Abstract
Ribosomopathies are diseases caused by alterations in the structure or function of ribosomal components. Progress in our understanding of the role of the ribosome in translational and transcriptional regulation has clarified the mechanisms of the ribosomopathies and the relationship between ribosomal dysfunction and other diseases, especially cancer. This review aims to discuss these topics with updated information.
Keywords: ribosomopathy, Diamond-Blackfan anemia (DBA), 5q-syndrome, Schwachman-Diamond syndrome (SDS), dyskeratosis congenita (DC), cartilage hair hypoplasia (CHH), Treacher Collins syndrome (TCS), p53
Introduction
As the apparatus of protein synthesis, the ribosome is one of the most precisely constructed and regulated molecular machines in the cell. Alterations in ribosomal components, or in the numerous cellular products with effects on ribosomal structure and function, can cause a heterogeneous class of diseases known as ribosomopathies. Although they all involve ribosomal dysfunction, these diseases differ significantly in mechanism, clinical presentation, and potential for treatment. This diversity corresponds to a developing understanding of the multiple specialized roles of the ribosome in normal function. Recent studies on ribosomopathies have yielded more insight into other diseases, including multiple cancers, and key cellular pathways, notably of the tumor suppressor p53. We will review these developments here, with an emphasis on the molecular mechanisms of disease, and discuss their implications for treatment and further research.
Ribosomes and Ribosomal Proteins: Structure and Function
The ribosome principally consists of ribosomal RNA (rRNA), ribosomal proteins (RPs), and small nucleolar RNAs (snoRNAs). rRNA catalyzes peptide bond formation during protein synthesis; RPs optimize rRNA processing and stabilize the ribosome’s final structure; and snoRNAs primarily regulate chemical modifications of other RNAs.1,2 rRNA transcription and assembly with RPs occur in the nucleolus, after which the ribosomal subunit is exported from the nucleus to the cytoplasm, the site of translation. In eukaryotes, regulation of translation occurs primarily at the initiation step and is mediated by eukaryotic initiation factors (eIFs).3 Most mRNAs are primed for translation via a mechanism by which eIFs recognize either the 7-methylguanylate cap at the 5′ end or the poly-A tail at the 3′ end of the mRNA, facilitating its binding to the translation preinitiation complex (PIC). However, some mRNAs undergo a cap-independent mechanism of initiation, in which the PIC is directly recruited by a nucleotide sequence called the internal ribosomal entry site (IRES) within the mRNA.4 IRESs were first discovered in viral mRNAs, which are often uncapped; IRESs facilitate translation of viral mRNAs even when eIFs are downregulated, as they commonly are in infected cells.5,6 Eukaryotic genes whose transcripts feature an IRES include those involved in the cellular stress response, and accordingly IRES-dependent translation can promote either apoptosis or cell survival.7
The ribosome itself has long been thought to play a primarily constitutive role in translation, rather than a regulatory one. However, studies of Escherichia coli in the 1960s showed that alterations in ribosomal structure, such as those induced by the antibiotic streptomycin, can affect translational fidelity.8,9 These findings have been corroborated by mutational analyses of E. coli ribosomes,10 as well as by structural studies.2,11 Furthermore, analysis of polypeptide chain elongation kinetics has shown that inhibiting the steps that precede mRNA binding preferentially blocks the translation of lower-quality mRNAs, which have smaller initiation rate constants. This represents another mechanism by which translational fidelity can be ensured.12
More recent investigations have shown that ribosomes and RPs selectively regulate the expression of specific mRNAs, an idea known as the ribosome filter hypothesis.13 In 2011, Kondrashov et al showed that skeletal patterning defects in tail-short (Ts/+) mice were caused by loss of function in the Rpl38, which resulted in decreased translation in a subset of homeobox proteins.14 In the same study, quantitative gene-expression profiling of 72 RPs showed heterogeneity of expression levels among different tissues during organogenesis, suggesting that RPs other than Rpl38 can regulate translation in a tissue-specific manner.
Other RPs regulate translation via extraribosomal mechanisms.15 One such RP, Rpl26, induces translation of p53 by binding to the 5′ and 3′ untranslated regions of p53 mRNA.16,17 It remains uncertain whether ribosomal or extraribosomal effects account for most translational regulations by RPs.
Given the ubiquity of ribosomes and RPs, a paradoxical feature of ribosomopathies is the variety of their phenotypic effects. This variety is explained in part by the emerging concept of “specialized ribosomes,” in which tissue-specific variations in ribosomal structure or function confer regulatory specificity in translation.18 These variations include the expression of RP paralogs in different tissues,19,20 heterogeneity of RP expression levels during embryogenesis14,21,22 and in adult life,23 differences in post-translational modifications,24–26 changes in expression of ribosome-associated factors other than RPs,27–30 and heterogeneity in rRNA structure.31–33 The composition of ribosomes and other elements of the translational apparatus can also vary within cells, especially in neurons, where certain RPs and RNAs are selectively enriched in axons or dendrites relative to the soma.34,35 These variations in turn affect the translation of specific subsets of mRNAs. For example, modifications to rRNAs36–39 and to RPs40–42 can selectively impair IRES-dependent translation, and certain RPs appear to be necessary for the translation of some mRNAs but not others.43 These variations are clinically significant in several ribosomopathies.
