Abstract
Maraviroc (MVC) is licensed in clinical practice for patients with R5 virus and virological failure; however, in anecdotal reports, dual/mixed viruses were also inhibited. We retrospectively evaluated the evolution of HIV-1 coreceptor tropism in plasma and peripheral blood mononuclear cells (PBMCs) of an infected adolescent with a CCR5/CXCR4 Trofile profile who experienced an important but temporary immunological and virological response during a 16-month period of MVC-based therapy. Coreceptor usage of biological viral clones isolated from PBMCs was investigated in U87.CD4 cells expressing wild-type or chimeric CCR5 and CXCR4. Plasma and PBMC-derived viral clones were sequenced to predict coreceptor tropism using the geno2pheno algorithm from the V3 envelope sequence and pol gene-resistant mutations. From start to 8.5 months of MVC treatment only R5X4 viral clones were observed, whereas at 16 months the phenotype enlarged to also include R5 and X4 clones. Chimeric receptor usage suggested the preferential usage of the CXCR4 coreceptor by the R5X4 biological clones. According to phenotypic data, R5 viruses were susceptible, whereas R5X4 and X4 viruses were resistant to RANTES and MVC in vitro. Clones at 16 months, but not at baseline, showed an amino acidic resistance pattern in protease and reverse transcription genes, which, however, did not drive their tropisms. The geno2pheno algorithm predicted at baseline R5 viruses in plasma, and from 5.5 months throughout follow-up only CXCR4-using viruses. An extended methodological approach is needed to unravel the complexity of the phenotype and variation of viruses resident in the different compartments of an infected individual. The accurate evaluation of the proportion of residual R5 viruses may guide therapeutic intervention in highly experienced patients with limited therapeutic options.
Introduction
HIV-1 infects target cells through the binding of the envelope protein gp120 to the main receptor CD4 and a coreceptor, mainly CCR5 or CXCR4, expressed on the cell surface.1 Viral isolates with unique use of CCR5 or CXCR4 are termed R5 and X4, respectively, while those capable of using both coreceptors are termed R5X4 or dual/mix. These latter ones can be composed of either dual-tropic variants and/or a mixture of pure R5- and X4-tropic variants, which can be revealed only if the viral population is analyzed at a clonal level.2,3 Today several methods based on genotyping of the viral env V3 region are used in clinical practice to predict viral chemokine receptor tropism instead of using the classical phenotyping approach. Among the available algorithms used to determine genotypic tropism, geno2pheno is currently used most often and is the most promising tool, due to its good concordance with phenotypic results in addition to its contained cost and the time needed to deliver the result.4–6
In children and in adults R5 variants are preferentially isolated during the early stage of the disease, whereas CXCR4-using viruses appear thereafter and have been associated with CD4+ T cell decline.7–10 R5 viruses can be further classified into R5 broad and narrow according to their ability to use a set of different CCR5/CXCR4 chimeric receptors in addition to wild-type CCR5 and sensitivity to inhibition by the CC-chemokine RANTES,11,12 demonstrating that R5 viruses are extremely heterogeneous. Furthermore, the presence of R5 broad viruses is prognostic of fast disease progression in adults and in children.11,12
The CCR5 molecule has become the target of a new class of inhibitors, with a novel drug, maraviroc (MVC), approved for clinical use.13,14 MVC has demonstrated potent activity in vitro and in vivo against R5 HIV-1, including variants that are resistant to multiple drug classes.13–17 Because MVC specifically targets CCR5 and has no inhibitory effect against CXCR4-using viruses, the drug is recommended only for patients harboring CCR5-dependent variants exclusively. Thus, a tropism test is mandatory before treatment to define the patient's viral tropism. Indeed, virological failure on MVC treatment was often accompanied by the appearance of non-R5 virus variants.17 Whether this is due to a switch in tropism from R5 to non-R5 or to the selection of preexisting CXCR4-using variants is still a matter of debate.
The addition of MVC to optimized background therapy in an extensively treatment-experienced patient population infected with dual- or mixed-tropic HIV-1 did not confer a significant additional reduction in plasma viremia, compared with placebo, over 24 weeks of treatment.18 This further supported the recommendation for the preferential use of MVC in patients with R5 viruses. Recently, it was suggested that the antiviral activity of MVC may extend to a broader range of HIV variants than previously suspected.19 Thus, there is the need for in vivo studies to appropriately define the activity of MVC.
Here we present a case report of a 17-year-old adolescent with a predicted dual/mix plasma HIV-1 population, who during MVC–raltegravir (RAL)-containing salvage therapy had an impressive although temporary immunological recovery with an important reduction in viral load, and displayed an enlargement of the PBMC-derived clonal viral population from R5X4 to also include R5 and X4 in parallel with the increase in the viral variability of the plasma.
Materials and Methods
Patient characteristics
A 17-year-old patient, infected with HIV-1 subtype B via mother-to-child transmission, was referred to the Department of Infectious Disease of Ospedale San Raffaele in Milan, Italy, in December 2007, where the patient is still followed today. The patient was treated for years with multiple antiretroviral regimens, including nucleoside reverse transcriptase inhibitors (NRTIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs), fusion inhibitors, and protease inhibitors (PIs). Sustained virological suppression was never achieved, immunological deterioration was observed, and failure of highly active antiretroviral therapy (HAART)-based regimens led to the selection of multidrug-resistant strains. On February 2008 (baseline, t=0) a new therapeutic regimen was introduced as described in the results section and whole blood samples were collected over a 16-month period for virological studies. Starting in July 2009 the patient was maintained on holding antiretroviral therapy (ARV) for 2 years, and was then subsequently treated with dolutegravir (50 mg bid), entravirine (200 mg bid), and darunavir/ritonavir (DRV/r) (600 and 100 mg bid).20 During this period the viral load was mostly undetectable and the last CD4+ T cell count (in April 2014) was 395 cells/ml.
