcription/translation STOP cassette. Upon mating of these mice with transgenic mice expressing Cre in hepatocytes under control of the albumin promoter (AlbCre mice) (Jackson Labs, Bar Harbor, Maine), the STOP cassette is excised and caNrf2 is expressed. The transgenic mice were shown to express the caNrf2 protein at similar levels as the endogenous protein in the liver (Kohler et al., 2014). Since the caNrf2 protein is constitutively present in the nucleus, it efficiently induces expression of Nrf2 target genes, resulting in their upregulation. Importantly, levels of caNrf2 and endogenous Nrf2 protein were similar in the livers of these mice, indicating that the induction of Nrf2 target gene expression by the transgene is comparable to the induction seen upon pharmacological activation of the endogenous protein (Kohler et al., 2014).
Transgenic mice (age 6–8 weeks) with hepatocyte-specific expression of caNrf2 (AlbCre+/caNrf2+) and littermate controls (AlbCre+/caNrf2−) were fed either standard chow (8604 Teklad Rodent Diet; Harlan Teklad, Madison, Wisconsin) or MCD diet (Cat No. 0296043910; MP Biomedical, Solon, Ohio). Among the AlbCre+/caNrf2− mice, 6 were fed standard chow and 7 were fed the MCD diet. Among the AlbCre+/caNrf2+ mice, 7 were fed standard chow and 8 were fed the MCD diet. All mice had access to diet and water ad libitum. After 28 days of diet, livers and serum were obtained for analyses. All experiments were performed according to the ethical guidelines outlined in the Guide for Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee at the University of Wisconsin School of Medicine and Public Health.
Histology
The murine livers were flushed with PBS at the time of sacrifice. Fresh liver specimens were then divided equally; snap freezing a portion in O.C.T for subsequent lipid staining, and the remaining tissue fixed in 10% neutral buffered formalin for further processing into paraffin blocks.
Oil Red O staining
Frozen mouse liver specimens were sectioned at a thickness of 5 µ, fixed with 10% neutral buffered formalin for 5 min and subsequently stained for lipid deposition. Lipids were detected using an Oil Red O (ORO) stain, consisting of 1 g ORO dye dissolved in 100 ml 70% EtOH/acetone mixture. Sections underwent an acclimation rinse in 70% EtOH, followed by 10 min incubation in working ORO solution. The sections were then differentiated in 70% EtOH before being counterstained with hematoxylin for contrast.
Immunohistochemistry
Paraffin-embedded mouse livers were sectioned at a thickness of 5 µ and either stained with routine Hematoxylin and eosin (H&E) or immunohistochemistry was performed using rabbit anti-α-SMA (Epitomics, Burlingame, California) or rabbit anti-F4/80 (eBioscience, San Diego, California) primary antibodies. For α-SMA staining, tissue sections underwent antigen retrieval using citrate buffer pH 6.0 in a decloaking chamber (Biocare Medical, Concord, California), followed by incubation with anti-α-SMA (1:8000) for 1 h at room temperature. For F4/80, enzyme-induced epitope retrieval using proteinase K was performed for 5 min at room temperature, followed by incubation with anti-F4/80 (1:40) for 1 h at room temperature. For both, antibody detection was conducted using ImmPRESS anti-rabbit Ig HRP (Vector Labs, Burlingame, California), the signal visualized with DAB, and sections counterstained with hematoxylin for contrast.
For α-CD3 and α-Ly6G immunohistochemistry, the sections were initially incubated in 12% bovine serum albumin in phosphate-buffered saline (PBS)/0.025% NP40 for 1 h for blocking. The sections were then incubated overnight at 4°C with primary antibodies α-Ly6G (BD Biosciences) and α-CD3 (DAKO) diluted in blocking solution. After washing in PBS, sections were stained using the ABC Vectastain Peroxidase Kit and the AEC Peroxidase Substrate Kit (Vector Laboratories) as described by the manufacturer. Sections were counterstained with hematoxylin and mounted with Mowiol.
Image analysis
The slides for each animal were visualized under light microscopy using Axiovert 200M microscope (Carl Zeiss, Gottingen, Germany). Four random high-powered fields of view were taken for each animal.
Serum analysis
Whole blood was collected at the time of sacrifice. Blood was allowed to coagulate at room temperature for at least 30 min and then spun down to collect the serum. Serum ALT and AST were measured using the IDEXX VetTest Chemistry Analyzer (IDEXX, Westbrook, Maine). Serum triglyceride content was measured using Wako L-Type TG M Assay kit (Wako Chemicals USA, Richmond, Virginia) following the protocol provided in the kit.
Tissue triglyceride
Liver tissue triglyceride was extracted using the Folch method (Folch et al., 1957). Briefly, approximately 25 mg of liver was homogenized in a 2:1 chloroform:methanol mixture. Water was then added to the mixture and phase extraction performed. The organic layer was isolated and subjected to speed vacuum. The resulting pellet was then resuspended in isopropanol. The triglyceride content was determined using Wako L-Type TG M Assay kit (Wako Chemicals USA) and the protocol provided in the kit. Triglyceride content was normalized to the weight of the liver tissue used.
Thiobarbituric acid reactive substances
Thiobarbituric acid reactive substances (TBARS) in the liver tissue were determined via commercially available TBARS Assay kit (Cayman, Ann Arbor, Michigan). Briefly, approximately 25 mg of liver was homogenized in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitor. The resulting supernatant was then used for the assay following the protocol provided in the kit. Fluorescence for the samples was read at an excitation wavelength of 530 nm and an emission wavelength of 550 nm using SpectraMax M2 Microplate Reader (Molecular Devices, Sunnyvale, California). TBARS content was normalized to the weight of the liver tissue used.
8-Isoprostane
The hepatic 8-isoprostane levels were determined using the 8-Isoprostane EIA kit (Cayman) and the protocol provided in the kit. Briefly, approximately 100 mg of liver tissue from each animal was homogenized in 0.1 M phosphate, 1 mM EDTA, and 0.005% butylated hydroxytoluene (BHT), pH 7.4 solution. Total 8-isoprostane was obtained by adding 1 vol of 15% KOH to each of the sample, incubated at 40°C for 60 min, followed by neutralization using 3 vol of 1 M potassium phosphate buffer, pH 7.4. Fifty microliter of each sample was used for the assay in duplicates. 8-Isoprostane AChE tracer was then added to the sample wells and allowed to incubate overnight at 4°C. After rinsing with Wash Buffer 5 times, Ellman’s Reagent was added to the wells, and the reaction mixture was allowed to develop at room temperature for 90 min. Absorbance at 412 nm was then read using SpectraMax M2 Microplate Reader (Molecular Devices). 8-Isoprostane levels were normalized to the weight of the liver tissue used.
Quantitative RT–PCR
A small piece of liver tissue was initially stored in RNAlater (Life Technologies, Carlsbad, California) at the time of sacrifice. RNA was subsequently extracted using TRIzol Reagent (Life Technologies) per commercial protocol. RNA quality and quantity was assessed on an Agilent 2100 Bioanalyzer. Samples with RNA integrity number greater than 8.0 were used in subsequent studies. Synthesis single-stranded cDNA was performed using the Promega Reverse Transcription System (Promega, Madison, Wisconsin). Quantitative RT-PCR was then performed using the Bullseye EvaGreen qPCR mix (MidSci, St Louis, Missouri) in Roche LightCycler 480 (Roche Diagnostics, Mannheim, Germany). The primer sequences used are listed in the Supplementary Material.
Western blot analysis
Three random animals from each experimental group were used for western analysis. Whole-cell protein was obtained from frozen liver tissue homogenized in RIPA buffer. Protein concentration was determined via BCA protein Assay (Thermo Scientific Pierce, Rockford, IL). Ten micrograms of protein was denatured in Laemmli loading buffer at 95°C for 5 min. The samples were then loaded on 12% acrylamide gels. Gels were transferred to PVDF membranes and probed overnight with anti-NQO1 (Epitomics), anti-GCLM (provided by Dr Terry Kavanagh, University of Washington, Seattle, WA), anti-GstA2, anti-MTTP (Abcam), anti α-SMA (Abcam), or anti-β-actin (Abcam) antibodies. Blots were probed with anti-rabbit HRP or anti-mouse HRP (KPL, Gaithersburg, Maryland) as appropriate for 1 h and developed with Pierce ECL Western Blotting Substrate (Thermo Scientific Pierce). Densitometry for bands of interest was performed using ImageJ (Rasband, 1997–2012).
Statistics
All data in figures are presented as mean ± SEM. Comparisons between the groups were performed with one-way ANOVA followed by Fisher’s LSD post hoc test, unless stated otherwise in the figure legends. Log transformation was performed as appropriate as determined by Levene’s test for equality of variance. P < 0.05 was used for statistical significance. All statistical tests were performed using IBM SPSS Statistics version 21.