Translational and Transcriptional Control: Ribosomes and p53
The transcription factor p53, a key tumor suppressor and regulator of cell fate, is particularly involved in the control of ribosomal function.44,45 p53 can activate or repress transcriptional targets to induce cell-cycle arrest or apoptosis,46,47 and indeed, both cell cycle arrest and apoptosis have been observed in impairments of ribosomal biogenesis. Studies of Bop1, a mouse protein involved in rRNA processing and production of the 60S ribosomal subunit, provided an early indication of the role of p53 in ribosomal dysfunction. A dominant-negative mutation of Bop1 led to p53-dependent cell-cycle arrest that could be relieved by inactivation of p53.48,49 Animal models have since provided further evidence for the connection between ribosomal insufficiency and p53 activation. For example, mice with T-cell-specific heterozygous deletion of the RP gene Rps6 exhibited decreased T-cell-mediated proliferation via a likely p53-dependent response.50 Mouse embryos with heterozygous Rps6 deletions died at gastrulation, when a p53-dependent checkpoint induced widespread apoptosis. Inactivation of p53 prolonged life briefly, until embryos appeared to die from placental abnormalities and impaired erythropoiesis.50,51 Mutations of other RPs in mice can cause less dramatic phenotypic abnormalities that are also specifically mediated by p53,52 and RP deficiencies in other animals can cause similar p53-dependent effects.53,54
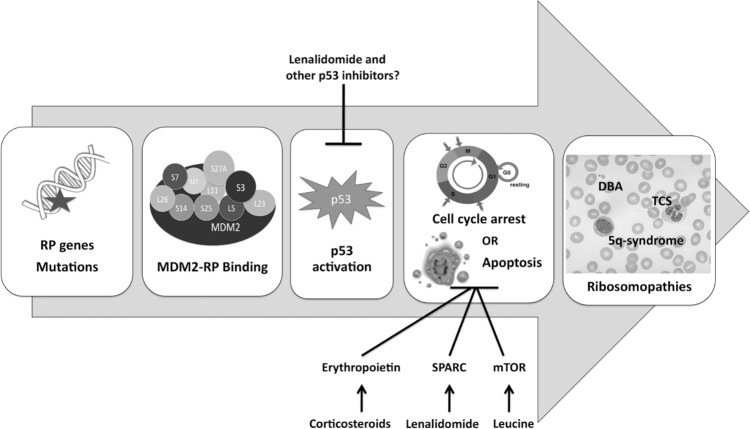
The mechanism by which impaired ribosomal biogenesis activates p53 has become increasingly clear in recent years.55 Individual RPs, including Rpl11,56,57 Rpl5,58 and Rpl23,59,60 can migrate from the nucleolus to the nucleoplasm; there, they bind to the protein MDM2, inhibiting its E3 ubiquitin ligase activity and preserving p53 from proteosomal degradation (Fig. 1). The precise effects, however, vary according to the perturbation, and we discuss them in the context of specific ribosomopathies below.
Figure 1.
The role of p53 activation in the pathogenesis of certain ribosomopathies. Bars indicate inhibition.
Ribosomopathies: Old and New Mechanisms
Clinical features of the ribosomopathies can include bone marrow failure, developmental abnormalities, and increased risk of cancer. However, ribosomal dysfunction can cause a wide range of signs and symptoms, and presentation and severity can differ dramatically even among patients with the same diagnosis (Table 1).
Table 1.
Clinical characteristics of ribosomopathies.
| DISEASE | GENE DEFECT | CLINICAL FEATURES | TREATMENT | REFERENCES |
|---|---|---|---|---|
| Diamond-Blackfan anemia (DBA) | 10–15 ribosomal proteins (esp. S19, S26, L5, L11) | Anemia Growth retardation Congenital (esp. craniofacial and thumb) abnormalities |
Corticosteroids Blood transfusions Hematopoietic stem cell transplantation (HSCT) Leucine (?) |
62,100,184 |
| 5q-syndrome | RPS14 | Anemia | Lenalidomide | 104,115 |
| Schwachman-Diamond syndrome (SDS) | SBDS | Exocrine pancreatic insufficiency Hematologic abnormalities (esp. neutropenia) Neurocognitive impairment Gl (esp. liver) abnormalities |
Pancreatic enzyme supplementation HSCT | 121,122 |
| X-linked dyskeratosis congenita (DC) | DKC1 | Mucocutaneous abnormalities (e.g. skin pigmentation and nail changes) Pulmonary fibrosis Bone marrow failure |
Oxymetholone HSCT | 135 |
| Cartilage-hair hypoplasia (CHH) | RMRP | Short stature Bone deformities Hair growth abnormalities |
Symptomatic | 149 |
| Treacher-Collins syndrome (TCS) | TCOF1 | Craniofacial abnormalities | Symptomatic | 156 |
| Bowen-Conradi syndrome | EMG1 | Severe growth retardation | None | 162–164 |
| North American Indian childhood cirrhosis | hUTP4/Cirhin | Biliary cirrhosis Portal hypertension |
Liver transplantation | 166–168 |
Diamond-Blackfan anemia (DBA)
This disease, a “founding member” of the class of ribosomopathies,61 presents with pure red cell aplasia in the first year of life as the cardinal symptom. In addition, approximately 40% of patients present with growth retardation or congenital abnormalities of the head, upper limb, kidney, or heart.62 In 1999, RPS19 was shown to be mutated in DBA, and RPS19 mutations have since been estimated to occur in approximately 25% of cases.63 Since then, the list of RPs implicated in DBA has expanded to include 10–15 additional candidates, although definitive roles in pathogenesis have not been established for all of them.62,64 Mutations in genes encoding RPs have been identified in 50–70% of patients with DBA.65 Not all cases of DBA can be attributed to RP dysfunction, however, and a recent study has identified a mutation in the hematopoietic transcription factor GATA1 (which has no known mechanistic links to ribosomes) in two DBA pedigrees.66 These uncertainties in classification may resolve with improved understanding of the molecular basis of the disease.
