Abstract
Genetic mutations are known to drive cancer progression and certain tumors have mutation signatures that reflect exposures to environmental carcinogens. Benzo[a]pyrene (BaP) has a known mutation signature and has proven capable of inducing changes to DNA sequence that drives normal pre-stasis human mammary epithelial cells (HMEC) past a first tumor suppressor barrier (stasis) and towards immortality. We analyzed normal, pre-stasis HMEC, three independent BaP-derived post-stasis HMEC strains (184Aa, 184Be, 184Ce) and two of their immortal derivatives(184A1 and 184BE1) by whole exome sequencing. The independent post-stasis strains exhibited between 93 and 233 BaP-induced mutations in exons. Seventy percent of the mutations were C:G>A:T transversions, consistent with the known mutation spectrum of BaP. Mutations predicted to impact protein function occurred in several known and putative cancer drivers including p16, PLCG1, MED12, TAF1 in 184Aa; PIK3CG, HSP90AB1, WHSC1L1, LCP1 in 184Be and FANCA, LPP in 184Ce. Biological processes that typically harbor cancer driver mutations such as cell cycle, regulation of cell death and proliferation, RNA processing, chromatin modification and DNA repair were found to have mutations predicted to impact function in each of the post-stasis strains. Spontaneously immortalized HMEC lines derived from two of the BaP-derived post-stasis strains shared greater than 95% of their BaP-induced mutations with their precursor cells. These immortal HMEC had 10 or fewer additional point mutations relative to their post-stasis precursors, but acquired chromosomal anomalies during immortalization that arose independent of BaP. The results of this study indicate that acute exposures of HMEC to high dose BaP recapitulate mutation patterns of human tumors and can induce mutations in a number of cancer driver genes.
Keywords: Benzo[a]pyrene, p16, HMEC, carcinogenesis
1. Introduction
Cancer is a disease of genes, with DNA sequencing data showing tens to thousands of somatic mutations per tumor [1,2]. Solid tumors of the breast, prostate and colon have an average of 33 to 66 somatic mutations that are expected to alter protein structure while tumors linked to known environmental mutagen exposures such as melanomas or lung cancers have averages of 135 and 163 [3]. It is thought that only a small portion of these mutations are cancer drivers.
Somatic mutations are caused by many distinct processes, and some tumors have signature mutation patterns which reflect the major mutational process at work in the development of that particular tumor [4]. For instance, lung cancers of smokers have a signature mutation pattern where C:G>A:T transversions predominate due to mutagenic polycyclic aromatic hydrocarbons such as benzo[a]pyrene (BaP). BaP is one of the most extensively studied mutagens, a potent carcinogen and is a cause of cancer driving mutations [5,6]. The mutagenic effects of BaP are preferentially targeted to epithelial cells due to exposure routes and specific metabolism pathways that convert BaP to DNA-reactive, diol epoxide intermediates [7-9]. BaP-derived epoxide intermediates primarily form covalent adducts at the N2 position of guanine, which can lead to C:G>A:T transversions if not appropriately repaired. Thus, BaP is a prototype complete carcinogen with a known mutation signature; however, the exome-wide mutation pattern it produces along with any potential cancer drivers it may generate, have not been studied in an in vitro human model system relevant to cancer [10].
We have used an in vitro human mammary epithelial cell (HMEC) model system that has proven useful for identifying and reflecting the genetic and epigenetic events involved in early human breast carcinogenesis [11-16]. In this model system, the transformation of normal finite lifespan HMEC to malignancy requires overcoming three distinct senescence-associated barriers to immortality [11,17]. The first barrier, termed stasis or stress-induced senescence, is characterized by elevated levels of the cyclin-dependent kinase inhibitor p16INK4A (gene CDKN2A) maintaining the retinoblastoma protein (RB) in an active state [11,13,18]. The stasis barrier has been overcome or bypassed in cultured normal HMEC by various means, such as exposure to BaP [12-14,17,19]. The resultant post-stasis cells frequently exhibit inactivation of p16 by gene mutation or by promoter hypermethylation [13,20]. HMEC that get past stasis proliferate further before encountering a second more stringent proliferation barrier, replicative senescence, resulting from critically shortened telomeres [11,14]. When approaching replicative senescence, HMEC exhibit increased chromosomal instability and a DNA damage response. Rare cells that gain telomerase expression may escape this barrier and acquire immortal potential; additional perturbations can confer malignant properties on the telomerase-expressing immortally transformed cells, which are no longer vulnerable to the third barrier, oncogene-induced senescence [21-24]. To broaden our understanding of BaP-induced mutagenesis that helps render HMEC capable of bypassing stasis and progressing to immortality, we performed whole exome sequencing on normal pre- stasis HMEC cells, and multiple isogenic BaP-induced post-stasis and immortal derivatives.
In this study we show that three independent post-stasis HMEC cultures generated by BaP exposure of normal primary HMEC, and their two immortal derivatives, have hundreds of BaP-linked mutations. C:G>A:T transversions are the predominant type of mutation followed by C:G<G:C and C:G>T:A in order of prevalence. The BaP-induced mutations occur across a broad array of genes including several known and putative cancer drivers. There is no overlap of putative driver gene mutations among the independent BaP-derived post-stasis cells, but the protein altering mutations do affect biological processes that typically harbor cancer drivers in each of the different post-stasis strains. The immortal lines have almost identical mutation patterns as their post-stasis precursors with 10 or fewer additional mutations. Our exome sequencing data also confirm that immortalized cells have gross copy number variations not present in their post-stasis precursors, indicating that the genomic instability occurred after generation of the BaP post-stasis strains. This study provides new insights and confirmatory evidence into the exome-wide mutational effects of an environmental, complete carcinogen in an established model system relevant to human carcinogenesis.