RESULTS
AlbCre+/caNrf2+ Mice Show Enhanced Nrf2-Dependent Gene and Protein Expression When Compared With Control Mice
Transgenic mice with hepatocyte-specific Nrf2 overactivation (AlbCre+/caNrf2+) and their littermate controls without the constitutively active Nrf2 construct (AlbCre+/caNrf2−) were fed a standard chow or MCD diet for 28 days. Six AlbCre+/caNrf2− mice received standard chow, and 7 AlbCre+/caNrf2− mice received MCD diet. Seven AlbCre+/caNrf2+ mice received standard chow, and 8 AlbCre+/caNrf2+ mice received MCD diet. Unless stated otherwise, all subsequent analyses were performed on these groups of animals.
To confirm that the AlbCre+/caNrf2+ mice had increased activity of Nrf2 at baseline compared with littermate controls, we examined the expression of Nrf2-dependent genes in the livers of the AlbCre+/caNrf2+ mice. RNA was isolated from the liver of the experimental animals, and qRT-PCR was performed for the prototypical Nrf2-dependent genes. The results are listed in Table 1. Baseline mRNA abundance of GSTM1, GSTA2, GSTA4, and NQO1 were significantly higher in AlbCre+/caNrf2+ animals on standard diet when compared with littermate controls, consistent with previously reported data (Kohler et al., 2014). The greatest increases were seen with NQO1 (11.4-fold) and GSTA2 (13.0-fold), while there was an approximate 50% decrease in GCLM message at baseline in the AlbCre+/caNrf2+ animals. Among the animals receiving MCD diet, there were significant increases in all of the Nrf2-dependent genes tested in the AlbCre+/caNrf2+ animals when compared with their littermate controls. When AlbCre+/caNrf2− animals were fed an MCD diet, there were significant increases in GSTA2 (2.56-fold) and NQO1 (2.06-fold). In comparison, for AlbCre+/caNrf2+ animals fed a MCD diet, there were significant increases in all of the prototypical Nrf2-dependent genes.
TABLE 1.
Nrf2-Dependent Gene Expression
| Genes |
AlbCre+/caNrf2- |
AlbCre+/caNrf2+ |
Dietary Effect |
Genetic Effect |
||||
|---|---|---|---|---|---|---|---|---|
| Std Chow | MCD | Std Chow | MCD | AlbCre+/caNrf2- | AlbCre+/caNrf2+ | Std Chow | MCD | |
| Columns | 1 | 2 | 3 | 4 | 1 versus 2 | 3 versus 4 | 1 versus 3 | 2 versus 4 |
| GCLC | 1.00 ± 0.08 | 0.55 ± 0.11* | 1.35 ± 0.23 | 3.67 ± 0.31*, ** | 0.55 | 2.72 | — | 6.66 |
| GCLM | 1.00 ± 0.10 | 0.48 ± 0.07* | 0.52 ± 0.08** | 0.96 ± 0.09*, ** | 0.48 | 1.85 | 0.52 | 1.97 |
| GSTM1 | 1.00 ± 0.11 | 0.69 ± 0.08 | 3.68 ± 1.47** | 7.38 ± 1.72*, ** | — | 3.26 | 2.26 | 10.7 |
| GSTA2 | 1.00 ± 0.21 | 2.56 ± 0.36* | 13.0 ± 3.87** | 32.7 ± 4.93*, ** | 2.56 | 2.51 | 13.0 | 12.8 |
| GSTA4 | 1.00 ± 0.16 | 1.32 ± 0.14 | 1.89 ± 0.16** | 3.38 ± 0.45*, ** | — | 1.79 | 1.89 | 2.57 |
| NQO1 | 1.00 ± 0.08 | 2.06 ± 0.22* | 11.4 ± 1.95** | 21.3 ± 2.91*, ** | 2.06 | 1.87 | 11.4 | 10.3 |
RNA was isolated from the collected liver sample, and qRT-PCR was performed for the prototypical Nrf2-dependent genes. The mRNA levels are first normalized to the β-actin mRNA content of each sample, then each group is normalized to the AlbCre+/caNrf2−, standard chow group. All data are presented as mean ± SEM. N = 6–8 for each of the 4 groups. The fold changes in mRNA abundance were compared between groups using one-way ANOVA with Fisher’s LSD post hoc test. Significant fold changes are presented on the right side of the table. *P < 0.05 when compared with standard chow of the same genotype. **P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
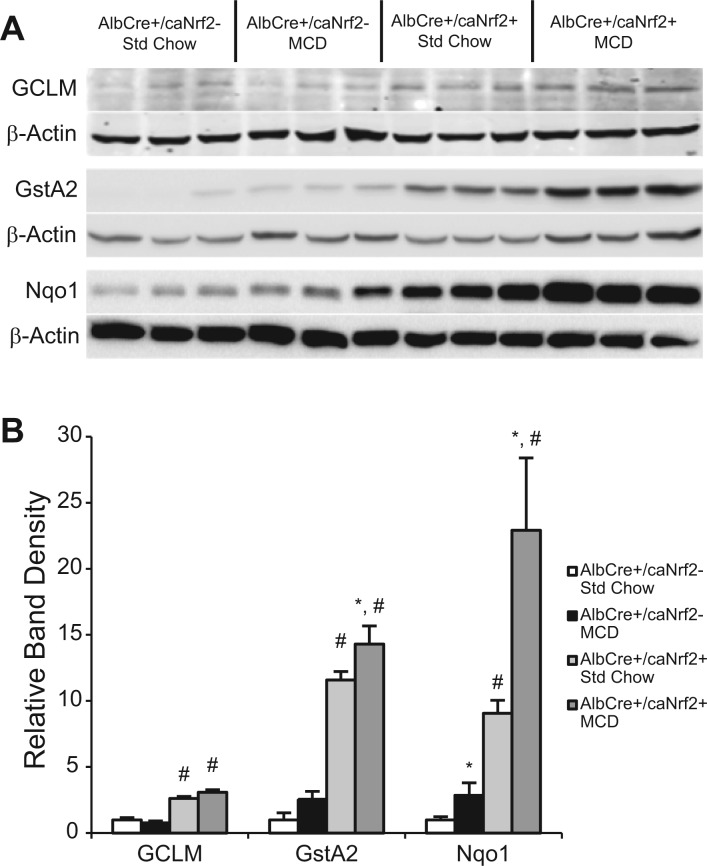
We then used 3 random animals from each experimental group for Western blot to determine the protein expression of the prototypical Nrf2-dependent genes (Fig. 1). At baseline with standard chow, there were significant increases in GCLM, GstA2, and Nqo1 in AlbCre+/caNrf2+ animals when compared with controls. This increase in GCLM protein was seen in spite of the lower abundance of GCLM mRNA. When AlbCre+/caNrf2− animals were fed MCD diet compared with standard chow, there was significant increase in Nqo1 protein, and no significant changes in GCLM and GstA2. For the AlbCre+/caNrf2+ animals, MCD feeding resulted in significant increases in GCLM, GstA2, and Nqo1 protein when compared with standard chow. Among the animals receiving a MCD diet, there were significant increases in GCLM, GstA2, and Nqo1 in the AlbCre+/caNrf2+ animals compared with littermate controls. Taken together, the mRNA and protein data suggest that there are increases in several Nrf2-dependent proteins in the AlbCre+/caNrf2+ animals at baseline without any stress stimulus. Independent of the mouse genotype, the MCD diet activates the Nrf2-ARE pathway, resulting in increased expression of some Nrf2-dependent proteins. However, in the AlbCre+/caNrf2+ animals, MCD feeding leads to further augmentation of Nrf2-dependent gene and protein expression when compared with their littermate controls.
FIG. 1.
Western blot analysis of Nrf2 target proteins. A, After 28 days of diet, Western blot of 3 random animals from each experimental group was performed for the prototypical Nrf2-dependent proteins: GCLM, GstA2, and Nqo1. B, Densitometry of the presented blot demonstrated significant increases in the examined Nrf2-dependent proteins in the AlbCre+/caNrf2+ animal when compared with AlbCre+/caNrf2− animals independent of diet. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Constitutive Activation of Nrf2 in Hepatocytes Decreases Hepatocellular Damage from MCD Diet
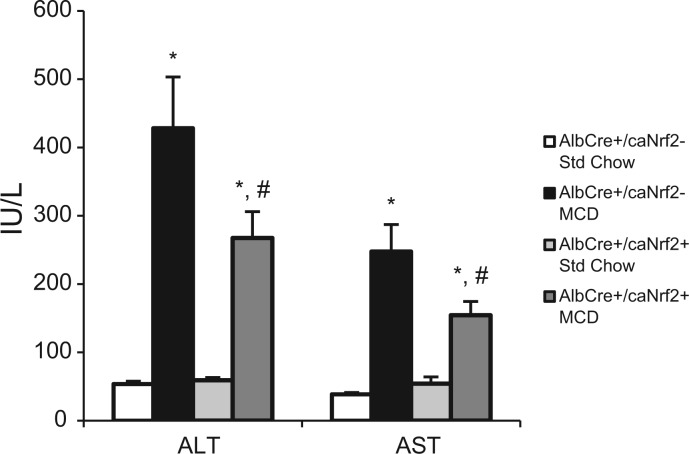
The transgenic mice were fed 28 days of standard chow versus MCD diet. Blood serum was collected at the time of sacrifice, and serum ALT and AST were measured to assess for clinical evidence of hepatocellular damage. After 28 days of MCD diet, AlbCre+/caNrf2− and AlbCre+/caNrf2+ mice had significantly elevated serum transaminases when compared with those on standard chow. However, AlbCre+/caNrf2+ mice had an approximate 40% decrease in serum transaminase levels when compared with AlbCre+/caNrf2− mice on MCD diet, which was statistically significant (Fig. 2). This suggests that constitutive activation of Nrf2 in hepatocytes alone offers hepatocellular protection from the stress of MCD diet.