All RP mutations observed in DBA are heterozygous, supporting the idea that homozygous RP mutations are lethal to embryos. This hypothesis has been confirmed in some animal models of DBA.52,67 Mutations in the RPS19 gene have been identified as a possible cause of hydrops fetalis, suggesting that more severe RP mutations than those observed in DBA patients may not come to clinical attention.68 More than 200 mutations in DBA-associated genes have been cataloged in DBA patients,69 and as a result, the phenotype of DBA patients is also highly variable. Most cases of DBA are sporadic in transmission, but incomplete penetrance and variable expressivity are observed even in familial cases associated with the same mutation.70 Array comparative genomic hybridization studies have also identified genomic deletions of RP loci in mutation-negative patients.71 Some genotype–phenotype correlations have been established by studies of patient cohorts; for example, mutations in RPL5 and RPL11 are associated with oral cleft and thumb abnormalities, respectively,72,73 while RPL35A mutations are associated with genitourinary malformations.69
Given the diverse etiologies and presentations of DBA, understanding of its pathogenesis is incomplete. This state of affairs is further complicated by the fact that animal models to date have not fully replicated the DBA phenotype.74–76 RP deficiencies are associated with global decreases in translation in cells of both hematopoietic and non-hematopoietic lineage, an observation that may account for general features of DBA such as small stature.77–79 RP “crosstalk” has been observed in some studies, where removal or knockdown of an RP promoted the recruitment of other RP transcripts to polyribosomes for translation, but this enhanced recruitment could not compensate for decreased translational efficacy.42,80
The role of p53 in the pathogenesis of DBA is also incompletely understood. In vitro RNAi knockdown of RPS19 in human CD34+ hematopoietic progenitor cells replicates the defective erythropoiesis of DBA81,82 and is associated with p53 accumulation in the erythroid lineage; in this setting, restoration of RPS19 expression and p53 inactivation both rescue the DBA phenotype.83 RPS19 knockdown interferes with ribosomal biogenesis in yeast84 and in mammalian cells, leading to p53 activation.85,86 More generally, knockdown of some RPs can relieve miRNA-mediated repression of translation, again via p53 activation.87 RPL5 and RPL11, the two RPs that are responsible for a plurality of observed non-erythroid phenotypes when mutated in DBA, have the specific function of transporting 5S rRNA to the developing 60S ribosomal subunit and forming a complex that inhibits Mdm2.88,89
It has been postulated that hypersensitivity of erythroblasts to p53 or high requirements for protein synthesis during erythropoiesis account for the central role of p53 in the pathogenesis of DBA78,83 (Fig. 1). Intriguingly, a recent study of CD34+ cells from DBA patients suggested that the effect of p53 activation can vary depending on the affected RP; RPS19 mutations decreased proliferation and cell-cycle arrest but had little effect on differentiation and apoptosis, while RPL11 mutations delayed differentiation and increased apoptosis.90 There is also evidence for p53-independent mechanisms in the pathogenesis of DBA and especially of the erythroid defect. In an RPS19-deficient zebrafish model of DBA, for example, inhibition of p53 rescued the morphological abnormalities, but not the erythropoietic defect.91 RP knockdown in a p53-deficient mouse erythroblast line increased transcription but decreased translation of Bag1 and Csde1, two proteins that are essential for erythroid differentiation.42 Translation of Bag1 and Csde1 is cap-dependent and IRES-mediated, and impaired IRES-mediated translation is also observed in models of other ribosomopathies.36,39 RPS19 is also thought to play a role in the regulation of alternative splicing;92 enhanced alternative splicing and decreased translation of FLVCR1, an erythroblastic heme exporter that plays a role in erythroid differentiation, have been observed in hematopoietic stem cells of RPS19-mutated DBA patients.93
Corticosteroids, which were first shown to be efficacious in DBA treatment in 1951, remain the first-line therapy today. Activation of the glucocorticoid receptor promotes erythroid proliferation,94,95 and it is believed that corticosteroids exert a general antiapoptotic effect among erythroid precursors and downregulate the expression of non-erythroid genes.82,95 Corticosteroids also increase sensitivity to erythropoietin, levels of which are often elevated in DBA patients.96 Leucine, which stimulates translation via the mTOR pathway, has been shown to improve hematopoiesis in some animal models of DBA,97,98 as well as in a small clinical trial;99 larger trials are underway. Hematopoietic stem cell transplantation, the only curative treatment for the anemia, is currently considered for certain patients and is likely to be further extended as outcomes improve.100,101
5q-syndrome
This disease, first reported as a refractory anemia with a distinct karyotype in 1974, shares clinical and pathologic characteristics with DBA. Now considered a subtype of myelodysplastic syndrome (MDS), it is defined cytogenetically, as a de novo deletion of the region between bands q21 and q32 of chromosome 5.102 Although 5q deletions are also observed in other cases of MDS and acute myeloid leukemia (AML), the region involved in 5q-syndrome is known to be distinct.103 Clinically, 5q-syndrome presents predominantly among females, with a more indolent course, lower risk of progression to AML, and more favorable prognosis than other subtypes of MDS. Blood findings include macrocytic anemia with hypolobulated megakaryocytes and normal or elevated quantities of platelets, with <5% blasts in both bone marrow and peripheral blood.104