2. Methods
2.1 Cell Culture
The HMEC cultures utilized were developed previously as described [12,19]. In brief, HMEC were isolated from the reduction mammoplasty tissue of a 21-year-old woman, specimen 184. In three separate experiments, primary cultures of pre-stasis 184 HMEC were exposed to 1μg/mL (4μM) BaP for two or three 24-hr periods, leading to clonal post-stasis populations that maintained growth after control populations ceased growth at stasis. Final concentration of DMSO in the BaP-containing culture medium was 0.05%. The first experiment produced one post-stasis clone, whereas the second and third experiments gave rise to at least two post-stasis clones each [19]. One post-stasis clone from each experiment was examined. Rare immortal cell lines emerged from post-stasis populations during the period of telomere dysfunction. DNA was isolated from sub-confluent cultures using the DNeasy blood and tissue kit (QIAgen).
2.2 Exome sequencing
Exome sequencing was performed using the Life Technologies AmpliseqTM Exome Kit (Cat. #4487084) according to the manufacturer’s protocols (MAN0008346). This kit uses a pool of 293,000 primer sets divided into 12 multiplexed PCR reactions to selectively amplify exons. In brief, 100 ng of genomic DNA was used to seed 12 exon amplification reactions of 10 cycles. After completing PCR cycling the 12 reactions were combined prior to primer digestion and adapter ligation. A unique barcoded adapter was used for each sample. After adapter ligation, the libraries were purified and then quantified by qPCR. Library concentrations were in the range of 150-600 pM. Pairs of barcoded libraries were diluted in equimolar concentrations for template preparation using the Ion OneTouch instrument and then sequenced on a single Ion Proton P1 chip. In total, exomes of 6 HMEC cultures were sequenced including pre-stasis 184, post-stasis 184Aa, post-stasis 184Be, post-stasis 184Ce, immortal 184A1 and immortal 184BE1. The data are publicly available through NCBI’s Sequence Read Archive (SRA) under project code SRP045165 (www.ncbi.nlm.nih.gov/sra/).
2.3 Data processing and analysis
Reads were aligned to the hg19 reference genome including unassigned contigs but without alternative haplotypes using TMAP aligner, included in the torrent suite software v4.0. An initial round of variant calling was performed using Torrent Variant Caller version 4.0-r76860 and hg19 reference using the parameters recommended by Life Technologies (Supplemental File 1). Variants from the first round of calling were merged into a master VCF file using VCFtools v0.1.7. This master VCF file was used in a second round of variant calling as a hotspots file. The second round of variant calling was run with higher stringency parameters (Supplemental File 2) and with the hotspots file, providing coverage statistics for all variants and all samples. Genomic positions that were unsuitable for variant calling due to low coverage or strand bias in any one of the samples were filtered out of the combined variant set, leaving a set of variants that exceeded the minimum coverage standard in each of the samples thus allowing even comparisons between samples. Variants within introns were filtered out of the variant files. Lists of BaP-induced mutations were analyzed using IntOGen-mutations software to annotate and predict variant effects [25]. Genes with mutations that were predicted to impact protein function were annotated to biological processes using DAVID [26]. IonReporter 4.0 software was used to find copy number variations in the post-stasis and immortal cells relative to pre-stasis, untreated 184 HMEC. Data from ten Affymetrix HT HG U133A gene expression arrays of pre-stasis 184 HMEC were downloaded from the Gene Expression Omnibus (Series GSE16058 and GSE37485) and normalized using the aroma.affymetrix package in R [18,27-29].
3. Results
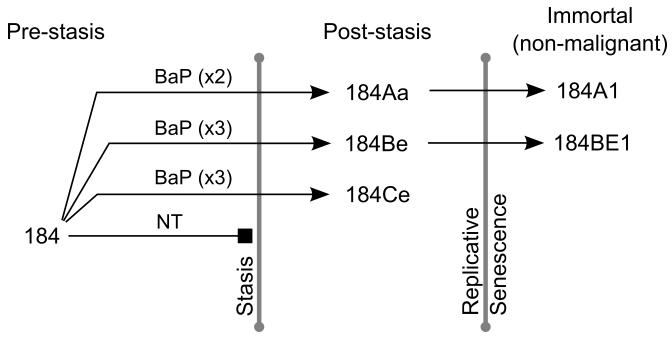
Primary cultures of pre-stasis 184 HMEC were exposed to 1μg/mL (4μM) BaP for two or three 24 hour periods, leading to the outgrowth of post-stasis cells from three independent experiments (Figure 1) [12,19]. The BaP post-stasis strains 184Aa, 184Be and 184Ce have been cultured until they either ceased growth at replicative senescence, or in rare instances, gave rise to immortal lines, such as the post-stasis strains 184Aa and 184Be giving rise to the immortal lines 184A1 and 184BE1 respectively. Thus far, the less extensively cultured post-stasis strain 184Ce has not produced an immortal line.
Figure 1.