FIG. 2.
Serum ALT and AST. Serum was collected from AlbCre+/caNrf2+ and AlbCre+/caNrf2− animals at the end of 28 days of diet, and serum transaminases were measured. Animals on MCD diet have significantly elevated serum transaminases compared with animals on standard chow for both genotypes. However, AlbCre+/caNrf2+ animals have significantly decreased serum transaminases when compared with the AlbCre+/caNrf2− animals on MCD. N = 6–8 for each of the four groups. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Constitutive Activation of Nrf2 in Hepatocytes Leads to Decreased Hepatic Lipid Accumulation from MCD Diet
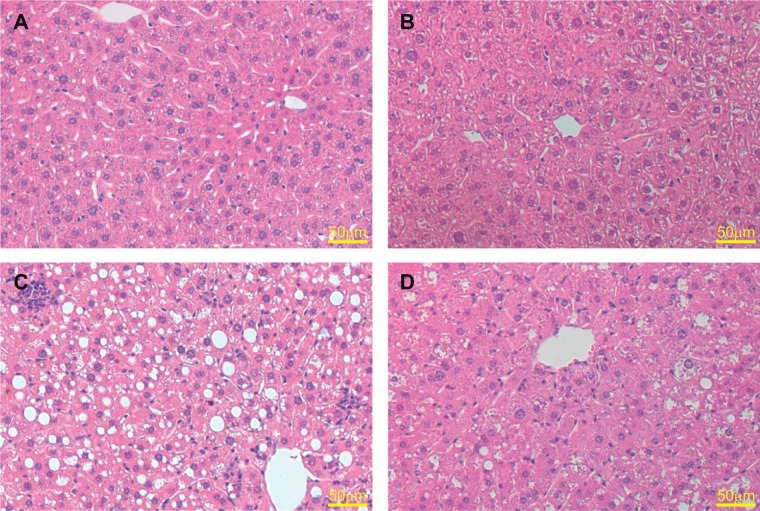
To further investigate the effect of constitutive activation of the Nrf2-ARE pathway on the development of steatosis, we performed histologic analyses on the liver tissue. H&E staining revealed normal appearing livers in mice receiving standard chow. We saw increased steatosis and inflammatory infiltration in the animals receiving MCD diet when compared with the standard chow animals. However, among the animals receiving MCD diet, marked increase in steatosis was seen in the livers of AlbCre+/caNrf2− mice (Fig. 3). There was a marked decrease in the percentage of hepatocytes with macrosteatosis in the AlbCre+/caNrf2+ animals on MCD diet in comparison. This protection against steatosis is seen most prominently around zone 3 on histology.
FIG. 3.
Representative hematoxylin and eosin stain of the livers after 28 days diet. Both A, AlbCre+/caNrf2− and B, AlbCre+/caNrf2+ animals on standard chow demonstrated normal liver histology. When fed an MCD diet, C, AlbCre+/caNrf2− animals exhibited widespread macrosteatosis, whereas D, AlbCre+/caNrf2+ animals demonstrated less steatosis in comparison. The area of protection against steatosis in AlbCre+/caNrf2+ animals appeared most prominently around zone 3.
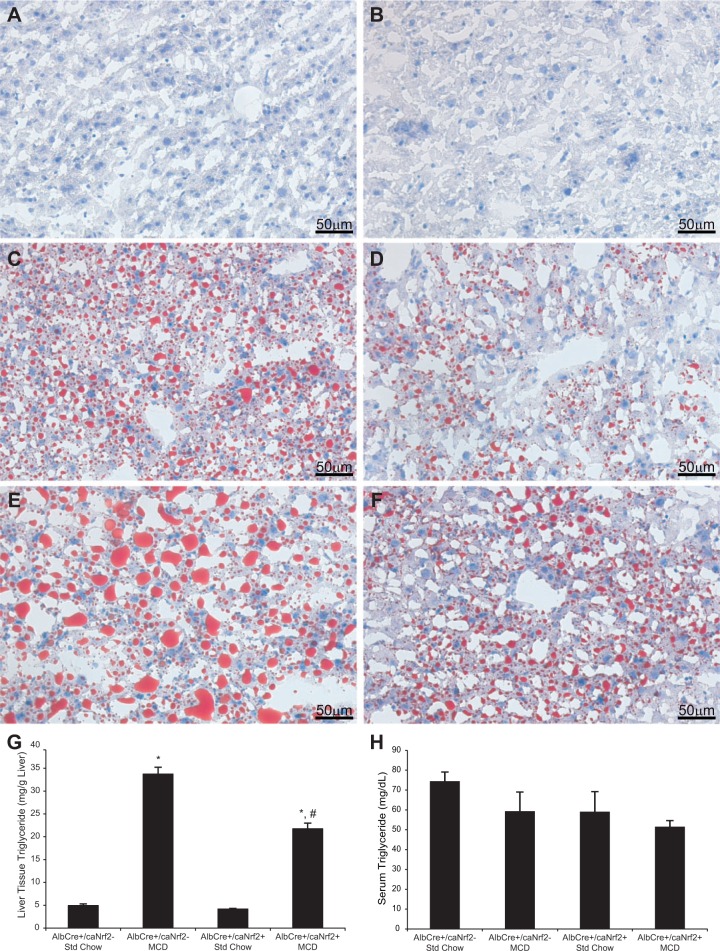
We then performed ORO staining to better characterize the extent of lipid deposition (Fig. 4). Although animals on standard chow have minimal lipid staining, there was increased lipid staining for animals on MCD diet. Among the animals receiving MCD diet, even though there was a range in the amount of steatosis present, there was in general a decrease in the amount of lipid staining in the livers from AlbCre+/caNrf2+ mice when compared with AlbCre+/caNrf2− animals.
FIG. 4.
Liver lipid deposition. Oil Red O staining was performed on the liver tissue at the end of the diet. AlbCre+/caNrf2− (A) and AlbCre+/caNrf2+ (B) animals on standard diet demonstrated minimal lipid staining. MCD diet increased lipid deposition in the livers. C, and E, demonstrate the range of lipid deposition in the AlbCre+/caNrf2− animals on MCD diet. D, and F, demonstrate the range of lipid deposition in the AlbCre+/caNrf2+ animals on MCD diet. In general, there was more lipid staining in the AlbCre+/caNrf2− animals on MCD diet when compared with the AlbCre+/caNrf2+ animals. G, Triglyceride was extracted from the liver tissue and its amount determined. MCD diet led to increased tissue triglyceride for both AlbCre+/caNrf2− and AlbCre+/caNrf2+ animals. However, the amount of triglyceride in AlbCre+/caNrf2+ animals on MCD is significantly decreased when compared with the AlbCre+/caNrf2− animals. This confirms the histological findings. H, Serum triglyceride level was obtained at the time of sacrifice. There were no differences in the serum triglyceride levels among the animals. N = 6–8 for each of the 4 groups. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
To quantify the amount of lipid deposition in the liver, we measured the amount of hepatic tissue triglyceride biochemically. Animals on MCD diet had significantly increased tissue triglyceride when compared with animals on standard chow. There was a significant, 35% reduction of hepatic tissue triglyceride in the AlbCre+/caNrf2+ animals when compared with littermate controls (Fig. 4G). We also measured the amount of serum triglyceride at the time of sacrifice. There were no significant differences in serum triglyceride levels among all 4 groups of animals (Fig. 4H).