Haploinsufficiency of RPS14 is critical to the pathogenesis of 5q-syndrome. shRNA knockdown of RPS14 in human CD34+ hematopoietic stem cells has been shown to replicate the erythroid defect, which could in turn be rescued by RPS14 overexpression.105 This finding has been confirmed in a mouse model of the disease, which further suggested a p53-dependent mechanism for the anemia,106 and recent studies in cancer cell lines have confirmed that RPS14 exerts both p53-dependent and p53-independent tumor suppressor effects107,108 (Fig. 1). Other genes in the deleted region also appear to play a role in the pathogenesis of 5q-syndrome. These include the tumor suppressor gene SPARC, which has antiproliferative and antiangiogenic effects;109 miR-145 and miR-146a, whose deletion may contribute to the thrombocytosis;110 and the candidate tumor suppressors EGR1, CTNNA1, and CDC25C, among other genes.111–114
5q-syndrome is characterized by a striking therapeutic response to lenalidomide, a thalidomide analog with fewer side effects and increased selectivity115,116 (Fig. 1). Lenalidomide is also used as a treatment for multiple myeloma; in this setting, it exerts immunomodulatory and antiangiogenic effects, in addition to directly inhibiting the proliferation of neoplastic cells.117 Although its mechanism of action in 5q-syndrome is incompletely understood, it has been shown to promote erythroid differentiation,118 upregulate SPARC,109 inhibit the cell-cycle regulators Cdc25C and PP2Acα,114 induce cell death by blocking cytokinesis,119 and promote degradation of p53.120
Shwachman-Diamond syndrome (SDS)
This rare disorder is clinically characterized by exocrine pancreatic insufficiency and hematologic abnormalities, most commonly neutropenia.121,122 Skeletal and neurocognitive abnormalities have also been observed, and there is increased risk of neoplastic transformation, particularly to MDS and AML. Although the diagnosis is usually made in the first few years of life, SDS can also present among older children and even adults. The presentation can vary widely, although confirmation of pancreatic and hematologic dysfunction is necessary to establish the clinical diagnosis.121
Approximately, 90% of patients with a clinical diagnosis of SDS carry biallelic mutations in the SBDS gene. The SBDS protein is highly conserved among archaea and eukaryotes; in mammals, it has been ascribed functions in key cellular pathways, including mitotic spindle stabilization,123,124 DNA metabolism and stress responses,125 reactive oxygen species regulation,126 and actin-dependent processes such as chemotaxis,127,128 in addition to ribosomal biogenesis.129,130 Mouse models have indicated roles for SBDS in both hematopoietic and stromal cells of the bone marrow.131,132 Because of this multiplicity of functions, the role of SBDS in the pathogenesis of SDS is unclear, and the classification of the disease as a ribosomopathy has been somewhat controversial. However, recent studies have established that the role of SBDS in ribosomal biogenesis is conserved in Saccharomyces cerevisiae and other eukaryotes as well as mammals. In particular, two recent studies showed that SBDS interacts with the GTPase EFL1 to catalyze the removal of eIF6 from the 60S ribosomal subunit, a key step in the formation of actively translating ribosomes.133,134 The latter of these studies also demonstrated defects in ribosomal subunit joining in lymphoblasts from SDS patients, supporting the idea that ribosomal dysfunction plays a role in the disease.134
Dyskeratosis congenita (DC)
This genetically and clinically heterogeneous disease is classically associated with mucocutaneous abnormalities, pulmonary fibrosis, bone marrow failure, and predisposition to cancer.135 DC can be caused by mutations in any of approximately 10 genes, each of which is associated with telomerase function or telomere integrity;136,137 in addition, Hoyeraal-Hreidarsson syndrome, a severe variant of DC whose symptoms include cerebellar hypoplasia, immunodeficiency, and enteropathy, has recently been shown to be caused by mutations in the regulator of telomere elongation helicase Rtel1.138,139 Because of the clear role of telomere defects in the pathogenesis of DC, authors have tended to classify it among other diseases that cause telomere shortening, including ataxia-telangiectasia, Bloom syndrome, and Fanconi anemia.136 Indeed, the clinical phenotype of DC is consistent with premature aging and loss of cells in high-turnover tissues.140
Defects in rRNA processing have been suggested to play a role in the X-linked form of DC, which is caused by mutations in the DKC1 gene. DKC1 encodes the dyskerin protein, which is found not only in the telomerase complex but also in H/ACA ribonucleoprotein complexes, where it is involved in rRNA pseudouridylation.141,142 The functional consequences of this ribosomal defect remain controversial, but there is evidence that it leads to decreased translational fidelity39,143 and impaired control of IRES-mediated translation.36,144,145 This dysregulation of translation may contribute to increased susceptibility to cancer in DC patients.146–148
Cartilage-hair hypoplasia (CHH)
This disorder, most commonly observed in Old Order Amish and Finnish populations, is clinically characterized by short stature, hair growth abnormalities, and bone deformities that can be detected radiographically.149 Anemia and immunodeficiency can also occur, among other symptoms. The disease is autosomal recessive in transmission, but the observed phenotypes are widely variable even within families. Some authors have described a spectrum of disorders including metaphyseal dysplasia without hypertrichosis, CHH, and anauxetic dysplasia (AD), ranging from least to most severe.149 There is increased risk of cancer, particularly non-Hodgkin lymphoma and basal cell carcinoma.150