Development of BaP-derived post-stasis and immortal HMEC cells. Untreated (NT) pre-stasis 184 HMEC display complete growth arrest known as stasis. Pre-stasis 184 HMEC were exposed to 1μg/mL (4μM) BaP for two or three 24 hour periods. From independent exposures, clones emerged that bypassed the stasis barrier; for instance 184Aa, 184Be and 184Ce. The post-stasis cells continued proliferating another 10-40 population doublings until the telomeres were depleted, inducing genomic instability, the DNA damage response and a p53- induced growth arrest. In rare instances a cell was able to escape replicative senescence and acquire immortality. The immortal lines 184A1 and 184BE1 emerged from the post-stasis strains 184Aa and 184Be respectively.
3.1 Exomic analysis of BaP-induced mutations
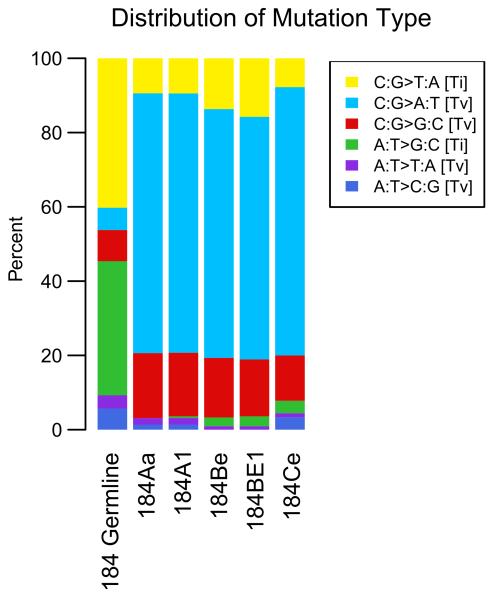
We performed exome sequencing on all the cells shown in Figure 1: pre-stasis HMEC 184, its derivatives post-stasis HMEC 184Aa, 184Be, 184Ce and the immortal HMEC 184A1 and 184BE1. As expected, the composition of variants in the untreated pre-stasis 184 HMEC relative to the hg19 reference was predominately transitions, which is in agreement with the typical range of 2.8-3.0 for transition to transversion ratios of germline variants observed in human exomes (Figure 2) [30].
Figure 2.
Relative distribution of single base substitutions by type in the BaP-derived post-stasis and immortal HMEC. The 184 germline variants bar represents the deviation from the reference genome hg19. All other bars show the deviation from 184 germline which is the somatic mutations after subtracting 184 germline variants.
Since all the samples used in this study were derived from a single genotype, the BaP-induced mutations were isolated by subtracting the germline variants from each of the post-stasis and immortal samples. The acute BaP exposures resulted in roughly 3 to 9 mutations per million base pairs of exome and almost 70% of the BaP-induced single nucleotide mutations were C:G>A:T transversions while the remaining 30% of the mutations were mostly C:G>G:C and C:G>T:A (Figure 2, Supplemental Table). This mutation rate and pattern is consistent with what has been described in lung cancers of smokers [31-33]. Greater than 90% of the BaP-induced mutations occurred at C:G base pairs which is in agreement with guanine as the major site of BaP-diol epoxide DNA adducts. Overall, both the number and type of mutations detected in the BaP-derived HMEC samples match the somatic mutation pattern of lung tumors from smokers, supporting the BaP-HMEC model system as an accurate representation of BaP- induced carcinogenesis.
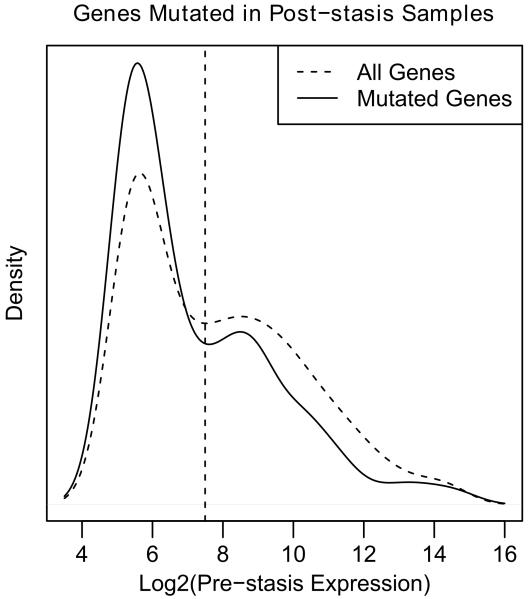
We next examined whether there was any correlation between a gene’s expression level and its susceptibility to BaP-induced mutation. We analyzed previously published data from ten pre-stasis, 184 HMEC gene expression arrays in order to estimate a gene expression profile for the untreated, parental cells. By combining the BaP-induced mutations with the pre-stasis gene expression profile we found that mutations of genes expressed in the top half prior to BaP exposure were significantly under- represented in post-stasis cells (hypergeometric test, p=1.05×10−6) (Figure 3). This under-representation of mutated genes amongst those that are expressed could be due to transcription coupled nucleotide excision repair. In support of this hypothesis, we found that of the mutated genes, only 13 of the78 (17%) G to T mutations in the top half of gene expression occurred on the transcribed strand. Conversely, 58 of 139 (42%) G to T mutations in the bottom half of gene expression occurred on the transcribed strand suggesting that the overrepresentation of mutations amongst the weakly expressed genes likely reflects less efficient repair in these genes. Another contributor to the uneven distribution of mutated genes is the possibility that expressed genes, when mutated, are more likely to cause a decrease in cellular fitness than non-expressed genes. Thus, unaffected cells would overgrow the cells with mutations in essential genes thereby preventing propagation and detection of these mutations. Taken together, these data suggest that expressed genes could be partially protected from BaP-induced mutagenesis due to targeted DNA repair mechanisms (reviewed in [34]).