Constitutive Activation of Nrf2 in Hepatocytes Alone Leads to Changes in the Expression Profile of Lipid Metabolism Genes When Fed a MCD Diet
To better understand the mechanisms underlying the effect of Nrf2-ARE activation on steatosis, we investigated the mRNA profiles of several lipid metabolism-related genes. Using RNA isolated from the animals’ liver samples, we performed qRT-PCR of lipid metabolic genes, and these results are presented in Table 2. At baseline with standard chow, there was a decrease in CPT1A mRNA in AlbCre+/caNrf2− animals. When fed a MCD diet, both groups of mice responded with significant decreases in almost all of the lipid metabolism genes tested. However, among the animals on MCD diet, AlbCre+/caNrf2+ mice demonstrated increased CPT2 and MTTP when compared with the AlbCre+/caNrf2− animals. There were no significant changes in mRNA abundance of the major lipid metabolic genes.
TABLE 2.
Expression of Genes Associated with Lipid Metabolism
| Genes | AlbCre+/caNrf2− |
AlbCre+/caNrf2+ |
Dietary Effect |
Genetic Effect |
||||
|---|---|---|---|---|---|---|---|---|
| Std Chow | MCD | Std Chow | MCD | AlbCre+/caNrf2- | AlbCre+/caNrf2+ | Std Chow | MCD | |
| Columns | 1 | 2 | 3 | 4 | 1 versus 2 | 3 versus 4 | 1 versus 3 | 2 versus 4 |
| Master regulators | ||||||||
| LXR | 1.00 ± 0.05 | 0.88 ± 0.16 | 0.90 ± 0.15 | 0.92 ± 0.17 | — | — | — | — |
| SREBF1 | 1.00 ± 0.14 | 0.50 ± 0.07* | 1.29 ± 0.24 | 0.54 ± 0.07* | 0.50 | 0.41 | — | — |
| PPARα | 1.00 ± 0.09 | 0.49 ± 0.08* | 0.85 ± 0.18 | 0.44 ± 0.09* | 0.49 | 0.52 | — | — |
| PPARγ | 1.00 ± 0.06 | 1.32 ± 0.21 | 1.35 ± 0.29 | 2.22 ± 0.39 | — | — | — | — |
| β-Oxidation | ||||||||
| ACOX1 | 1.00 ± 0.11 | 0.33 ± 0.05* | 0.82 ± 0.12 | 0.35 ± 0.04* | 0.33 | 0.43 | — | — |
| CPT1A | 1.00 ± 0.12 | 0.41 ± 0.02* | 0.68 ± 0.06** | 0.52 ± 0.06* | 0.41 | 0.76 | 0.68 | — |
| CPT2 | 1.00 ± 0.06 | 0.46 ± 0.03* | 0.94 ± 0.08 | 0.77 ± 0.05*, ** | 0.46 | 0.81 | — | 1.66 |
| Triglyceride export | ||||||||
| MTTP | 1.00 ± 0.17 | 0.38 ± 0.04* | 1.00 ± 0.15 | 0.61 ± 0.07*, ** | 0.38 | 0.61 | — | 1.62 |
| APOB | 1.00 ± 0.05 | 0.70 ± 0.06* | 1.05 ± 0.09 | 0.85 ± 0.06* | 0.70 | 0.80 | — | — |
qRT-PCR was performed for lipid metabolism genes of AlbCre+/caNrf2− versus AlbCre+/caNrf2+ animals on MCD diet after 28 days. The mRNA levels are first normalized to the β-actin mRNA content of each sample, and each experimental group is then normalized to the AlbCre+/caNrf2−, standard chow group. All data are presented as mean ± SEM. N = 6–8 for each of the 4 groups. The fold changes in message abundance were compared between groups using one-way ANOVA with Fisher’s LSD post hoc test. Significant fold changes are presented on the right side of the table. *P < 0.05 when compared with standard chow of the same genotype. **P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Western blot of MTTP was performed using three random animals from each experimental group. Although MTTP protein level did not appear to change based on diet, there was significantly increased MTTP protein in AlbCre+/caNrf2+ animals when compared with the littermate controls independent of the diet given (Fig. 5).
FIG. 5.
Microsomal triglyceride transfer protein (MTTP) Western blot. A, Western blot for MTTP was performed on 3 random animals from each experimental group. B, Densitometry of the MTTP blot. Although there was no difference due to MCD diet, there was significant increase of MTTP protein in the AlbCre+/caNrf2+ animals when compared with AlbCre+/caNrf2− animals independent of diet. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Constitutive Activation of Nrf2 in Hepatocytes Alone Does Not Affect Hepatic Inflammation Resulting from MCD Diet
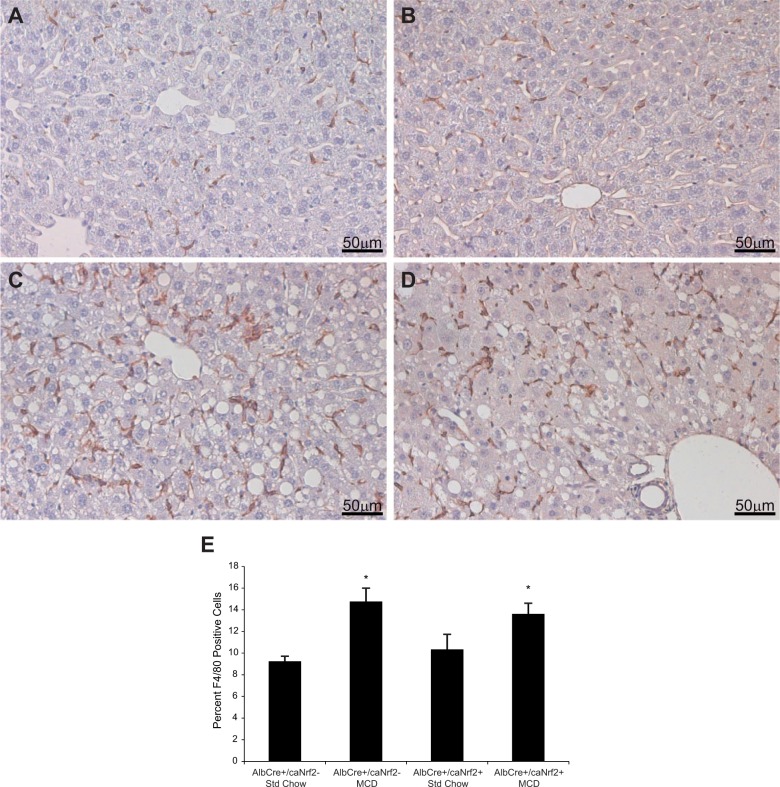
To more specifically look at inflammation, we first performed immunohistochemistry using F4/80 antibody on the liver tissue to detect the number of macrophages in each of the 4 groups of mice. Feeding the MCD diet led to significant increase in the number of infiltrating macrophages when compared with standard chow. However, among the animals fed a MCD diet, there was no difference in the number of macrophages in the AlbCre+/caNrf2+ mice when compared with the AlbCre+/caNrf2− mice (Fig. 6).
FIG. 6.
F4/80 staining. Immunohistochemistry staining was performed with α-F4/80 antibody to determine the number of macrophages present as a marker of inflammation. A, AlbCre+/caNrf2− and B, AlbCre+/caNrf2+ animals on standard chow demonstrated baseline amount of F4/80 staining. There was increased F4/80 staining for both C, AlbCre+/caNrf2− and D, AlbCre+/caNrf2+ animals after 28 days of MCD diet. E, Counting the number of F4/80 positive cells demonstrated that MCD diet led to increased inflammation in mice of both genotypes. However, there was no difference between the two genotypes on MCD diet. N = 6–8 for each of the 4 experimental groups, with 4 different fields of views for each N. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Quantitative RT-PCR was also performed for proinflammatory genes, including IL1B, IL6, IL12B, TNF, ICAM1, CCL2, and CXCL10. We did not see changes in the mRNA abundance in the majority of the proinflammatory genes (Table 3). MCD diet did lead to significant increases in CCL2 gene transcript levels compared with standard chow. However, there was no significant difference in CCL2 mRNA abundance in AlbCre+/caNrf2+ mice versus AlbCre+/caNrf2− mice fed MCD diet.
TABLE 3.