CHH and other disorders on the CHH-AD spectrum are caused by mutations in the untranslated RMRP gene, which encodes the RNA component of the RNase mitochondrial RNA-processing (MRP) complex.151 RNase MRP RNA, which is classified as a snoRNA, plays several roles in the normal cell, and as with dyskeratosis congenita, the role of ribosomal dysfunction in the pathogenesis of CHH is uncertain. Nme1, the yeast homolog of RMRP, is involved in rRNA processing.152,153 On the other hand, in mammalian cells, RNase MRP RNA forms complexes with the catalytic subunit of telomerase reverse transcriptase—one of the genes mutated in some cases of DC—to produce double-stranded RNA that can be processed into small interfering RNAs (siRNAs).154 Targets of these siRNAs include genes involved in skeletal development, hair development, and hematopoietic differentiation, among others, suggesting that disturbed siRNA production may be the primary pathogenic mechanism in CHH.155
Treacher-Collins syndrome (TCS)
This disease, which is autosomal dominant in transmission, is characterized by craniofacial defects involving structures derived from the first and second branchial arches.156 Most cases of TCS are caused by mutations in the TCOF1 gene, whose protein product, named treacle, is involved in rRNA transcription and processing and is most strongly expressed in neural crest cells of the branchial arches during the embryonic period.157–159 As in other ribosomopathies, neural crest cell hypoplasia in TCS has been attributed to apoptosis caused by impaired ribosomal biogenesis.158 In a TCOF1-haploinsufficient mouse model, inactivation of p53 was sufficient to prevent craniofacial abnormalities160 (Fig. 1). Mutations in subunits of RNA polymerases I and III, which transcribe distinct rRNA subunits, have also been identified among TCS patients without TCOF1 mutations.161 Unlike other ribosomopathies, TCS is not associated with hematologic abnormalities, in agreement with the idea that ribosomes carry out specialized activities in different tissues but leaving open the question of how these activities are regulated.
Other ribosomopathies
Ribosomal dysfunction has been implicated in the pathogenesis of several other diseases. Bowen-Conradi syndrome, an extremely rare disorder that causes severe growth retardation and death in early childhood, is caused by mutations in EMG1, which encodes a pseudouridine methyltransferase involved in ribosomal biogenesis.162–165 North American Indian childhood cirrhosis, which affects children in a small population in Northwestern Quebec, is caused by mutations in hUTP4/Cirhin, which is involved in the synthesis of 18S rRNA.166–168 Haploinsufficiency of ribosomal protein SA, a component of the 40S subunit, was recently demonstrated in a majority of studied cases of isolated congenital asplenia.169 Impaired ribosomal biogenesis has been proposed to play a role in certain cases of common variable immunodeficiency, possibly as an atypical presentation of more canonical ribosomopathies such as DBA and SDS.170,171 Mutations in eIFs that may disrupt translational machinery and stress responses are observed in vanishing white matter disease172–174 and some cases of autism spectrum disorders.175–177
Ribosomal Dysfunction and Cancer
As our survey indicates, susceptibility to cancer is a common symptom of ribosomopathies. Indeed, at the cellular level, dysregulation of translation has been proposed as a common pathway for cancer progression.178 The mechanistic link may appear paradoxical, since cellular proliferation in tumors is generally associated with an increase in ribosomal biogenesis and translation, and several cancers induce overexpression of RPs.179 In the case of X-linked DC, the selective impairment of IRES-mediated translation results in decreased expression of tumor suppressors, such as p53, promoting proliferation and tumor growth.36,37,144 However, evidence for increased IRES-mediated translation of other proteins in X-linked DC suggests that this description may be too straightforward.145 Putative mechanisms of cancer progression in other ribosomopathies are not as well studied, but may involve regulatory crosstalk between RPs and other components of the translational apparatus with known oncogenic pathways, including those involving c-Myc180–182 and mTOR.183 The widespread activation of apoptosis observed in several ribosomopathies raises the possibility of selective pressure in favor of cellular clones in which p53 or other regulatory effectors are mutated or otherwise dysregulated. These explanations are preliminary, however, and the role of ribosomes in disease progression remains a largely open problem in cancer research.
Conclusion
With the notable exception of 5q-syndrome, treatment for most ribosomopathies has been symptomatic, with hematopoietic stem cell transplantation ultimately indicated in cases of progression to cancer or severe bone marrow failure. The possibility of treatment via novel pathways, such as inactivation of p53, has been proposed by some authors,45 but the potential of such therapies may be limited by the heterogeneity of disease mechanisms, as well as by the counterbalancing risk of cancer owing to the loss of p53 surveillance (Fig. 1). In spite of these challenges, the emerging diversity of ribosomal functions sheds light on the molecular basis of the ribosomopathies and informs the development of potential treatments. This improved understanding, as well as improved outcomes in established treatment protocols, is a sign of clinical progress against this complex class of diseases.
Acknowledgments
We thank Ramzi Nakhoul for editorial assistance and all the members of the Lu laboratory for active discussion.