Ethical statement
Due to extremely limited treatment options the patient received MVC and RAL with a strict follow-up in the context of compassionate use after signing an informed consent approved by the local ethical committee, according to national rules.
Isolation of biological HIV-1 clones and phenotype determination in U87.CD4 cells
The HIV-1 quasispecies were cloned biologically from peripheral blood mononuclear cells (PBMCs) by a previously published method.21 Virus was assumed to derive from a single infected cell if the fraction of positive cultures did not exceed 33% of the total number of cultures. Positive cultures were expanded for 7–10 days for viral stock preparation by coculture with phytohemagglutinin (PHA)-stimulated PBMCs.
Virus stocks were used to infect human glioma U87.CD4 cells stably expressing the wild-type chemokine receptor CCR5 or CXCR4 or the five CCR5/CXCR4 chimeric receptors FC-2, FC-4b, FC-5, FC-6, and FC-7, as previously described.11,22,23 Chimeric receptors were obtained by replacing successively larger parts of CCR5 with corresponding parts of CXCR4 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/aid). In the resulting chimeras CXCR4 comprised gradually larger parts: the N-terminal tail and the first transmembrane portion (FC-2) and the first (FC-4b), second (FC-5 and FC-6), and third (FC-7) extracellular loops. R5 viruses able to exclusively use the wild-type CCR5 are defined as R5narrow, whereas those able to also use one chimeric receptor or different combinations of chimeric receptors are defined as R5broad. Methodological details are available as Supplementary Materials and Methods.
RANTES and MVC sensitivity assay
PHA-activated PBMCs derived from two HIV-seronegative blood donors were infected with viral supernatant in the presence or absence of RANTES (R&D Systems, Minneapolis, MN) or MVC (Pfizer, New York, NY) as previously described.24 Methodological details are available as Supplementary Materials and Methods.
Sequencing of env V3 domain and genotypic determination of coreceptor usage
HIV-1 RNA and DNA were extracted from plasma and PBMCs, respectively, by means of a commercially available kit (QIAamp RNA Viral Mini kit, Qiagen, QIAamp DNA Viral Mini kit, Qiagen). Amplification and sequencing of the V3 region of the env gene [nucleotides (nt) 630–1,310 of HIV-1 HXB2] were performed as previously described.4,25
HIV-1 coreceptor usage was inferred from the V3 loop nucleotide sequence by the geno2pheno algorithm available at the following website: http://coreceptor.bioinf.mpi-inf.mpg.de/. Based on the analysis of clinical data from MOTIVATE and MERIT studies, HIV-1 coreceptor usage was set at a false-positive rate (FPR) of 10% when analyzing plasma-derived viral sequences and 2–5.75% when analyzing PBMC-derived sequences from biological viral clones.26,27
Sequencing of the reverse transcriptase (RT)-protease (PR) region
The RT-PR region, a 1,244-base pair fragment of HIV-1 pol, was amplified from plasma with an in-house nested polymerase chain reaction (PCR) method with primer 5′prot 1 (nt: 2,082–2,106 of HIV-1 HXB2) and RT4R (nt: 3,348–3,370) using Super-Script III RT-PCR with Platinum Taq (Invitrogen). The inner amplification step was performed with the forward primer F1 (nt: 2,170–2,189) and the reverse primer 3R (nt: 3,304–3,326). Sequencing was done with an automated sequencer (ABI-3130) using the amplification primers with the BigDye terminator v3.1 cycle sequencing kit (Applied-Biosystems). Drug resistance mutations have been identified using the Stanford Genotypic Resistance Interpretation Algorithm (http://hivdb.stanford.edu/pages/algs/HIVdb.html).
Phylogenetic analysis
Nucleotide sequences of the V3 loop and RT and PR regions were aligned by Clustal X (1.81), and phylogenetic analysis was performed using MEGA 5 software, estimating tree and branch support with a bootstrapped neighbor-joining method.