Expression of Genes Associated with Inflammation
| Genes | AlbCre+/caNrf2− |
AlbCre+/caNrf2+ |
Dietary Effect |
Genetic Effect |
||||
|---|---|---|---|---|---|---|---|---|
| Std Chow | MCD | Std Chow | MCD | AlbCre+/caNrf2− | AlbCre+/caNrf2+ | Std Chow | MCD | |
| Columns | 1 | 2 | 3 | 4 | 1 versus 2 | 3 versus 4 | 1 versus 3 | 2 versus 4 |
| IL1B | 1.00 ± 0.25 | 0.61 ± 0.09 | 1.36 ± 0.52 | 0.54 ± 0.08 | — | — | — | — |
| IL6 | 1.00 ± 0.46 | 0.84 ± 0.18 | 0.73 ± 0.27 | 0.74 ± 0.13 | — | — | — | — |
| IL12B | 1.00 ± 0.43 | 8.13 ± 0.88* | 5.21 ± 2.10 | 5.63 ± 1.26 | 8.13 | — | — | — |
| TNF | 1.00 ± 0.13 | 0.71 ± 0.12 | 0.82 ± 0.12 | 0.54 ± 0.10 | — | — | — | — |
| ICAM1 | 1.00 ± 0.22 | 0.58 ± 0.09 | 0.64 ± 0.11 | 0.88 ± 0.19 | — | — | — | — |
| CCL2 | 1.00 ± 0.10 | 8.61 ± 1.79* | 1.78 ± 0.41 | 7.18 ± 1.05* | 8.61 | 4.03 | — | — |
| CXCL10 | 1.00 ± 0.13 | 2.66 ± 0.73 | 1.24 ± 0.25 | 1.61 ± 0.27 | — | — | — | — |
qRT-PCR was performed for genes associated with inflammation using RNAs from AlbCre+/caNrf2− versus AlbCre+/caNrf2+ animals on MCD diet after 28 days. The mRNA levels are first normalized to the β-actin mRNA content of each sample, and each experimental group is then normalized to the AlbCre+/caNrf2−, standard chow group. All data are presented as mean ± SEM. N = 6–8 for each of the 4 groups. The fold changes in mRNA abundance were compared between groups using one-way ANOVA with Fisher’s LSD post hoc test. Significant fold changes are presented on the right side of the table. *P < 0.05 when compared with standard chow of the same genotype. **P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Constitutive Activation of Nrf2 in Hepatocytes Alone Does Not Affect Oxidative Stress from MCD Diet
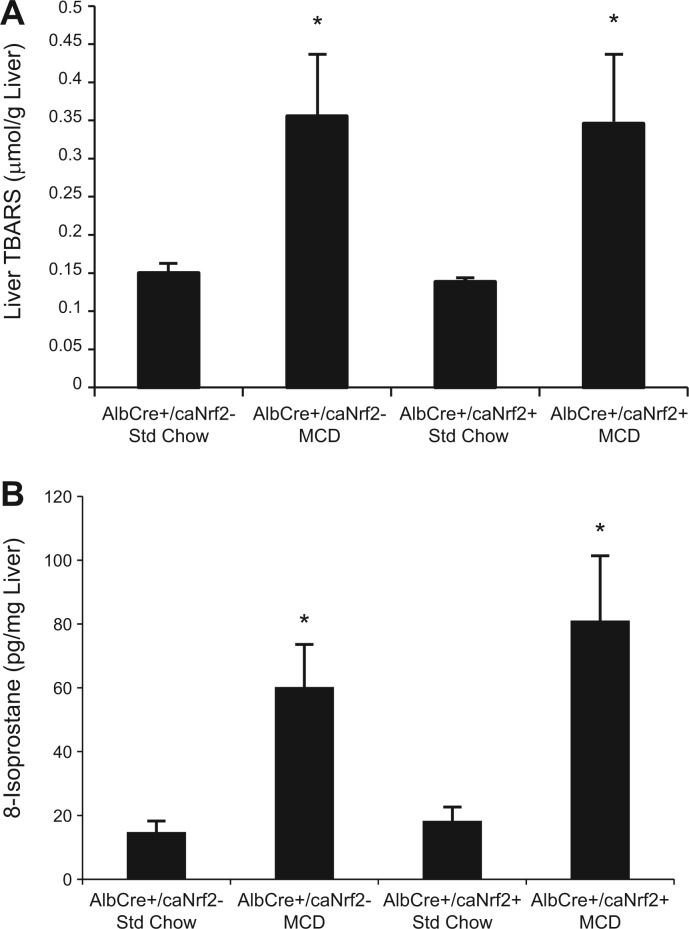
Seeing that Nrf2 overactivation in hepatocytes led to a reduction in steatosis with MCD diet, we decided to examine its impact on hepatic oxidative stress. We measured the amount of TBARS in the livers among the different groups of mice. TBARS is formed as a byproduct of lipid peroxidation, and it is routinely measured as a marker of oxidative stress (Armstrong and Browne, 1994). The amount of TBARS was significantly elevated in mice receiving MCD diet versus standard chow. However, there was no difference in TBARS in the AlbCre+/caNrf2+ mice when compared with AlbCre+/caNrf2− mice receiving MCD diet (Fig. 7A).
FIG. 7.
Oxidative stress. The amounts of (A) thiobarbituric acid reactive substances (TBARS) and (B) 8-isoprostane in the liver tissues collected were measured as markers for oxidative stress. MCD diet led to increased oxidative stress in mice of both genotypes, but there was no difference between the AlbCre+/caNrf2− and the AlbCre+/caNrf2+ animals on MCD diet. N = 6–8 for each of the 4 groups. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
We also measured the levels of 8-isoprostane as another marker of lipid peroxidation and oxidative stress (Morrow and Roberts, 1996). All mice receiving MCD diet demonstrated significantly increased hepatic levels of 8-isoprostane when compared with those receiving standard chow. However, there was no difference in 8-isoprostane level in the AlbCre+/caNrf2+ mice compared with AlbCre+/caNrf2− mice receiving MCD diet (Fig. 7B).
Constitutive Activation of Nrf2 in Hepatocytes Alone Does Not Affect Stellate Cell Activation in Response to MCD Diet
Hepatic stellate cell (HSC) activation is a well-described process contributing to the generation of fibrosis in NASH. We investigated the impact of Nrf2 overexpression in hepatocytes on the degree of HSC activation in this dietary model of NASH. To assess HSC activation, we performed qRT-PCR on fibrogenic genes, including COL1A1, TIMP1, TIMP2, MMP2, and MMP9. We did not see significant changes in most of the fibrogenic genes with feeding MCD diet for 28 days. There was significant increase in TIMP1 mRNA abundance with MCD diet feeding in both groups of transgenic mice when compared with standard chow. However, there was no difference in mRNA abundance of TIMP1 in the AlbCre+/caNrf2+ mice when compared with AlbCre+/caNrf2− mice receiving MCD diet (Table 4).
TABLE 4.
Expression of Genes Associated with Fibrosis
| Genes | AlbCre+/caNrf2− |
AlbCre+/caNrf2+ |
Dietary Effect |
Genetic Effect |
||||
|---|---|---|---|---|---|---|---|---|
| Std Chow | MCD | Std Chow | MCD | AlbCre+/caNrf2- | AlbCre+/caNrf2+ | Std Chow | MCD | |
| Columns | 1 | 2 | 3 | 4 | 1 versus 2 | 3 versus 4 | 1 versus 3 | 2 versus 4 |
| COL1A1 | 1.00 ± 0.11 | 0.80 ± 0.11 | 1.45 ± 0.29 | 1.13 ± 0.22 | — | — | — | — |
| TIMP1 | 1.00 ± 0.09 | 5.25 ± 0.82* | 1.75 ± 0.50 | 3.74 ± 0.53* | 5.25 | 2.14 | — | — |
| TIMP2 | 1.00 ± 0.08 | 1.05 ± 0.12 | 1.12 ± 0.13 | 1.03 ± 0.09 | — | — | — | — |
| MMP2 | 1.00 ± 0.14 | 0.99 ± 0.11 | 1.39 ± 0.10 | 1.26 ± 0.20 | — | — | — | — |
| MMP9 | 1.00 ± 0.09 | 0.91 ± 0.09 | 1.27 ± 0.28 | 0.96 ± 0.11 | — | — | — | — |
qRT-PCR was performed for genes associated with fibrosis using RNAs from AlbCre+/caNrf2− versus AlbCre+/caNrf2+ animals on MCD diet after 28 days. The mRNA levels are first normalized to the β-actin mRNA content of each sample, and each experimental group is then normalized to the AlbCre+/caNrf2−, standard chow group. All data are presented as mean ± SEM. N = 6–8 for each of the 4 groups. The fold changes in mRNA abundance were compared between groups using one-way ANOVA with Fisher’s LSD post hoc test. Significant fold changes are presented on the right side of the table. *P < 0.05 when compared with standard chow of the same genotype. **P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
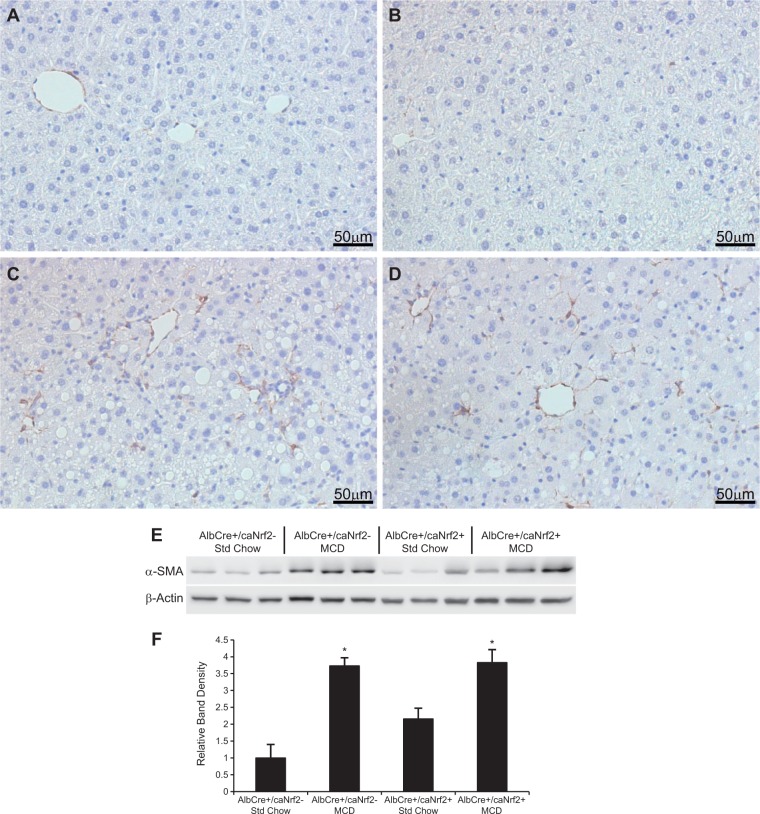
Immunohistochemistry for α-SMA was also performed to determine the extent of HSC activation (Fig. 8). There was increased α-SMA staining in the livers of animals fed an MCD diet compared with control diet. However, there was no difference in HSC activation between the AlbCre+/caNrf2+ and AlbCre+/caNrf2− animals on MCD diet. Western blot was then performed to determine the amount of α-SMA protein. Three random animals from each experimental group were used for protein analysis. Although there was significantly increased level of α-SMA protein in the animals fed an MCD diet when compared with the standard chow animals, there was no significant difference between the AlbCre+/caNrf2+ and AlbCre+/caNrf2− animals on MCD diet (Figs. 8E and 8F).