Figure 3.
Distribution of pre-stasis expression levels for all genes and genes that were found mutated in post-stasis cell. The dashed curve shows the expression profile and the dashed vertical line the median of all genes on the Affymetrix HT HG U133A array (14732). The solid curve shows the genes that are mutated in post-stasis cells (348 with expression data).
3.2 BaP-derived post-stasis cells harbor unique mutations that affect cancer drivers and biological processes linked to cancer
HMEC undergo stasis through the induction of p16 expression and can become post-stasis by defects in the RB pathway leading to RB inactivation. Getting past stasis is often mediated by functional inactivation of p16, generally by deletion, mutation, or epigenetic silencing. Previous work identified a nonsense p16 mutation (E88*) in the immortal line 184A1 suggesting that the mutation was also present in the precursor cells [20]. In 184Aa we found the expected nonsense mutation in the p16 coding region (E88*) which rationally drives the functional inactivation of one allele while the other allele is silenced by DNA methylation [16]. Although p16 expression is absent in each of the post-stasis strains examined, it is not known how this silencing event was attained in 184Be, 184BE1 and 184Ce. p16 inactivation could be achieved through distinct mutations to p16 in the other post-stasis strains or other genes in this signaling pathway. In 184Be and 184BE1, we found a novel C to A transversion at the codon for amino acid 70 of p16 (NM_000077), but this mutation is silent (P70=, Supplemental Table). The alternate codon has comparable usage in the human genome as the original and is therefore unlikely to affect protein translation. Therefore, in some instances such as 184Aa, BaP exposure contributes to the bypass of stasis by directly causing p16 mutations, but in the other post-stasis strains there are likely to be different defects that affect the RB pathway or cell cycle.
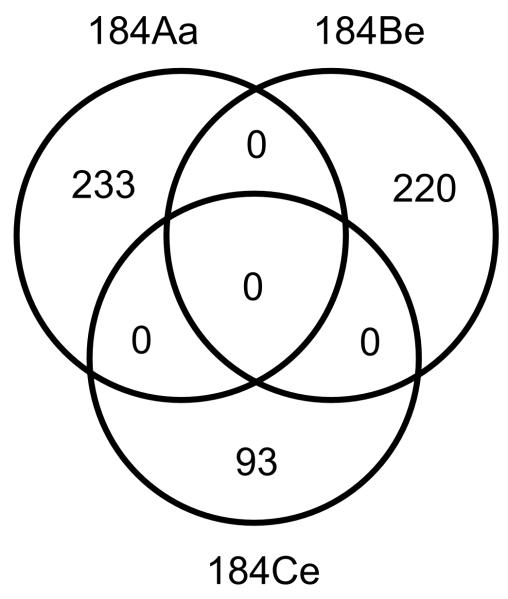
In addition to p16 mutations, more than 200 other BaP-induced mutations were found in the post-stasis strains 184Aa and 184Be whereas 184Ce had 93 (Figure 4). When the specific nucleotide positions mutated by BaP were considered, each of the independently obtained post-stasis strains had a completely unique set of mutations (Figure 4). When the genes mutated by BaP were considered, five genes with mutations predicted to alter protein function were common in two of the three post-stasis strains; the mutations occurring in different codons; no genes were common to all three post-stasis strains (Table 1).
Figure 4.
Venn diagram of the BaP-induced mutations in post-stasis HMEC. Each post-stasis strain has a completely unique set of mutations.
Table 1.
(Top) Genes that have mutations predicted to alter protein function in more than one post-stasis strain. (Bottom) Putative cancer drivers that have mutations predicted to alter protein function in post-stasis cells.
| Overlapping Genes |
184Aa | 184Be | 184Ce |
|---|---|---|---|
| XCR1 | G94C†† | A33P† | - |
| RGN | G111W†† | G116A†† | - |
| KIAA2018 | C1257S† | S1323T† | - |
| CROCC | A1156S† | - | D394H† |
| GAS6 | - | W379L†† | R397S† |
|
| |||
|
Putative Cancer
Drivers |
184Aa | 184Be | 184Ce |
|
| |||
| p16 | E88*††† | - | - |
| PLCG1 | D342Y††† | - | - |
| MED12 | M880I†† | - | - |
| TAF1 | D333H†† | - | - |
| MLL2 | M1417I† | - | - |
| ARHGAP32 | R1886S† | - | - |
| KALRN | A212D† | - | - |
| PIK3CG | - | R472S††† | - |
| HSP90AB1 | - | R168L††, D170H†† | - |
| WHSC1L1 | - | Q1013H†† | - |
| LCP1 | - | D557Y†† | - |
| FANCA | - | - | R600H†† |
| LPP | - | - | R418P†† |
Predicted Variant Impact: High
Medium
Low
Each of the post-stasis strains had at least two mutated genes classified as high-confidence cancer drivers (Table 1). A previous study analyzed over 3000 tumors and combined several complementary methods to generate a list of 291 high-confidence cancer driver genes that mostly correspond to 5 broad biological modules [25]. Seven genes from this list had protein altering mutations in 184Aa including p16, PLCG1, MED12, TAF1, MLL2, ARHGAP32 and KALRN (Table 1). 184Be had 4 cancer drivers with protein altering mutations including PIK3CG, HSP90AB1 (one allele with two mutations), WHSC1L1 and LCP1. 184Ce had two cancer drivers with predicted protein altering mutations, FANCA and LPP.