Results
Clinical and immunovirological characteristics of the patient at baseline
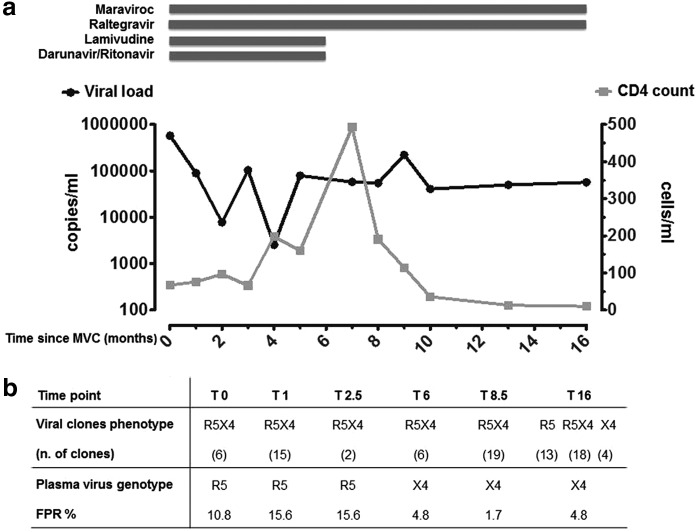
Here we report a case study of a 17-year-old HIV-1 patient infected since birth, who on February 2008 (baseline, t=0) presented with clinical features of Center for Disease Control (CDC) stage B3 with severe lipodystrophy, CD4+ T cell count of 67 cell/ml, and HIV-RNA plasma load of 578.938 copies/ml as well as an extensive resistance profile to ARV drugs. At baseline resistance mutations were D30N, M46L, I54V, N88D, L90M for PIs, M41L, E44D, D67N, L74V, V118I, M184V, L210W, T215Y, K219E for NRTIs, V179I, and Y181C for NNRTIs and none for integrase inhibitors (InIs). Efforts to design an effective ARV regimen included the determination of viral coreceptor tropism using the phenotypic assay Trofile (Monogram Biosciences, USA), which revealed a dual/mix viral population, and thus capable of using CXCR4 and CCR5 as coreceptors. Due to a lack of alternative treatment options, the ongoing therapeutic regimen DRV/r, ETR, TDF, and FTC was replaced with DRV/r (600 and 100 mg bid), 3TC (300 mg qd), MVC (150 mg bid), and RAL (400 mg bid) (Fig. 1a). The CD4+ T cell count increased to 493 cells/ml and the viral load decreased to 2,554 copies/ml; however, this was only temporary; afterward the CD4+ T cells declined (114 cells/ml) and the viral load rebounded to remain stable until the end of virological follow-up in June 2009 (t=16 months). At further Trofile phenotypic assay determinations the patient harbored dual/mix virus in the plasma during the entire follow-up.
FIG. 1.
(a) Antiretroviral therapy, plasma HIV RNA load, and CD4+ T cell count during maraviroc (MVC) treatment. (b) Comparison of phenotype of peripheral blood mononuclear cell (PBMC)-derived biological viral clones and plasma virus genotype, as assessed by geno2pheno relative false-positive rate (FPR), during 16 months of follow-up. The phenotype of PBMC-derived biological viral clones was determined in U87.CD4 cell lines expressing CCR5 and CXCR4 coreceptors. Geno2pheno was set at an FPR of 10% when analyzing plasma-derived viral sequences and 2–5.75% when analyzing PBMC-derived sequences from biological viral clones.
Phenotypic evolution of biological viral clones
The virus was biologically cloned from the patient's PBMC and the phenotype of each biological viral clone was tested in U87.CD4 cells expressing CCR5, CXCR4, or the chimeric receptors FC-2, 4b, 5, 6, and 7. From baseline to 8.5 months of follow-up only R5X4 variants were detected among the viral clones (Fig. 1b). Moreover, the 48 R5X4 clones isolated during this time frame used all FC-4b, of which 47 clones also used FC-7, 42 also used FC-6, and 11 also used FC-5, whereas only 4 of 48 clones were able to use the chimeric receptor FC-2, suggesting that they were more heterogeneous and flexible in the use of CXCR4 than CCR5 (Table 1).
Table 1.
Viral Phenotype of the Biological Viral Clones Obtained During 16 Months of Follow-Up
| Chimeric receptor use combination (no. of clones) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Time point (months) | Viral phenotype (no. of clones) | FC 2 | FC 4b | FC 6 | FC 4b-6 | FC 4b-7 | FC 4b-6-7 | FC 2-4b-6-7 | FC 4b-5-6-7 | FC 2-4b-5-6-7 |
| 0 | R5X4 (6) | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 2 | 0 |
| 1 | R5X4 (15) | 0 | 0 | 0 | 0 | 3 | 7 | 0 | 3 | 2 |
| 2,5 | R5X4 (2) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| 6 | R5X4 (6) | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 |
| 8,5 | R5X4 (19) | 0 | 0 | 0 | 0 | 0 | 16 | 0 | 3 | 0 |
| 16 | R5N (3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| R5B (10) | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| R5X4 (18) | 0 | 0 | 1 | 0 | 1 | 15 | 0 | 1 | 0 | |
| X4 (4) | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | |
Phenotype of each biological viral clone was determined in the cell line U87.CD4 expressing the chemochine receptors CCR5 or CXCR4 or the chimeric receptors FC-2, 4b, 5, 6, and 7. R5N stays for narrow, which means that the viral clones were able to use only the wild-type CCR5 as coreceptor, whereas R5B stays for broad, which means that the virus was able to use at least one of the chimeric receptors in addition to CCR5.
After 16 months of MVC treatment, only R5 (n=13) and X4 (n=4) clonal variants rose in addition to R5X4 (n=18) (Fig. 1b). Three of the R5 clones were R5narrow as they used only the wild-type CCR5; nine also used FC2 and one FC4 chimeric receptor and, thus, had a flexible use of CCR5 and were classified as R5broad (Table 1). The R5X4 viral clones were not flexible in their CCR5 usage, as none of these was able to use the FC2 receptor, whereas 1 clone used FC5 and 17 used FC4b, FC6, and FC7.
To gain further insight into coreceptor preference, we investigated RANTES and MVC susceptibility in a PBMC-based assay of two R5X4 clones (CL_0.03, CL_16.08), with one X4 (CL_16.10) and one R5 (CL_16.01) as control. The CXCR4-using clones were resistant to both CCR5 inhibitors, whereas the R5 clone was sensitive to RANTES and MVC (ID50 7.8 ng/ml and 14 nM, respectively) (Table 2). This further supports the prevalent CXCR4-using nature of the R5X4 clones.