Footnotes
Author Contributions
Wrote the first draft of the manuscript: HN. Contributed to the writing of the manuscript: HN, JK, XZ, WL, SXZ, HL. Agree with manuscript results and conclusions: HN, JK, XZ, WL, SXZ, HL. Jointly developed the structure and arguments for the paper: HN, JK, HL. Made critical revisions and approved final version: HL. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Robert E. Richard, Editor in Chief
FUNDING: H.L. was supported by NIH-NCI grants CA095441, CA079721, CA129828, and CA172468.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 3.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 6.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 8.Gorini L, Kataja E. Phenotypic repair by streptomycin of defective genotypes in E coli. Proc Natl Acad Sci U S A. 1964;51:487–493. doi: 10.1073/pnas.51.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies J, Gilbert W, Gorini L. Streptomycin, suppression, and the code. Proc Natl Acad Sci U S A. 1964;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triman KL. Mutational analysis of the ribosome. Adv Genet. 2007;58:89–119. doi: 10.1016/S0065-2660(06)58004-6. [DOI] [PubMed] [Google Scholar]
- 11.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 12.Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 13.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondrashov N, Pusic A, Stumpf CR, et al. Ribosome-mediated specificity in hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Kastan MB. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24:2146–2156. doi: 10.1101/gad.1968910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem. 2012;287:16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher EMC, Beer-Romero P, Brown LG, et al. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 20.Lopes AM, Miguel RN, Sargent CA, Ellis PJ, Amorim A, Affara NA. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills AA, Mills MJ, Gardiner DM, Bryant SV, Stanbridge EJ. Analysis of the pattern of QM expression during mouse development. Differentiation. 1999;64:161–171. doi: 10.1046/j.1432-0436.1999.6430161.x. [DOI] [PubMed] [Google Scholar]
- 22.Green H, Canfield AE, Hillarby MC, et al. The ribosomal protein QM is expressed differentially during vertebrate endochondral bone development. J Bone Miner Res. 2000;15:1066–1075. doi: 10.1359/jbmr.2000.15.6.1066. [DOI] [PubMed] [Google Scholar]
- 23.Sahin F, Qiu W, Wilentz RE, Iacobuzio-Donahue CA, Grosmark A, Su GH. RPL38, FOSL1, and UPP1 are predominantly expressed in the pancreatic ductal epithelium. Pancreas. 2005;30:158–167. doi: 10.1097/01.mpa.0000151581.45156.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg J, Hofsteenge J, Thomas G. Identification of the 40S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J Biol Chem. 1988;263:11473–11477. [PubMed] [Google Scholar]
- 25.Ruvinsky I, Sharon N, Lerer T, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeidan Q, Wang Z, De Maio A, Hart GW. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol Biol Cell. 2010;21:1922–1936. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colon-Ramos DA, Shenvi CL, Weitzel DH, et al. Direct ribosomal binding by a cellular inhibitor of translation. Nat Struct Mol Biol. 2006;13:103–111. doi: 10.1038/nsmb1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs G, Diges C, Kohlstaedt LA, Wehner KA, Sarnow P. Proteomic analysis of ribosomes: translational control of mRNA populations by glycogen synthase GYS1. J Mol Biol. 2011;410:118–130. doi: 10.1016/j.jmb.2011.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 32.Castle JC, Armour CD, Löwer M, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using PolyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higa-Nakamine S, Suzuki T, Uechi T, et al. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012;40:391–398. doi: 10.1093/nar/gkr700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroz LL, Edwards JR, Puthanveettil SV, et al. Neuronal transcriptome of aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 36.Yoon A, Peng G, Brandenburger Y, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 37.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bellodi C, Krasnykh O, Haynes N, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jack K, Bellodi C, Landry DM, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muhs M, Yamamoto H, Ismer J, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horos R, IJspeert H, Pospisilova D, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 43.Lee AS-Y, Burdeinick-Kerr R, Whelan SPJ. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci U S A. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 45.Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Semin Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 47.Carvajal LA, Manfredi JJ. Another fork in the road—life or death decisions by the tumour suppressor p53. EMBO Rep. 2013;14:414–421. doi: 10.1038/embor.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strezoska Ž, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20:5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pestov DG, Strezoska Ž, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Panic L, Montagne J, Cokaric M, Volarevic S. S6-haploinsufficiency activates the p53 tumor suppressor. Cell Cycle. 2007;6:20–24. doi: 10.4161/cc.6.1.3666. [DOI] [PubMed] [Google Scholar]
- 52.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 55.Zhou X, Liao J-M, Liao W-J, Lu H. Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer. 2012;3:298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohrum MAE, Ludwig RL, Kubbutat MHG, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang YP, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 59.Dai M-S, Zeng SX, Jin Y, Sun X-X, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horos R, von Lindern M. Molecular mechanisms of pathology and treatment in Diamond Blackfan anaemia. Br J Haematol. 2012;159:514–527. doi: 10.1111/bjh.12058. [DOI] [PubMed] [Google Scholar]