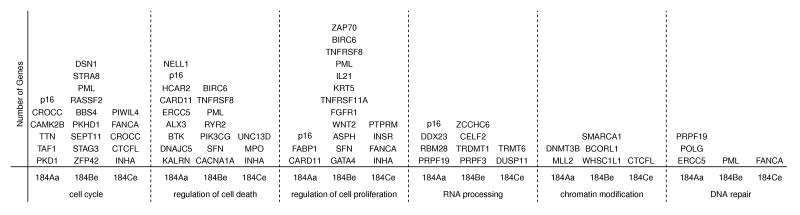
We chose six biological processes from the gene ontology database to represent those processes typically affected by cancer driver mutations and found that each of the post-stasis strains has at least one mutation predicted to alter protein function in each of these processes (Figure 5). These results identify potentially important BaP-induced protein altering mutations in genes and pathways that are known to be preferentially disrupted in human tumors and suggest that defects in these processes can confer survival and proliferation advantages.
Figure 5.
Genes with mutations predicted to alter protein function in post-stasis cells that affect biological processes known to be perturbed by cancer driver mutations.
3.3 Characterization of the genetic alterations unique to immortalized cells
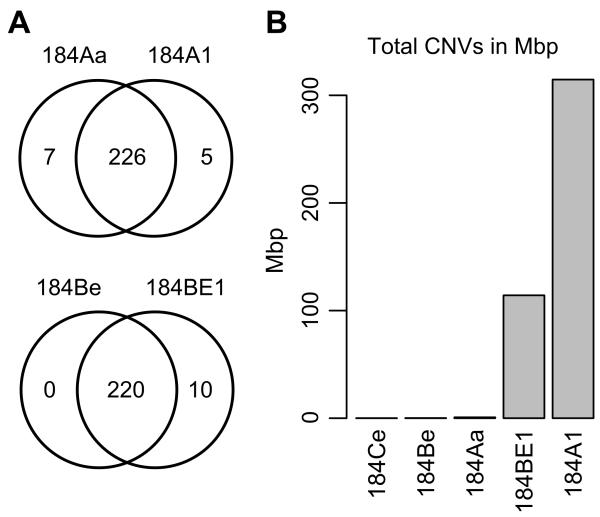
Point mutations in the immortal cells have a high degree of overlap with their post-stasis precursors (Figure 6A). Overall 184Aa and 184A1 share 226 BaP-induced mutations, many of which are predicted to alter protein function (Figure 6A). One of these overlapping mutations is the previously described nonsense mutation in p16 (E88*) [20]; a known driver mutation. Our sequencing data confirmed the presence of this mutation in 184A1 and here extend the finding to the 184Aa post-stasis strain. Similarly, 184BE1 also shares the vast majority of its mutations with its post-stasis precursor 184Be. The immortal lines 184A1 and 184BE1 have additional point mutations that appear to occur during or after immortalization and are therefore not likely to be directly caused by BaP-induced DNA adducts (Figure 6A). This interpretation is supported by the fact that only 5 of the additional 15 mutations seen in 184A1 and 184BE1 are the BaP signature C:G>A:T transversion. Of the 5 unique mutations in 184A1, 3 are predicted to impact protein function and only the A to G transition at the codon for amino acid 239 of NKRF is predicted to have a medium impact (Table 2). Six of the 10 mutations unique to 184BE1 are predicted to impact protein function (Table 2). 184Aa has mutations that were not detected in its immortal derivative 184A1, suggesting that they were lost during immortalization, most likely due to chromosomal deletions (as described below). In summary, the majority of the mutations detected in the immortal lines were directly inherited from the post-stasis precursors (>95%); however, a small portion of mutations that could possibly influence the immortal lines’ phenotype appear to have been acquired during immortalization.
Figure 6.
(A) Venn diagrams showing the overlaps of point mutations between BaP-derived post-stasis and immortal HMEC. (B) Cumulative copy number variation as measured by total base pairs covered. The genomic distance between consecutive amplicons is included in the calculation even though those genomic regions were not sequenced.
Table 2.
Genes with mutations that are unique to the immortal lines and are predicted to alter protein function.