FIG. 8.
Hepatic stellate cell activation. Immunohistochemistry was performed with an α-SMA antibody to determine stellate cell activation. There appeared to be minimal α-SMA staining in both (A) AlbCre+/caNrf2− and (B) AlbCre+/caNrf2+ animals on standard chow. There was increased α-SMA staining for both (C) AlbCre+/caNrf2− and (D) AlbCre+/caNrf2+ animals after 28 days of MCD diet. E, Western blot of α-SMA was performed on 3 random animals of each experimental group. F, Densitometry of the Western blot demonstrated that MCD diet led to increased HSC activation for both genotypes. However, there was no difference between the two genotypes on MCD diet. *P < 0.05 when compared with standard chow of the same genotype. #P < 0.05 when compared with AlbCre+/caNrf2− on the same diet.
Constitutive Activation of Nrf2 in Hepatocytes Alone Does Not Affect Liver Homeostasis in Young and Aged Mice
In addition to feeding our transgenic animals with MCD diet, we also looked at the effect of the transgenes themselves on the animal livers beyond their experimental age. Four to five animals in each of the AlbCre+/caNrf2− and AlbCre+/caNrf2+ group were aged up to 18 months old on standard chow. They were then sacrificed and their livers examined. The results are shown in Supplementary Data.
By 12 months of age, there was no significant difference in the liver-to-body weight ratio between the two genotypes. ORO was performed on the animals’ livers. Although there appeared to be some ORO staining demonstrating development of steatosis due to aging, there was no difference between the genotypes. There was also no significant difference in hepatic inflammation as determined by immunohistochemistry using α-CD3 and α-Ly6G. Blood serum was also collected at 48- and 72-weeks of age, which demonstrated no significant differences in serum glucose and LDL-cholesterol levels, suggesting no difference in hepatic metabolism.
DISCUSSION
The Nrf2-ARE pathway is an endogenous antioxidant pathway that provides cellular defense from oxidative damage. NAFLD and NASH are pathologies with increasing relevance due to the obesity epidemic. Although oxidative stress has been implicated in the development of NASH from NAFLD, little is known regarding the cell-specific effects of the Nrf2-ARE pathway on the development of NAFLD and NASH. To the best of our knowledge, this is the first study that has investigated the effect of hepatocyte-specific activation of the Nrf2-ARE pathway in the development of dietary-induced NASH. Here, we show that selective expression of caNrf2 in hepatocytes leads to decreased steatosis in mice fed a MCD diet. However, despite this significant reduction on steatosis, the chronic, cell-specific activation of Nrf2-mediated gene expression in hepatocytes did not lead to a reduction in the other characteristics of NASH development, including inflammation, oxidative stress, and fibrogenesis.
In this study, we used the MCD diet model to assess the impact of cell-specific Nrf2 activation on liver pathology most commonly seen in human NASH. We recognize that the MCD diet is not a true representation of the clinical situation whereby patients develop NASH in the settings of obesity, insulin resistance, and metabolic syndrome. We chose this diet because the pathologic manifestations of clinical NASH, including steatosis, ballooning degeneration, inflammation, and fibrosis, are all present in the livers of mice receiving the MCD diet (Larter and Yeh, 2008). Numerous groups have studied the effects of systemic Nrf2 deficiency on the development of NASH using a variety of steatosis inducing diets, including high fat diet (HFD), MCD diet, or even an atherogenic plus HFD. In all these studies, independent of the diets utilized, the absence of Nrf2 led to increased steatosis and oxidative stress (Chowdhry et al., 2010; Okada et al., 2013; Sugimoto et al., 2010; Tanaka et al., 2008, 2012).
However, published studies using models of Nrf2 overactivation revealed a more heterogeneous view on Nrf2’s effect on hepatic steatosis. In these studies, systemic Nrf2 overactivation was achieved with a genetic Keap1-knock down (K1kd) model or through pharmacologic induction with CDDO-Im. Studies from Shin et al. (2009) using CDDO-Im with HFD for up to 95 days and Zhang et al. (2010) using K1kd mice with MCD diet for 5 days, both demonstrated attenuation of NASH development. However, Xu et al. (2012) using leptin-deficient (Lepob/ob) K1kd mice up to 16 weeks of age and More et al. (2013) using K1kd mice with HFD for 24 weeks, both demonstrated increased hepatic steatosis with Nrf2 overactivation. Although our reduction in steatosis corresponds with the former results, the contradictory result with the latter remains difficult to reconcile. One noticeable difference is that in the studies demonstrating a protective effect of Nrf2, the diets were used for a relative short term, up to 4 weeks. However, More et al. used 24 weeks of diet, and Xu et al. used 16 week old Lepob/ob mice that likely began obesity development early in life during preweaning (Friedman and Halaas, 1998; Thurlby and Trayhurn, 1978). In fact, Xu et al. noted in their study that there was no difference in hepatic steatosis between Lepob/ob and Lepob/ob K1kd at 8 weeks of age. They also noted that K1kd inhibited lipid accumulation in Lepob/ob skeletal muscle at 8 weeks of age, but promoted lipid accumulation by 16 weeks of age. Therefore, it is possible that the protection of Nrf2 on hepatic steatosis may be a time limited effect. Alternatively, the alterations seen in K1kd mice may result from alterations in other pathways, since Keap1 may target other proteins in addition to Nrf2.
The novelty of our study resides in the fact that we are the first group to investigate the impact of cell-specific induction of Nrf2 in NASH development using a well-characterized caNrf2 construct (Kohler et al., 2014; Schafer et al., 2012). One advantage of our model is that it allows us to look at the effect of chronic hepatocyte-specific Nrf2 activation on NASH development. By using the caNrf2 construct, which leads to direct activation of the Nrf2-ARE pathway, we can better assess the effects that are specifically due to induction of the Nrf2-ARE pathway. This is as opposed to the limitations of pharmacological activation, where the extent of Nrf2 activation may be lower than in the genetic model. In particular, Nrf2-independent effects of the compounds used cannot be excluded (Aleksunes and Klaassen, 2012). In the K1kd model, Nrf2 is overexpressed via repression of the Keap1 repressor. Although K1kd does increase Nrf2-ARE pathway activation in all cells, it is uncertain if Keap1 plays a role in Nrf2-independent pathways. Keap1-knock down (K1kd) mice were initially generated due to the global Keap1-knockout causing postnatal lethality at weaning, secondary to hyperkeratosis of the foregut leading to obstruction. Although K1kd mice survive the weaning age, they have also been shown to develop hyperkeratosis of their foregut, leading to poorer feeding and lower body weight (Taguchi et al., 2010). This malnutrition may lead to derangement in the metabolism of K1kd mice globally, which can complicate the interpretation of experiments studying NASH development using the K1kd genetic model. Finally, it has been shown that in autophagy-deficient mice, constitutive accumulation of Nrf2 is the dominant cause for liver injury, and the injury is worsened with Keap1 deficiency (Taguchi et al., 2012). However, in our model of hepatocyte-specific Nrf2 activation using the caNrf2 construct, we have shown that by 18 months of age, which is beyond our experimental period, there are no demonstrable histological abnormalities and no obvious inflammation in these livers when compared with controls. In addition, there are no apparent physiologic differences either in terms of glucose and cholesterol metabolism.