Table 2.
Phenotypic and Genotypic Characterization of the Biological Viral Clones Obtained at Baseline and End of Maraviroc Treatment
| Genotypeb | |||||
|---|---|---|---|---|---|
| Biological viral clones | Phenotypea | geno2pheno | PSSM | FPR %c | Amino acid sequence of the env V3 loop |
| Baseline | CTRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAYCd | ||||
| CL_0.01 | R5X4 | R5 | R5 | 15.6 | .............L.L..RW.A.-N......K... |
| CL_0.02 | R5X4 | R5 | R5 | 15.6 | .............L.L..RW.A.-N......K... |
| CL_0.03e | R5X4 | R5 | R5 | 15.6 | .............L.L..RW.A.-N......K... |
| CL_0.04 | R5X4 | R5 | R5 | 15.6 | .............L.L..RW.A.-N......K... |
| T=16 months | |||||
| CL_16.01e | R5N | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.02 | R5N | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.03 | R5N | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.04 | R5B (FC2) | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.05 | R5B (FC2) | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.06 | R5B (FC4) | R5 | R5 | 9 | .I........G........W.A.-D......K... |
| CL_16.07 | R5X4 | X4 | X4 | 1.7 | .........RD..L.L..RW.A.-K.V....K... |
| CL_16.08e | R5X4 | X4 | X4 | 1.7 | .........RD..L.L..RW.A.-K.V....K... |
| CL_16.09 | X4 | X4 | X4 | 1.7 | .........RD..L.L..RW.A.-K.V....K... |
| CL_16.10e | X4 | X4 | X4 | 1.7 | .........RD..L.L..RW.A.-K......K... |
| CL_16.11 | X4 | X4 | X4 | 1.7 | .........RD..L.L..RW.A.-K.V....K... |
Phenotype of each biological viral clone was determined in the cell line U87.CD4 expressing the chemochine receptors CCR5 or CXCR4 or the chimeric receptors FC 2, 4b, 5, 6, and 7. R5N stays for narrow, which means that the viral clones were able to use only the wild-type CCR5 as coreceptor, whereas R5B stays for broad, which means that the virus was able to use at least one of the chimeric receptors in addition to CCR5. The use of the chimeric receptor is indicated in parentheses.
The predicted phenotype was obtained by the geno2pheno approach setting the FPR cut off at 2–5.75% and by position-specific scoring matrices (PSSM).
FPR means false positive rate and is expressed in %.
Amino acid sequences are aligned to the HIV-1 subtype B consensus sequence. (.) means identical amino acid, (-) lacking amino acid, or otherwise the alternative amino acid is indicated.
Sensitivity to RANTES and MVC of the indicated viral clones has been evaluated in a PBMC-based assay. Clone CL_16.01 is the only one sensitive to RANTES (ID50 7.8 ng/ml) and MVC (ID50 14 nM).
Resistance pattern to drugs targeting PR-RT
To determine whether the observed viral tropism could be driven by a specific resistance pattern to drugs targeting genes other than env, the coding sequence of PR-RT from four R5X4 biological clones at baseline and 11 clones at 16 months representing the different phenotypes (three R5narrow and three R5broad, three X4, and two R5X4) was sequenced. Surprisingly, the four clones at baseline were wild-type for all PR and RT mutations characteristic of drug resistance. Instead a multiresistance pattern was present in viral variants sequenced from plasma samples obtained immediately before and after the baseline time point (data not shown), also including the mutation M184V, a critical marker of low fitness that was already present at baseline. At 16 months of follow-up all viral clones showed several markers of drug resistance (cumulative resistance profile: PR: D30N, M46L, I54V, L90M, L10F, V11I, N88D; NRT: M41L, D67N, L74V M184V; NNRT: V90I, Y181C), independent of coreceptor tropism, thus suggesting that the selection of the viral tropism was not due to a pressure induced by NRT, NNRT, or PI drugs.
A phylogenetic analysis was performed with the PR and RT sequences of the 15 biological clones, which also included four plasma samples spanning a period beyond the virological follow-up from December 2007 to October 2009. The phylogenetic tree in Fig. 2 shows a clear separation into two clusters, with one cluster including the four plasma samples and the biological clones of the end of follow-up (June 2009) and the other cluster including the four PR and RT wild-type clones at baseline (February 2008).
FIG. 2.