| Gene | 184A1 | 184BE1 | dbSNP ID |
|---|---|---|---|
| NKRF | I239T†† | - | - |
| IL10RA | V233M† | - | rs41354146 |
| SLC13A4 | Q237E† | - | - |
| SORBS2 | - | S50F†† | - |
| GLTSCR1L | - | T162M†† | - |
| MMP19 | - | P288S† | - |
| CYP2B6 | - | R487L† | rs149386398 |
| MIOX | - | T49K† | - |
| CDYL | - | S308L† | - |
Predicted Variant Impact: High
Medium
Low
Ongoing culture of the BaP post-stasis HMEC eventually leads to replicative senescence resulting from telomere erosion [11,17,18]. HMEC that have overcome this barrier, 184A1 and 184BE1, express telomerase activity and display genomic rearrangements [14,35,36]. Using sequencing read depths, copy number variations (CNVs) were estimated in each of the post-stasis and immortal cultures relative to untreated, pre-stasis HMEC (Figure 6B). Overall, the post-stasis 184Aa, 184Be and 184Ce populations displayed few, very small CNVs (Supplemental Figs. S1-S3). In contrast, the immortal lines 184A1 and 184BE1 showed dramatically more CNVs than their post-stasis precursors (Figure 6B, Supplemental Figs. S1-S5). CNV analysis of the immortal 184A1 exome-sequencing data detected three copies of chromosome 20, previously observed in late passage 184A1 as well as in several other cell lines cultured for long periods (Walen & Stampfer, 1989). Immortal 184A1 cells also have previously reported large single copy deletions on chromosomes 3, 6 and 12 which account for all 7 mutations that are present in 184Aa but not 184A1 (Figure 6A, Supplemental Figure S4). These deletions also cause the loss of heterozygosity for five mutations that are predicted to alter protein function including NMBR, KLHL32, COL12A1, SLC17A4 and HCAR2, associating loss of the wild type alleles with immortalization (Supplemental Figure S4). Immortal 184BE1 cells have an amplification on chromosome 2, approximately 4 copies of the long arm of chromosome 8 and a single copy deletion on chromosome 13 (Supplemental Figure S5). Interestingly, amplification of the long arm of chromosome 8 is common in immortal cell lines and encompasses the MYC oncogene, a known genetic and etiologic event in breast cancer. To summarize, the sequencing data showed that BaP-derived immortal cells have additional point mutations and substantially more CNVs than their post-stasis precursor cells, providing significant evidence that the cells experienced a period of genomic instability during their progression to immortality, but after and independent of their exposure to BaP.
4. Discussion
The present work shows that acute exposures of normal finite pre-stasis HMEC to the complete carcinogen BaP produces hundreds of mutations that reflect both the number and signature of mutations observed in human lung cancers of smokers. Several studies have found p53 mutations in lung cancers of smokers with a BaP signature, but none of the post-stasis and immortal HMEC examined in this study had protein altering p53 mutations. The absence of p53 mutations in the post-stasis and immortal HMEC is consistent with the absence of p53 mutations in most human breast cancers and the enforcement of stasis in cultured HMEC by p16 and not p53-dependent p21 [5,6,17]. The 1μg/mL (4 μM) dosage of BaP used for the original experiments produced 80% cell kill following the 24 hour dosing periods [12]. Although this dose is a thousand times higher than what has been measured in serum from smokers, it is important to note that acute, high doses of BaP generate a similar mutation rate and pattern as are found in tumors [37]. This might suggest that for mutagenic carcinogens, the cumulative dose is a determinant of cancer risk.
Since stasis in normal HMEC is a p16 mediated process acting on the RB pathway, we do not expect any of the mutated cancer drivers reported here to be able to drive cells past stasis independent of p16/RB pathway inactivation. Although none of the mutations predicted to alter protein function that we report here are direct regulators of p16 or the RB pathway (with the exception of p16 E88*), it is possible that one or some of these defects indirectly contribute to p16 inactivation. In normal pre-stasis HMEC, p16 maintains an inducible bivalent epigenetic state thus allowing induction of p16 and consequently stasis. In post-stasis cells and human tumors, p16 is commonly inactivated via aberrant DNA methylation; therefore it would be interesting to test whether any combination of these putative cancer driver mutations or their experimental surrogates can lead to epigenetic inactivation of p16 in pre-stasis cells, thereby contributing to the bypass of stasis. Even if these putative cancer driver mutations turn out to be ineffective for the bypass of stasis, they are likely to contribute to malignancy in other ways once the cells achieve immortality.
Replicative senescence occurs in post-stasis cells when telomeres become critically short, leading to genomic instability, a DNA damage response and growth arrest. In rare instances, this senescence barrier can be overcome by the reactivation of telomerase. 184BE1 contains an amplification of the long arm of chromosome 8, suggesting that MYC deregulation could have contributed to their overcoming replicative senescence (Supplemental Figure S5). On the other hand, 184A1 appears to have no chromosomal abnormalities involving MYC, indicating that other defects were important for its bypass of replicative senescence (Supplemental Figure S4). In this study we identified 3 mutations predicted to alter protein function that occur in 184A1 but not in their post-stasis precursor cells 184Aa (Table 2). One of these mutated genes, NKRF (I239T), seems an interesting candidate for further experimentation based on its repressor function to NFKB. Other potentially important candidates associated with the immortal phenotype are those mutations that undergo loss of heterozygosity in 184A1, since loss of heterozygosity is a typical identifier of a tumor suppressor gene.
Inhalation of polycyclic aromatic hydrocarbons such as BaP is clearly a risk factor for lung cancer, but ingestion of BaP might be a risk factor for breast cancer. BaP is a well-known mammary carcinogen in rodents. Several studies have shown that environmental factors contribute to breast cancer risk [38-41]. Some studies have provided evidence of aromatic DNA adducts in tissues of breast cancer patients suggesting that BaP exposure could be involved in a portion of breast cancers which provides rationale for the study of BaP in this HMEC model system [42,43].