Although we do see protection against hepatic steatosis by Nrf2 activation in our model, we did not see obvious improvements in terms of inflammation or oxidative stress. When Zhang et al. (2010) investigated the effects of global Nrf2 overexpression with 5 days of MCD diet feeding, they demonstrated decreased hepatic triglycerides, as well as attenuation of oxidative damage and fibrosis as expected. In their study, the Nrf2-ARE pathway was augmented globally with a 4-fold increase in mRNA abundance and a 2-fold increase in prototypical Nrf2-dependent proteins at baseline. This modest increase led to a mitigation of NASH development with MCD feeding. In our study with more prolonged feeding and Nrf2 activation in the hepatocytes alone, we were able to generate 10- to 12-fold increases in mRNA and protein levels of Nrf2-dependent genes at baseline. Despite this increase, we did not see an obvious reduction in inflammation or oxidative stress when compared with controls. Given that Nrf2 overactivation is only present in the hepatocytes in our model, it may be possible that Nrf2-ARE activation in other cell types in the liver, ie, HSCs or Kupffer cells, are required to affect these endpoints in this model of NASH. It may be that Nrf2 overactivation in hepatocytes is not sufficient to convey protection, or that there are certain substances in the NASH environment that neutralizes the potential protective effects of Nrf2 overactivation in hepatocytes alone.
Upon liver injury, HSCs are activated and become myofibroblastic in phenotype. As a result of activation, HSCs begin to proliferate and deposit collagen, leading to fibrosis. Since the stellate cells have been demonstrated to express α-SMA upon activation both in vivo and in vitro (Rockey et al., 1992), α-SMA expression has long been a marker for stellate cell activation to suggest subsequent fibrosis. Other published studies using human liver tissue have demonstrated that α-SMA expression correlates with and predicts fibrosis (Akpolat et al., 2005; Carpino et al., 2005). Tissue inhibitor of metalloproteinase 1 (TIMP1) is expressed by activated stellate cells and contributes to the development of fibrosis by preventing degradation of collagen and other matrix proteins. Increased TIMP1 message expression has shown to be increased in fibrotic human livers when compared with normal human livers (Benyon et al., 1996). In our study, we saw increases in α-SMA expression and TIMP1 gene expression as a result of the MCD diet, suggesting HSC activation. However, among the animals that received MCD diet, we did not see changes between the genotypes, suggesting there are no changes in terms of fibrogenic potential. Interestingly, we did not see any increases in collagen mRNA as a result of diet, which may be due to the relative early time point of our experiment when the actual collagen deposition of the fibrosis process has yet to set in. Thus, although long-term Nrf2 activation in hepatocytes alone is enough for protection against steatosis, it is not sufficient to provide protection against oxidative stress and the early development of fibrosis in our model. Thus, it is possible that Nrf2 induction in the non-parenchymal cell population is required for further protection against NASH development.
To better understand how cell-specific activation of the Nrf2-ARE pathway in hepatocytes leads to a decrease in steatosis, we examined the fatty acid metabolism genetic expression profile for our transgenic animals on MCD diet. When we compared AlbCre+/caNrf2+ with AlbCre+/caNrf2− animals on MCD diet, we did not see any genotype-dependent alterations in the major lipid metabolism regulator genes tested, including LXR, PPARα, PPARγ, and SREBF1. This is contrary to previous studies where SREBF1 mRNA was increased in Nrf2 KO mice on HFD (Okada et al., 2013; Tanaka et al., 2008). Previous studies have also suggested PPARγ mRNA to be affected by changes in Nrf2 expression. However, the findings have been inconsistent: Sugimoto et al. (2010) showed that PPARγ increased in Nrf2 KO mice on MCD diet, and this was associated with increased steatosis; yet, Tanaka et al. (2008) showed that PPARγ decreased in Nrf2 KO mice on HFD, and this was also associated with worsened steatosis; finally, More et al. (2013) showed that PPARγ increased in K1Kd mice on HFD, and this was associated with worsened steatosis. Given that we were not able to detect any differences in the lipid regulators in our study, we suspect that changes in the expression of individual lipid metabolism genes may be directly due to changes in Nrf2 expression.
We did see significant changes in the gene expression of MTTP and CPT2 between AlbCre+/caNrf2− and AlbCre+/caNrf2+ animals on MCD diet. Microsomal triglyceride transfer protein, or MTTP, is a protein that is important in the assembly of VLDL, which is important for export of triglyceride from hepatocytes (Blasiole et al., 2007; Wang et al., 1997). In our study, we found that while MCD diet reduced MTTP mRNA in mice of both genotypes, this reduction in AlbCre+/caNrf2+ animals was significantly less than the reduction in AlbCre+/caNrf2− animals. This results in a relative increase in the expression of MTTP mRNA in the AlbCre+/caNrf2+ animals on MCD diet. On the protein level, we observed an increase in MTTP in AlbCre+/caNrf2+ animals regardless of diet type when compared with the AlbCre+/caNrf2− animals. The discrepancy we see between the decrease in gene expression and increase in protein expression may be due to prolonged MCD diet stimulation, leading to increased protein expression with negative regulation of the gene expression. In another study by Tanaka et al. (2008), they found decreased MTTP mRNA in the Nrf2 KO animals at baseline, and a further decrease in the Nrf2 KO animals on HFD diet. These findings together suggest that MTTP may be a potential Nrf2-dependent mechanism of protection against steatosis. Given this, we expected the serum triglycerides to be significantly elevated in the AlbCre+/caNrf2+ animals receiving MCD diet. Instead, we saw no differences in the serum triglycerides among all the experimental groups. This may be due to increased peripheral uptake of triglycerides in adipose tissue. Unfortunately, based on our study design, we were unable to assess this potential mechanism of serum triglyceride level modulation.
We also looked into the expression of genes related to β-oxidation, notably CPT1A and CPT2. CPT codes for carnitine palmitoyltransferase, which is an essential enzyme for fatty acid β-oxidation. Although CPT1A is the liver isoform located on the outer mitochondrial membrane, CPT2 is located on the inner mitochondrial membrane. In our study, we saw decreased CPT1A at baseline with Nrf2 overactivation. Although there appeared to be a trend toward increase in CPT1A expression in the AlbCre+/caNrf2+ when fed a MCD diet when compared with AlbCre+/caNrf2− mice on MCD diet, it was not statistically significant. On the other hand, the gene expression of CPT2 behaved similarly as MTTP. Although there was no difference at baseline, CPT2 was decreased in mice of both genotypes when subjected to MCD diet. However, in the animals with Nrf2 overactivation, the extent of decrease was significantly less than in the control animals. This relative increase in β-oxidation gene expression may be contributing to the decreased steatosis that is seen in AlbCre+/caNrf2+ animals on MCD diet.
Although not many published studies have looked at the expression of CPT2, some studies have looked at the gene expression of CPT1. In comparing WT to Nrf2 KO animals at baseline, Tanaka et al. (2012) showed increased CPT1 in the Nrf2 KO animals, consistent with our CPT1A finding. However, when the mice were fed their NASH inducing diet, which is a combination of HFD with carbonyl iron, Tanaka et al. demonstrated that Nrf2 KO animals have significantly decreased CPT1 expression when compared with WT animals, suggesting decreased β-oxidation gene expression in the Nrf2 KO animals under stress. Our findings in combination with other published data suggest that Nrf2 may affect β-oxidation via changes in the expression of CPTs.
CONCLUSION
In this study, we utilized transgenic mice with hepatocyte-specific expression of a constitutively active Nrf2 mutant (Kohler et al., 2014) to study its impact on NASH development using MCD diet. We found that hepatocyte-specific activation of the Nrf2-ARE pathway leads to decreased hepatic steatosis and decreased hepatocellular damage without affecting other aspects of NASH development including inflammation, oxidative stress, or HSC activation. Although the Nrf2-ARE pathway is a key antioxidant pathway, the overexpression of Nrf2 target genes in hepatocytes alone did not alter the degree of oxidative stress within the liver in our model. We suspect that induction of Nrf2 in non-parenchymal cells in the liver may be critical in decreasing oxidative stress, inflammation, and HSC activation that are seen with NASH generated by MCD diet.