- 63.Gazda HT, Zhong R, Long L, et al. RNA and protein evidence for haploinsufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- 64.Farrar JE, Dahl N. Untangling the phenotypic heterogeneity of Diamond Blackfan anemia. Semin Hematol. 2011;48:124–135. doi: 10.1053/j.seminhematol.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerrard G, Valgañón M, Foong HE, et al. Target enrichment and high-throughput sequencing of 80 ribosomal protein genes to identify mutations associated with Diamond-Blackfan anaemia. Br J Haematol. 2013;162:530–536. doi: 10.1111/bjh.12397. [DOI] [PubMed] [Google Scholar]
- 66.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest. 2012;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsson H, Davey EJ, Draptchinskaia N, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Da Costa L, Chanoz-Poulard G, Simansour M, et al. First de novo mutation in RPS19 gene as the cause of hydrops fetalis in Diamond-Blackfan anemia. Am J Hematol. 2013;88:160. doi: 10.1002/ajh.23366. [DOI] [PubMed] [Google Scholar]
- 69.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum Mutat. 2010;31:1269–1279. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Campagnoli MF, Garelli E, Quarello P, et al. Molecular basis of Diamond-Blackfan anemia: new findings from the Italian registry and a review of the literature. Haematologica. 2004;89:480–489. [PubMed] [Google Scholar]
- 71.Farrar JE, Vlachos A, Atsidaftos E, et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118:6943–6951. doi: 10.1182/blood-2011-08-375170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pospisilova D, Cmejlova J, Ludikova B, et al. The Czech National Diamond-Blackfan anemia registry: clinical data and ribosomal protein mutations update. Blood Cells Mol Dis. 2012;48:209–218. doi: 10.1016/j.bcmd.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 74.McGowan KA, Mason PJ. Animal models of Diamond Blackfan anemia. Semin Hematol. 2011;48:106–116. doi: 10.1053/j.seminhematol.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor AM, Zon LI. Modeling Diamond Blackfan anemia in the zebrafish. Semin Hematol. 2011;48:81–88. doi: 10.1053/j.seminhematol.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 76.Terzian T, Box N. Genetics of ribosomal proteins: “curiouser and curiouser”. PLoS Genet. 2013;9:e1003300. doi: 10.1371/journal.pgen.1003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gazda HT, Kho AT, Sanoudou D, et al. Defective ribosomal protein gene expression alters transcription, translation, apoptosis, and oncogenic pathways in Diamond-Blackfan anemia. Stem Cells. 2006;24:2034–2044. doi: 10.1634/stemcells.2005-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, Cmejla R. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 79.Avondo F, Roncaglia P, Crescenzio N, et al. Fibroblasts from patients with Diamond-Blackfan anaemia show abnormal expression of genes involved in protein synthesis, amino acid metabolism and cancer. BMC Genomics. 2009;10:442. doi: 10.1186/1471-2164-10-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fumagalli S, Di Cara A, Neb-Gulati A, et al. Absence of nucleolar disruption after impairment of 40s ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–U350. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flygare J, Kiefer T, Miyake K, et al. Deficiency of ribosomal protein S19 in CD34(+) cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 82.Ebert BL, Lee MM, Pretz JL, et al. An RNA interference model of RPS 19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Léger-Silvestre I, Caffrey JM, Dawaliby R, et al. Specific role for yeast homologs of the Diamond Blackfan anemia-associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;280:38177–38185. doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 85.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flygare J, Aspesi A, Bailey JC, et al. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109:980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Janas MM, Wang E, Love T, et al. Reduced expression of ribosomal proteins relieves microRNA-mediated repression. Mol Cell. 2012;46:171–186. doi: 10.1016/j.molcel.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Harnpicharnchai P, Jakovljevic J, et al. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Donati G, Peddigari S, Mercer CA, Thomas G. 5S Ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4:87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moniz H, Gastou M, Leblanc T, et al. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356. doi: 10.1038/cddis.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Torihara H, Uechi T, Chakraborty A, Shinya M, Sakai N, Kenmochi N. Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br J Haematol. 2011;152:648–654. doi: 10.1111/j.1365-2141.2010.08535.x. [DOI] [PubMed] [Google Scholar]
- 92.Orrù S, Aspesi A, Armiraglio M, et al. Analysis of the ribosomal protein S19 interactome. Mol Cell Proteomics. 2007;6:382–393. doi: 10.1074/mcp.M600156-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Rey MA, Duffy SP, Brown JK, et al. Enhanced alternative splicing of the FLVCR1 gene in Diamond Blackfan anemia disrupts FLVCR1 expression and function that are critical for erythropoiesis. Haematologica. 2008;93:1617–1626. doi: 10.3324/haematol.13359. [DOI] [PubMed] [Google Scholar]
- 94.Wessely O, Deiner E-M, Beug H, von Lindern M. The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. EMBO J. 1997;16:267–280. doi: 10.1093/emboj/16.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Von Lindern M, Zauner W, Mellitzer G, et al. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-Kit to enhance and sustain proliferation of erythroid progenitors in vitro. Blood. 1999;94:550–559. [PubMed] [Google Scholar]
- 96.Nathan DG, Clarke BJ, Hillman DG, Alter BP, Housman DE. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J Clin Invest. 1978;61:489–498. doi: 10.1172/JCI108960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Payne EM, Virgilio M, Narla A, et al. l-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood. 2012;120:2214–2224. doi: 10.1182/blood-2011-10-382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jaako P, Debnath S, Olsson K, Bryder D, Flygare J, Karlsson S. Dietary l-leucine improves the anemia in a mouse model for Diamond-Blackfan anemia. Blood. 2012;120:2225–2228. doi: 10.1182/blood-2012-05-431437. [DOI] [PubMed] [Google Scholar]