In addition to this work, another recent study showed that exposure of murine fibroblasts to BaP produced a mutation pattern that also resembled the mutation signature of lung cancers, supporting in vitro systems as models to study genome-wide mutational processes that drive cancer progression [44]. On the other hand, there are many critical differences relevant to carcinogenesis between mouse fibroblasts and human epithelial cells that warrant investigation of BaP-induced carcinogenesis in human epithelial cells [8,18,45-47]. The model system used in this study is an accurate representation of normal epithelial tissue that progresses through defined stages of tumor development upon BaP exposure. An important feature of these cells is that they have not been virally immortalized, allowing the study of the normal protective mechanisms that prevent tumor development, whereas most cell lines have these protective pathways artificially abrogated to facilitate in vitro culture. This study provides additional evidence to the body of literature supporting the BaP driven mutagenic mechanism of carcinogenesis and highlights potentially important candidate driver gene mutations that are possible contributors to overcoming the normal proliferation barriers.
Supplementary Material
Highlights.
We examine BaP-induced mutations in three post-stasis HMECs by exome sequencing.
70% of the mutations are C:G>A:T transversions, matching the profile of lung tumors.
Known and putative cancer driver mutations occurred in several genes.
Immortalized HMEC have additional, non-BaP-induced chromosomal abnormalities.
Acknowledgments
The authors would like to thank Gregory Metzger and the Genomics Shared Services Laboratory at the University of Arizona Cancer Center for their assistance with template preparation and sequencing. This work was supported by the National Institutes of Health (P42 ES04940, ES006694, CA23074, 5T32ES16652-5) and the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 (MRS).
Abbreviations
- (BaP)
Benzo[a]pyrene
- (p16)
p16INK4A
- (HMEC)
human mammary epithelial cells
Footnotes
Disclosure Statement. The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- [2].Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain. Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- [6].Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- [7].Stampfer MR, Bartholomew JC, Smith HS, Bartley JC. Metabolism of benzo[a]pyrene by human mammary epithelial cells: Toxicity and DNA adduct formation. Proc. Natl. Acad. Sci. U. S. A. 1981;78:6251–6255. doi: 10.1073/pnas.78.10.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bartley J, Bartholomew JC, Stampfer MR. Metabolism of benzo(a)pyrene by human epithelial and fibroblastic cells: Metabolite patterns and DNA adduct formation. J. Cell. Biochem. 1982;18:135–148. doi: 10.1002/jcb.1982.240180202. [DOI] [PubMed] [Google Scholar]
- [9].Bartley JC, Stampfer MR. Factors influencing benzo[a]pyrene metabolism in human mammary epithelial cells in culture. Carcinogenesis. 1985;6:1017–1022. doi: 10.1093/carcin/6.7.1017. [DOI] [PubMed] [Google Scholar]
- [10].Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, Varela I, Nik-Zainal S, Davies HR, Ordonez GR, Mudie LJ, Latimer C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J, Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF, Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA, Campbell PJ. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Garbe JC, Holst CR, Bassett E, Tlsty T, Stampfer MR. Inactivation of p53 function in cultured human mammary epithelial cells turns the telomere-length dependent senescence barrier from agonescence into crisis. Cell. Cycle. 2007;6:1927–1936. doi: 10.4161/cc.6.15.4519. [DOI] [PubMed] [Google Scholar]
- [12].Stampfer MR, Bartley JC. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc. Natl. Acad. Sci. U. S. A. 1985;82:2394–2398. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brenner AJ, Stampfer MR, Aldaz CM. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- [14].Romanov SR, Kozakiewicz BK, Holst CR, Stampfer MR, Haupt LM, Tlsty TD. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- [15].Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, Miranda M, Krig S, Garbe J, Stampfer M, Yaswen P, Gray JW, Lockett SJ. In situ analyses of genome instability in breast cancer. Nat. Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- [16].Novak P, Jensen TJ, Garbe JC, Stampfer MR, Futscher BW. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009;69:5251–5258. doi: 10.1158/0008-5472.CAN-08-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stampfer M, LaBarge M, Garbe J. An integrated human mammary epithelial cell culture system for studying carcinogenesis and aging. In: Schatten H, editor. Cell and Molecular Biology of Breast Cancer. Humana Press; 2013. pp. 323–361. [Google Scholar]
- [18].Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS, Wyrobek AJ, Stampfer MR. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stampfer MR, Bartley JC. Human mammary epithelial cells in culture: Differentiation and transformation. Cancer Treat. Res. 1988;40:1–24. doi: 10.1007/978-1-4613-1733-3_1. [DOI] [PubMed] [Google Scholar]
- [20].Brenner AJ, Aldaz CM. Chromosome 9p allelic loss and p16/CDKN2 in breast cancer and evidence of p16 inactivation in immortal breast epithelial cells. Cancer Res. 1995;55:2892–2895. [PubMed] [Google Scholar]
- [21].Clark R, Stampfer MR, Milley R, O’Rourke E, Walen KH, Kriegler M, Kopplin J, McCormick F. Transformation of human mammary epithelial cells by oncogenic retroviruses. Cancer Res. 1988;48:4689–4694. [PubMed] [Google Scholar]
- [22].Olsen CL, Gardie B, Yaswen P, Stampfer MR. Raf-1-induced growth arrest in human mammary epithelial cells is p16-independent and is overcome in immortal cells during conversion. Oncogene. 2002;21:6328–6339. doi: 10.1038/sj.onc.1205780. [DOI] [PubMed] [Google Scholar]
- [23].Cipriano R, Kan CE, Graham J, Danielpour D, Stampfer M, Jackson MW. TGF-beta signaling engages an ATM-CHK2-p53-independent RAS-induced senescence and prevents malignant transformation in human mammary epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8668–8673. doi: 10.1073/pnas.1015022108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sherman MY, Meng L, Stampfer M, Gabai VL, Yaglom JA. Oncogenes induce senescence with incomplete growth arrest and suppress the DNA damage response in immortalized cells. Aging Cell. 2011;10:949–961. doi: 10.1111/j.1474-9726.2011.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, Santos A, Lopez-Bigas N. IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods. 2013;10:1081–1082. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [27].Garbe JC, Pepin F, Pelissier FA, Sputova K, Fridriksdottir AJ, Guo DE, Villadsen R, Park M, Petersen OW, Borowsky AD, Stampfer MR, Labarge MA. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 2012;72:3687–3701. doi: 10.1158/0008-5472.CAN-12-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bengtsson H, Simpson K, Bullard J, Hansen K. Aroma.affymetrix: A generic framework in R for analyzing small to very large affymetrix data sets in bounded memory. 2008.