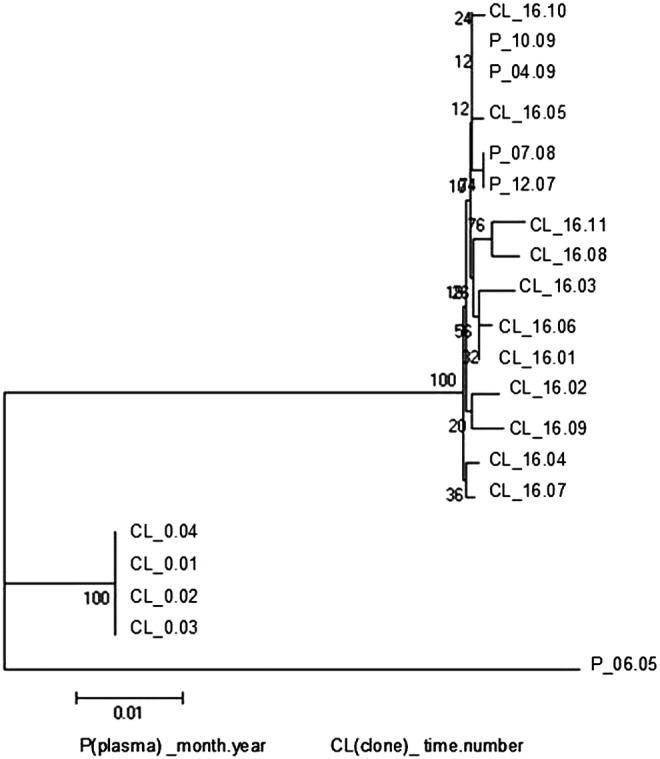
Phylogenetic analysis of reverse transcriptase (RT) and protease (PR) sequences from PBMC-derived biological clones as compared to those obtained from plasma. Biological clones from baseline (CL_0.number) and from the end (CL_16.number) of the MVC-containing regimen (obtained February 2008 and June 2009) were analyzed together with four plasma-derived viral sequences from December 2007 (P_12.07) to October 2009 (P_10.09). An unrelated sequence was used as an out-group. Bootstrap values are indicated at the nodes.
Tropism determination by env V3 loop sequencing
The same biological clones were sequenced to determine the tropism through the geno2pheno algorithm with the FPR set at 2–5.75% (Table 2). At baseline all the R5X4 biological variants showed an R5 sequence (FPR=9%). At the end of follow-up genotypic and phenotypic results were concordant for all but two viruses, which had an R5X4 phenotype but an X4 genotype (FPR=1.7%). The same tropism was also predicted by position-specific scoring matrices (PSSM).28 Thus, the inferred genotype can sometimes predict R5 or X4 instead of the R5X4 phenotype.
The alignment of the sequences of viral clones showed that the four clones at baseline had identical V3 loop sequences with an amino acid S in position 11 and N in position 25; thus, according to the 11–25 rule, they were defined as R5, but had an overall V3 loop net charge of 7, which is more characteristic of X4 viruses (Table 2). Moreover, all these clones showed a deletion of G at position 24, which may be indicative of preferential CXCR4 usage.29 However, at 16 months of follow-up a different amino acidic pattern characterized the R5 and CXCR4-using viruses. Amino acid sequences and FPR were identical within each phenotype and did not distinguish R5narrow from R5broad or R5X4 from X4.
The tropism of the virus isolated from plasma samples collected throughout the 16 months of follow-up was determined by analysis of the V3 loop sequence through the geno2pheno algorithm set at an FPR of 10%. The analysis predicted CCR5 usage at baseline with an FPR close to cut-off (10.8%) and a shift to X4 (FPR 4.8%) after 6 months, which persisted throughout the virological follow-up (Fig. 1b). Related to this shift, the number of viral quasispecies increased with multiple residues at positions 2, 10, 11, 14, 16, 19, 25, and 27, as revealed from the fact that the sequencing reached a maximum at 16 months after the start of MVC administration (data not shown). Thereafter the viral population tended to again narrow its variability. A phylogenetic analysis of the V3 loop sequences of the viral clones and of those obtained from plasma at 16 months showed that the consensus sequence obtained by direct sequencing from plasma was the overall expression of the two phenotypic population R5 and X4, as represented by its position dividing the phylogenetic tree into two distinct branches (Fig. 3).
FIG. 3.
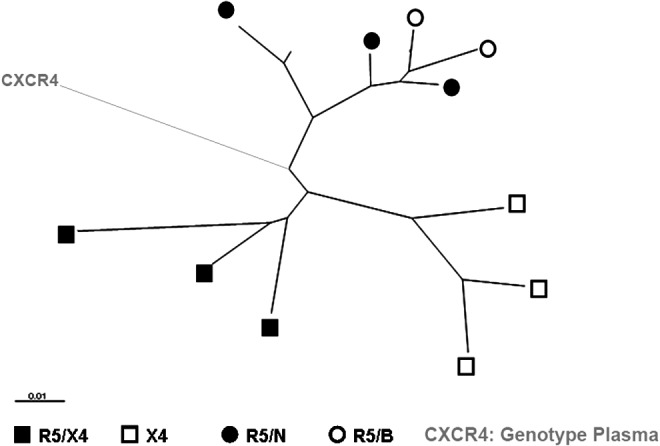
Phylogenetic analysis of HIV-1 env V3-loop sequences from PBMC-derived biological clones and from plasma after 16 months of an MVC-containing antiretroviral (ARV) regimen. Symbols indicate the viral phenotype of the biological viral clones as determined in U87.CD4 cells expressing CCR5, CXCR4, or the chimeric receptors. R5/N indicates a virus with an R5narrow phenotype able to use only wild-type CCR5 as the coreceptor. R5/B indicates a virus with an R5broad phenotype able to use one or more chimeric receptors other than wild-type CCR5. CXCR4 corresponds to the sequence derived from plasma, which has a predicted genotype of X4, as determined by the geno2pheno algorithm.
In summary, although there were no variations in the therapeutic regimen, an enlargement of the PBMC-derived viral population occurred in parallel with the increase in viral variability in the plasma population.
Discussion
Here we show that a prolonged MVC-containing ARV regimen in a multiexperienced and multiresistant HIV-1-infected adolescent with an R5X4 virus can induce enlargement of the virus population so as to also contain pure R5 and X4 viral clones. We decided to treat this adolescent with a salvage therapy containing MVC due to the extensive ARV resistance profile and limited treatment options despite the presence of a dual/mixed virus population; we retrospectively studied the viral evolution due to the observed immunological and virological response.