In addition, the reduction of hepatic steatosis was associated with increased expression of genes involved in triglyceride export (MTTP) and β-oxidation of fatty acids (CPT2). The changes in these lipid metabolic genes raise the possibility for novel roles of Nrf2 in lipid metabolism. Further experiments would be required to elucidate the mechanistic role of Nrf2 in lipid metabolism.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
The Clinical and Translational Science Award (CTSA) program, through the National Institutes of Health National Center for Advancing Translational Sciences (NCATS) (UL1TR000427 to D.P.F.); the American Society of Transplant Surgeons-Pfizer Mid-Level Faculty Award (to D.P.F.), National Institutes of Health (T32 CA090217 to L.-Y.L.), the American College of Surgeons Resident Research Scholarship (to L.-Y.L.), the Swiss National Science Foundation (310030_132884 to S.W.); and a predoctoral fellowship from the Boehringer Ingelheim Fonds (U.A.K.).
Supplementary Material
ACKNOWLEDGMENT
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Department of Veterans Affairs, or the US Government.
REFERENCES
- Adams L. A., Lindor K. D. (2007). Nonalcoholic fatty liver disease. Ann. Epidemiol. 17, 863–869. [DOI] [PubMed] [Google Scholar]
- Akpolat N., Yahsi S., Godekmerdan A., Yalniz M., Demirbag K. (2005). The value of alpha-SMA in the evaluation of hepatic fibrosis severity in hepatitis B infection and cirrhosis development: a histopathological and immunohistochemical study. Histopathology 47, 276–280. [DOI] [PubMed] [Google Scholar]
- Aleksunes L. M., Klaassen C. D. (2012). Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metabolism and Disposition: The Biological Fate of Chemicals 40, 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D., Browne R. (1994). The analysis of free radicals, lipid peroxides, antioxidant enzymes and compounds related to oxidative stress as applied to the clinical chemistry laboratory. Adv Exp Med Biol. 366, 43–58. [DOI] [PubMed] [Google Scholar]
- Benyon R. C., Iredale J. P., Goddard S., Winwood P. J., Arthur M. J. (1996). Expression of tissue inhibitor of metalloproteinases 1 and 2 is increased in fibrotic human liver. Gastroenterology 110, 821–831. [DOI] [PubMed] [Google Scholar]
- Blasiole D. A., Davis R. A., Attie A. D. (2007). The physiological and molecular regulation of lipoprotein assembly and secretion. Mol. Biosyst. 3, 608–619. [DOI] [PubMed] [Google Scholar]
- Caballero F., Fernandez A., Matias N., Martinez L., Fucho R., Elena M., Caballeria J., Morales A., Fernandez-Checa J. C., Garcia-Ruiz C. (2010). Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: impact on mitochondrial S-adenosyl-L-methionine and glutathione. J. Biol. Chem. 285, 18528–18536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpino G., Morini S., Ginanni Corradini S., Franchitto A., Merli M., Siciliano M., Gentili F., Onetti Muda A., Berloco P., Rossi M., et al. (2005). Alpha-SMA expression in hepatic stellate cells and quantitative analysis of hepatic fibrosis in cirrhosis and in recurrent chronic hepatitis after liver transplantation. Dig. Liver Dis. 37, 349–356. [DOI] [PubMed] [Google Scholar]
- Chowdhry S., Nazmy M. H., Meakin P. J., Dinkova-Kostova A. T., Walsh S. V., Tsujita T., Dillon J. F., Ashford M. L., Hayes J. D. (2010). Loss of Nrf2 markedly exacerbates nonalcoholic steatohepatitis. Free Radic. Biol. Med. 48, 357–371. [DOI] [PubMed] [Google Scholar]
- Farrell G. C., Larter C. Z. (2006). Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology 43(Suppl. 1), S99–S112. [DOI] [PubMed] [Google Scholar]
- Flegal K. M., Carroll M. D., Kit B. K., Ogden C. L. (2012). Prevalence of obesity and trends in the distribution of body mass index among US adults, 1999–2010. JAMA 307, 491–497. [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509. [PubMed] [Google Scholar]
- Friedman J. M., Halaas J. L. (1998). Leptin and the regulation of body weight in mammals. Nature 395, 763–770. [DOI] [PubMed] [Google Scholar]
- Kohler U. A., Kurinna S., Schwitter D., Marti A., Schafer M., Hellerbrand C., Speicher T., Werner S. (2014). Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell cycle control and apoptosis. Hepatology 60, 670–678. [DOI] [PubMed] [Google Scholar]
- Larter C. Z., Yeh M. M. (2008). Animal models of NASH: getting both pathology and metabolic context right. J. Gastroenterol. Hepatol. 23, 1635–1648. [DOI] [PubMed] [Google Scholar]
- Lee J. M., Calkins M. J., Chan K., Kan Y. W., Johnson J. A. (2003). Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 278, 12029–12038. [DOI] [PubMed] [Google Scholar]
- More V. R., Xu J., Shimpi P. C., Belgrave C., Luyendyk J. P., Yamamoto M., Slitt A. L. (2013). Keap1 knockdown increases markers of metabolic syndrome after long-term high fat diet feeding. Free Radic. Biol. Med. 61C, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. D., Roberts L. J., 2nd (1996). The isoprostanes. Current knowledge and directions for future research. Biochem. Pharmacol 51, 1–9. [DOI] [PubMed] [Google Scholar]
- Okada K., Warabi E., Sugimoto H., Horie M., Gotoh N., Tokushige K., Hashimoto E., Utsunomiya H., Takahashi H., Ishii T., et al. (2013). Deletion of Nrf2 leads to rapid progression of steatohepatitis in mice fed atherogenic plus high-fat diet. J. Gastroenterol. 48, 620–632. [DOI] [PubMed] [Google Scholar]
- Rasband W. S. (1997–2012). ImageJ. Available at: http://imagej.nih.gov/ij/. Accessed October 23, 2012. [Google Scholar]
- Rinella M. E., Elias M. S., Smolak R. R., Fu T., Borensztajn J., Green R. M. (2008). Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J. Lipid Res. 49, 1068–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockey D. C., Boyles J. K., Gabbiani G., Friedman S. L. (1992). Rat hepatic lipocytes express smooth muscle actin upon activation in vivo and in culture. J. Submicrosc. Cytol. Pathol. 24, 193–203. [PubMed] [Google Scholar]
- Rolo A. P., Teodoro J. S., Palmeira C. M. (2012). Role of oxidative stress in the pathogenesis of nonalcoholic steatohepatitis. Free Radic. Biol. Med. 52, 59–69. [DOI] [PubMed] [Google Scholar]
- Schafer M., Farwanah H., Willrodt A. H., Huebner A. J., Sandhoff K., Roop D., Hohl D., Bloch W., Werner S. (2012). Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol. Med. 4, 364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Wakabayashi J., Yates M. S., Wakabayashi N., Dolan P. M., Aja S., Liby K. T., Sporn M. B., Yamamoto M., Kensler T. W. (2009). Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur. J. Pharmacol. 620, 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandidos A., Wang X., Wang H., Seed B. (2010). PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38(Database issue), D792–D799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Okada K., Shoda J., Warabi E., Ishige K., Ueda T., Taguchi K., Yanagawa T., Nakahara A., Hyodo I., et al. (2010). Deletion of nuclear factor-E2-related factor-2 leads to rapid onset and progression of nutritional steatohepatitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G283–G294. [DOI] [PubMed] [Google Scholar]
- Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. (2012). Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl Acad. Sci. U. S. A. 109, 13561–13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K., Maher J. M., Suzuki T., Kawatani Y., Motohashi H., Yamamoto M. (2010). Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 30, 3016–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Aleksunes L. M., Yeager R. L., Gyamfi M. A., Esterly N., Guo G. L., Klaassen C. D. (2008). NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J. Pharmacol. Exp. Ther. 325, 655–664. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Ikeda T., Yamamoto K., Ogawa H., Kamisako T. (2012). Dysregulated expression of fatty acid oxidation enzymes and iron-regulatory genes in livers of Nrf2-null mice. J. Gastroenterol. Hepatol. 27, 1711–1717. [DOI] [PubMed] [Google Scholar]
- Thurlby P. L., Trayhurn P. (1978). The development of obesity in preweaning obob mice. Br. J. Nutr. 39, 397–402. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N., Dinkova-Kostova A. T., Holtzclaw W. D., Kang M. I., Kobayashi A., Yamamoto M., Kensler T. W., Talalay P. (2004). Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl Acad. Sci. U. S. A. 101, 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., McLeod R. S., Yao Z. (1997). Normal activity of microsomal triglyceride transfer protein is required for the oleate-induced secretion of very low density lipoproteins containing apolipoprotein B from McA-RH7777 cells. J. Biol. Chem. 272, 12272–12278. [DOI] [PubMed] [Google Scholar]
- Xu J., Kulkarni S. R., Donepudi A. C., More V. R., Slitt A. L. (2012). Enhanced Nrf2 activity worsens insulin resistance, impairs lipid accumulation in adipose tissue, and increases hepatic steatosis in leptin-deficient mice. Diabetes 61, 3208–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. D., Hannink M. (2003). Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23, 8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. K., Yeager R. L., Tanaka Y., Klaassen C. D. (2010). Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol. Appl. Pharmacol. 245, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.