- [29].R_Development_Core_Team. R A language and environment for statistical computing. 2014.
- [30].DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next- generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba II, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, Ha C, Johnson S, Kennemer MI, Mohan S, Nazarenko I, Watanabe C, Sparks AB, Shames DS, Gentleman R, de Sauvage FJ, Stern H, Pandita A, Ballinger DG, Drmanac R, Modrusan Z, Seshagiri S, Zhang Z. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- [33].Sritharan N. Genomic landscape of non-small-cell lung cancer in smokers and never-smokers. Thorax. 2013 [Google Scholar]
- [34].Scicchitano DA. Transcription past DNA adducts derived from polycyclic aromatic hydrocarbons. Mutat. Res. 2005;577:146–154. doi: 10.1016/j.mrfmmm.2005.03.015. [DOI] [PubMed] [Google Scholar]
- [35].Walen KH, Stampfer MR. Chromosome analyses of human mammary epithelial cells at stages of chemical-induced transformation progression to immortality. Cancer Genet. Cytogenet. 1989;37:249–261. doi: 10.1016/0165-4608(89)90056-3. [DOI] [PubMed] [Google Scholar]
- [36].Stampfer MR, Garbe J, Nijjar T, Wigington D, Swisshelm K, Yaswen P. Loss of p53 function accelerates acquisition of telomerase activity in indefinite lifespan human mammary epithelial cell lines. Oncogene. 2003;22:5238–5251. doi: 10.1038/sj.onc.1206667. [DOI] [PubMed] [Google Scholar]
- [37].Neal MS, Zhu J, Foster WG. Quantification of benzo[a]pyrene and other PAHs in the serum and follicular fluid of smokers versus non-smokers. Reprod. Toxicol. 2008;25:100–106. doi: 10.1016/j.reprotox.2007.10.012. [DOI] [PubMed] [Google Scholar]
- [38].Buell P. Changing incidence of breast cancer in japanese-american women. J. Natl. Cancer Inst. 1973;51:1479–1483. doi: 10.1093/jnci/51.5.1479. [DOI] [PubMed] [Google Scholar]
- [39].Ziegler RG, Hoover RN, Pike MC, Hildesheim A, Nomura AM, West DW, Wu-Williams AH, Kolonel LN, Horn-Ross PL, Rosenthal JF, Hyer MB. Migration patterns and breast cancer risk in asian-american women. J. Natl. Cancer Inst. 1993;85:1819–1827. doi: 10.1093/jnci/85.22.1819. [DOI] [PubMed] [Google Scholar]
- [40].Deapen D, Liu L, Perkins C, Bernstein L, Ross RK. Rapidly rising breast cancer incidence rates among asian-american women. Int. J. Cancer. 2002;99:747–750. doi: 10.1002/ijc.10415. [DOI] [PubMed] [Google Scholar]
- [41].Singletary SE. Rating the risk factors for breast cancer. Ann. Surg. 2003;237:474–482. doi: 10.1097/01.SLA.0000059969.64262.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Perera FP, Estabrook A, Hewer A, Channing K, Rundle A, Mooney LA, Whyatt R, Phillips DH. Carcinogen-DNA adducts in human breast tissue. Cancer Epidemiol. Biomarkers Prev. 1995;4:233–238. [PubMed] [Google Scholar]
- [43].Li D, Wang M, Dhingra K, Hittelman WN. Aromatic DNA adducts in adjacent tissues of breast cancer patients: Clues to breast cancer etiology. Cancer Res. 1996;56:287–293. [PubMed] [Google Scholar]
- [44].Olivier M, Weninger A, Ardin M, Huskova H, Castells X, Vallee MP, McKay J, Nedelko T, Muehlbauer KR, Marusawa H, Alexander J, Hazelwood L, Byrnes G, Hollstein M, Zavadil J. Modelling mutational landscapes of human cancers in vitro. Sci. Rep. 2014;4:4482. doi: 10.1038/srep04482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- [47].Vrba L, Garbe JC, Stampfer MR, Futscher BW. Epigenetic regulation of normal human mammary cell type-specific miRNAs. Genome Res. 2011;21:2026–2037. doi: 10.1101/gr.123935.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.