Indeed, MVC is currently licensed in clinical practice for patients with R5 virus and virological failure; however, in anecdotal reports the dual/mixed viruses were inhibited by MVC.30 This may be explained by the fact that dual/mixed virus populations may also contain R5 viruses in addition to R5X4. Furthermore, the prevalence of R5 viruses in multitreated patients supports the use of MVC.18 On the other hand, in ARV-experienced children and adolescents with vertical HIV-1 infection CXCR4-using variants are frequent, which would instead suggest a limited role for CCR5 antagonists.24,31 However, we previously showed that pure X4 clonal viruses are rare in children with an R5X4 bulk virus population.3 Therefore it is of great importance to extensively characterize the evolution of the viral population in R5X4-infected individuals during MVC treatment.
During the first 6 months of MVC treatment, the adolescent showed a steep increase in CD4+ T cell count accompanied by a decrease in viral load. The CD4+ T cell recovery was probably due to MVC, as in our clinical experience as well as that of other groups the use of MVC in multiexperienced patients is safe and efficacious, inducing an immunological recovery despite the presence of CXCR4-using viruses.30,32,33 The absence of resistant mutations to InIs suggests that RAL was prevalently responsible for the concomitant decrease in viral load observed. This is in line with the long-term data of BENCHMRK studies that show the virological efficacy of RAL, suggesting that the use of antiretroviral treatment containing InIs is associated with a fast viral decrease in the first weeks of treatment.34
The subsequent virological rebound observed in our patient is difficult to explain. It is apparently not due to the appearance of resistance mutations to RAL, which our patient never harbored. However, it has to be mentioned that a rebound after a switch to RAL/MVC therapy is not surprising, as was also described in R5-infected and virologically suppressed patients.35 The effect on patients carrying dual-tropic viruses, such as the adolescent described here, has not yet been investigated.
The impressive CD4+ T cell recovery of this patient suggests that MVC possibly also had an effect on R5X4 viruses. However, the R5X4 PBMC-derived biological clone obtained at baseline was resistant to RANTES and MVC when tested in vitro. Interestingly, the predicted phenotype of the plasma-derived bulk virus population showed an R5 phenotype with an FPR of 10.8–15.6%. Thus, possibly MVC was acting on viral populations still circulating in the plasma more than on the R5X4 viruses of the PBMCs.
Although there was no change in MVC treatment after 6 months, the CD4+ T cell counts dropped when the viral load stabilized. During this period the plasma-derived viral population changed to an X4 predicted phenotype, providing evidence of resistance to MVC of the circulating viral population. However, the PBMC-derived clonal population was not affected during this same period, but underwent an enlargement to later include R5 and X4 virus clones. A greater prevalence of X4 variants in PBMCs than in plasma has already been observed for patients in both early36 and chronic phases37,38 of HIV-1 infection, and this is consistent with the observation that the archived virus population may not correspond to the most prevalent variant in plasma.39–41
It has been reported that certain dual-tropic viruses prefer usage of the CCR5 and are thus defined as R5>X4 or dual-R5-tropic, whereas others use the CXCR4 coreceptor more efficiently (X4>R5 or dual-X4).42,43 In our case, the viral chimeric receptor usage suggested that the dominant viral population during the first 8 months of follow-up of the patient could be considered an X4>R5 phenotype since only a minority (4 out of 48) of the biological clones could utilize the chimeric receptor FC-2, in which the three extracellular loops are derived from CCR5. Analogously, for the R5X4 population at 16 months of follow-up, the first extracellular loop of the CXCR4, represented by chimera FC-4b, is enough to allow infection. Furthermore, entry of the R5X4 viruses from both baseline and 16 months of follow-up was not inhibited by RANTES and MVC in vitro, demonstrating their preferential usage of CXCR4. Interestingly, pure R5 biological clones, of which 77% have a flexible use of the CCR5 (R5broad), rose at 16 months of follow-up. Thus, R5 viral variants with an altered receptor interaction may emerge as a consequence of the changed CCR5 conformation due to MVC binding.
The geno2pheno prediction misplaced some of the R5X4 biological viral clones and predicted an R5 phenotype. Indeed, the FPR cut-off was set at 10% and the actual FPR of the clones was 15.6%, thus being close to an X4 predictive FPR. Analogously, the algorithm wrongly recognized as X4 the R5X4 clones at t=16 months. However, it did not misplace those clones that had a monotropic phenotype, i.e., R5 or X4. Further studies may be needed to determine if specific mutations, such as the deletion at position 24, are indicative of an X4 phenotype in subtype C HIV-1 or if others may affect the geno2pheno readout.29 Among amino acid mutations known to relate to MVC escape only the K305R (position 10 of the V3 loop) mutation was detected, but only in the CXCR4-using clones obtained at month 16.44
Interestingly at position 25 all the R5 plasma samples had the uncharged amino acid asparagine (N), whereas at 16 months the plasma sample had four predicted amino acids, one of them being the positive charged lysine (K), which was strictly correlated with CXCR4 tropism. The N-coding sequence in the R5 plasma samples (AAC) can shift to the K-coding sequence (AAA) seen in X4 viruses by a single nucleotide change. Indeed, the four R5 clones obtained at baseline also had an N in position 25, while among the 11 clones at t=16 months, those with CCR5 tropism had an aspartic acid (GAC) and those with X4 tropism had a lysine (AAA), indicating a divergence from the baseline genotype due to mutations AAC-AAA (X4 shift) and AAC-GAC (R5 stable).
We have addressed the issue of whether the different tropisms of the clones could be linked to a specific pol genotype and thus whether a “carryover” mechanism could be identified as a driving force for the change in tropism during follow-up45; however, no correlation was seen between specific mutations and the tropism of the clones. The four clones at baseline did not carry the mutations related to RT or PR inhibitors, suggesting that they are probably the expression of an archived minor viral population, which could not actively replicate and spread because of its sensitivity to drugs. Although we were able to recover them in an in vitro culture devoid of ARV pressure, they did not take over during 16 months of treatment of the adolescent.
In conclusion, our findings may provide a guide for therapeutic choices in multiexperienced HIV-infected patients with limited therapeutic options, and may emphasize the importance of extensively characterizing the evolution of the viral population in infected individuals.
Supplementary Material
Acknowledgments
We thank the patient and the family. This work was supported by the European Community funded projects EUROPRISE-Network of Excellence Grant LSHP CT-2006-037611 and by the Melissa & Bill Gates Foundation OPP38580. Lara Mainetti and Angela Rosa Pignataro contributed equally to this work.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Deng H, Liu R, Ellmeier W, et al. : Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996;381:661–666 [DOI] [PubMed] [Google Scholar]
- 2.Berger EA, Doms RW, Fenyo EM, et al. : A new classification for HIV-1. Nature 1998;391:240. [DOI] [PubMed] [Google Scholar]
- 3.Trouplin V, Salvatori F, Cappello F, et al. : Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J Virol 2001;75:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svicher V, D'Arrigo R, Alteri C, et al. : Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: Results from the OSCAR Study Group. New Microbiol 2010;33:195–206 [PubMed] [Google Scholar]
- 5.Prosperi MC, Bracciale L, Fabbiani M, et al. : Comparative determination of HIV-1 co-receptor tropism by Enhanced Sensitivity Trofile, gp120 V3-loop RNA and DNA genotyping. Retrovirology 2010;7:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon B, Grabmeier-Pfistershammer K, Rieger A, et al. : HIV coreceptor tropism in antiretroviral treatment-naive patients newly diagnosed at a late stage of HIV infection. AIDS 2010;24:2051–2058 [DOI] [PubMed] [Google Scholar]
- 7.Scarlatti G, Hodara V, Rossi P, et al. : Transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child correlates with viral phenotype. Virology 1993;197:624–629 [DOI] [PubMed] [Google Scholar]
- 8.Schuitemaker H, Kootstra NA, de Goede RE, et al. : Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol 1991;65:356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tersmette M, Gruters RA, de Wolf F, et al. : Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: Studies on sequential HIV isolates. J Virol 1989;63:2118–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koot M, Keet IP, Vos AH, et al. : Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med 1993;118:681–688 [DOI] [PubMed] [Google Scholar]
- 11.Cavarelli M, Karlsson I, Zanchetta M, et al. : HIV-1 with multiple CCR5/CXCR4 chimeric receptor use is predictive of immunological failure in infected children. PloS One 2008;3:e3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsson I, Antonsson L, Shi Y, et al. : Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J Virol 2004;78:11807–11815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulick RM, Lalezari J, Goodrich J, et al. : Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med 2008;359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorr P, Westby M, Dobbs S, et al. : Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 2005;49:4721–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fatkenheuer G, Pozniak AL, Johnson MA, et al. : Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med 2005;11:1170–1172 [DOI] [PubMed] [Google Scholar]
- 16.Cooper DA, Heera J, Goodrich J, et al. : Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis 2010;201:803–813 [DOI] [PubMed] [Google Scholar]
- 17.Fatkenheuer G, Nelson M, Lazzarin A, et al. : Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med 2008;359:1442–1455 [DOI] [PubMed] [Google Scholar]
- 18.Saag M, Goodrich J, Fatkenheuer G, et al. : A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J Infect Dis 2009;199:1638–1647 [DOI] [PubMed] [Google Scholar]
- 19.McGovern RA, Symons J, Poon AF, et al. : Maraviroc treatment in non-R5-HIV-1-infected patients results in the selection of extreme CXCR4-using variants with limited effect on the total viral setpoint. J Antimicrob Chemother 2013;68:2007–2014 [DOI] [PubMed] [Google Scholar]
- 20.Castagna A, Maggiolo F, Penco G, et al. : Dolutegravir in antiretroviral-experienced patients with raltegravir- and/or elvitegravir-resistant HIV-1: 24-week results of the phase III VIKING-3 study. J Infect Dis 2014;210:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuitemaker H, Koot M, Kootstra NA, et al. : Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol 1992;66:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavarelli M, Karlsson I, Ripamonti C, et al. : Flexible use of CCR5 in the absence of CXCR4 use explains the immune deficiency in HIV-1 infected children. AIDS 2010;24:2527–2533 [DOI] [PubMed] [Google Scholar]
- 23.Karlsson I, Antonsson L, Shi Y, et al. : HIV biological variability unveiled: Frequent isolations and chimeric receptors reveal unprecedented variation of coreceptor use. AIDS 2003;17:2561–2569 [DOI] [PubMed] [Google Scholar]
- 24.Scarlatti G, Tresoldi E, Bjorndal A, et al. : In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med 1997;3:1259–1265 [DOI] [PubMed] [Google Scholar]
- 25.Svicher V, Alteri C, Montano M, et al. : Performance of genotypic tropism testing on proviral DNA in clinical practice: Results from the DIVA study group. New Microbiol 2012;35:17–25 [PubMed] [Google Scholar]
- 26.McGovern RA, Thielen A, Mo T, et al. : Population-based V3 genotypic tropism assay: A retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 2010;24:2517–2525 [DOI] [PubMed] [Google Scholar]
- 27.McGovern RA, Thielen A, Portsmouth S, et al. : Population-based sequencing of the V3-loop can predict the virological response to maraviroc in treatment-naive patients of the MERIT trial. J Acquir Immune Defic Syndr 2012;61:279. [DOI] [PubMed] [Google Scholar]
- 28.Jensen MA, Li FS, van ’t Wout AB, et al. : Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol 2003;77:13376–13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coetzer M, Cilliers T, Ping LH, et al. : Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology 2006;356:95–105 [DOI] [PubMed] [Google Scholar]
- 30.Symons J, van Lelyveld SF, Hoepelman AI, et al. : Maraviroc is able to inhibit dual-R5 viruses in a dual/mixed HIV-1-infected patient. J Antimicrob Chemother 2011;66:890–895 [DOI] [PubMed] [Google Scholar]
- 31.Briz V, Garcia D, Mendez-Lagares G, et al. : High prevalence of X4/DM-tropic variants in children and adolescents infected with HIV-1 by vertical transmission. Pediatr Infect Dis J 2012;31:1048–1052 [DOI] [PubMed] [Google Scholar]
- 32.Bon I, Clo A, Borderi M, et al. : Prevalence of R5 strains in multi-treated HIV subjects and impact of new regimens including maraviroc in a selected group of patients with CCR5-tropic HIV-1 infection. Int J Infect Dis 2013;17:e875–e882 [DOI] [PubMed] [Google Scholar]
- 33.Nozza S, Galli L, Visco F, et al. : Raltegravir, maraviroc, etravirine: An effective protease inhibitor and nucleoside reverse transcriptase inhibitor-sparing regimen for salvage therapy in HIV-infected patients with triple-class experience. AIDS 2010;24:924–928 [DOI] [PubMed] [Google Scholar]
- 34.Eron JJ, Cooper DA, Steigbigel RT, et al. : Efficacy and safety of raltegravir for treatment of HIV for 5 years in the BENCHMRK studies: Final results of two randomised, placebo-controlled trials. Lancet Infect Dis 2013;13:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nozza S, Bigoloni A, Calcagno A, et al. : Viral rebound after switch to maraviroc/raltegravir dual therapy in highly experienced and virologically suppressed patients with HIV-1 infection. J Antimicrob Chemother 2014;69:1436–1439 [DOI] [PubMed] [Google Scholar]
- 36.Frange P, Galimand J, Goujard C, et al. : High frequency of X4/DM-tropic viruses in PBMC samples from patients with primary HIV-1 subtype-B infection in 1996–2007: The French ANRS CO06 PRIMO Cohort Study. J Antimicrob Chemother 2009;64:135–141 [DOI] [PubMed] [Google Scholar]
- 37.Baroncelli S, Galluzzo CM, Weimer LE, et al. : Evolution of proviral DNA HIV-1 tropism under selective pressure of maraviroc-based therapy. J Antimicrob Chemother 2012;67:1479–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhofstede C, Vandekerckhove L, Eygen VV, et al. : CXCR4-using HIV type 1 variants are more commonly found in peripheral blood mononuclear cell DNA than in plasma RNA. J Acquir Immune Defic Syndr 2009;50:126–136 [DOI] [PubMed] [Google Scholar]
- 39.Lehmann C, Daumer M, Boussaad I, et al. : Stable coreceptor usage of HIV in patients with ongoing treatment failure on HAART. J Clin Virol 2006;37:300–304 [DOI] [PubMed] [Google Scholar]
- 40.Noe A, Plum J, and Verhofstede C: The latent HIV-1 reservoir in patients undergoing HAART: An archive of pre-HAART drug resistance. J Antimicrob Chemother 2005;55:410–412 [DOI] [PubMed] [Google Scholar]
- 41.Turriziani O, Bucci M, Stano A, et al. : Genotypic resistance of archived and circulating viral strains in the blood of treated HIV-infected individuals. J Acquir Immune Defic Syndr 2007;44:518–524 [DOI] [PubMed] [Google Scholar]
- 42.Svicher V, Balestra E, Cento V, et al. : HIV-1 dual/mixed tropic isolates show different genetic and phenotypic characteristics and response to maraviroc in vitro. Antiviral Res 2011;90:42–53 [DOI] [PubMed] [Google Scholar]
- 43.Toma J, Whitcomb JM, Petropoulos CJ, et al. : Dual-tropic HIV type 1 isolates vary dramatically in their utilization of CCR5 and CXCR4 coreceptors. AIDS 2010;24:2181–2186 [DOI] [PubMed] [Google Scholar]
- 44.Berro R, Klasse PJ, Jakobsen MR, et al. : V3 determinants of HIV-1 escape from the CCR5 inhibitors maraviroc and vicriviroc. Virology 2012;427:158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeri E, Gianotti N, Canducci F, et al. : Evolutionary characteristics of HIV type 1 variants resistant to protease inhibitors in the absence of drug-selective pressure. AIDS Res Hum Retroviruses 2003;19:1151–1153 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.