- 99.Pospisilova D, Cmejlova J, Hak J, Adam T, Cmejla R. Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica. 2007;92:e66–e67. doi: 10.3324/haematol.11498. [DOI] [PubMed] [Google Scholar]
- 100.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Narla A, Vlachos A, Nathan DG. Diamond Blackfan anemia treatment: past, present, and future. Semin Hematol. 2011;48:117–123. doi: 10.1053/j.seminhematol.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boultwood J, Fidler C, Strickson AJ, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q-syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 103.Wang L, Fidler C, Nadig N, et al. Genome-wide analysis of copy number changes and loss of heterozygosity in myelodysplastic syndrome with del(5q) using high-density single nucleotide polymorphism arrays. Haematologica. 2008;93:994–1000. doi: 10.3324/haematol.12603. [DOI] [PubMed] [Google Scholar]
- 104.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 105.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q(-) syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q-syndrome. Nat Med. 2010;16:59–U93. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou X, Hao Q, Liao J, Liao P, Lu H. Ribosomal protein S14 negatively regulates c-Myc activity. J Biol Chem. 2013;288:21793–21801. doi: 10.1074/jbc.M112.445122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pellagatti A, Jädersten M, Forsblom A-M, et al. Lenalidomide inhibits the malignant clone and up-regulates the SPARC gene mapping to the commonly deleted region in 5q-syndrome patients. Proc Natl Acad Sci U S A. 2007;104:11406–11411. doi: 10.1073/pnas.0610477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 111.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Min IM, Pietramaggiori G, Kim FS, Passegue E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 113.Liu TX, Becker MW, Jelinek J, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding α-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 114.Wei S, Chen X, Rocha K, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci U S A. 2009;106:12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Padron E, Komrokji R, List AF. Biology and treatment of the 5q-syndrome. Expert Rev Hematol. 2011;4:61–69. doi: 10.1586/ehm.11.2. [DOI] [PubMed] [Google Scholar]
- 116.Voutsadakis IA, Cairoli A. A critical review of the molecular pathophysiology of lenalidomide sensitivity in 5q—myelodysplastic syndromes. Leuk Lymphoma. 2012;53:779–788. doi: 10.3109/10428194.2011.623255. [DOI] [PubMed] [Google Scholar]
- 117.Quach H, Ritchie D, Stewart AK, et al. Mechanism of action of immunomodulatory drugs (IMiDS) in multiple myeloma. Leukemia. 2009;24:22–32. doi: 10.1038/leu.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Narla A, Dutt S, McAuley JR, et al. Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood. 2011;118:2296–2304. doi: 10.1182/blood-2010-11-318543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsuoka A, Tochigi A, Kishimoto M, et al. Lenalidomide induces cell death in an MDS-derived cell line with deletion of chromosome 5q by inhibition of cytokinesis. Leukemia. 2010;24:748–755. doi: 10.1038/leu.2009.296. [DOI] [PubMed] [Google Scholar]
- 120.Wei S, Chen X, McGraw K, et al. Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32:1110–1120. doi: 10.1038/onc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dror Y, Donadieu J, Koglmeier J, et al. Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome. Ann N Y Acad Sci. 2011;1242:40–55. doi: 10.1111/j.1749-6632.2011.06349.x. [DOI] [PubMed] [Google Scholar]
- 122.Myers KC, Davies SM, Shimamura A. Clinical and molecular pathophysiology of Shwachman–Diamond syndrome: an update. Hematol Oncol Clin North Am. 2013;27:117–128. doi: 10.1016/j.hoc.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Austin KM, Gupta ML, Jr, Coats SA, et al. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008;118:1511–1518. doi: 10.1172/JCI33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Orelio C, Verkuijlen P, Geissler J, van den Berg TK, Kuijpers TW. SBDS expression and localization at the mitotic spindle in human myeloid progenitors. PLoS One. 2009;4:e7084. doi: 10.1371/journal.pone.0007084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ball HL, Zhang B, Riches JJ, et al. Shwachman-Bodian Diamond syndrome is a multi-functional protein implicated in cellular stress responses. Hum Mol Genet. 2009;18:3684–3695. doi: 10.1093/hmg/ddp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ambekar C, Das B, Yeger H, Dror Y. SBDS-deficiency results in deregulation of reactive oxygen species leading to increased cell death and decreased cell growth. Pediatr Blood Cancer. 2010;55:1138–1144. doi: 10.1002/pbc.22700. [DOI] [PubMed] [Google Scholar]
- 127.Orelio C, Kuijpers TW. Shwachman-Diamond syndrome neutrophils have altered chemoattractant-induced F-actin polymerization and polarization characteristics. Haematologica. 2009;94:409–413. doi: 10.3324/haematol.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leung R, Cuddy K, Wang Y, Rommens J, Glogauer M. Sbds is required for Rac2-mediated monocyte migration and signaling downstream of RANK during osteoclastogenesis. Blood. 2011;117:2044–2053. doi: 10.1182/blood-2010-05-282574. [DOI] [PubMed] [Google Scholar]
- 129.Austin KM, Leary RJ, Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ganapathi KA, Austin KM, Lee C-S, et al. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110:1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Raaijmakers MHGP, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sezgin G, Henson AL, Nihrane A, et al. Impaired growth, hematopoietic colony formation, and ribosome maturation in human cells depleted of Shwachman-Diamond syndrome protein SBDS. Pediatr Blood Cancer. 2013;60:281–286. doi: 10.1002/pbc.24300. [DOI] [PubMed] [Google Scholar]
- 133.Finch AJ, Hilcenko C, Basse N, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 135.Mason PJ, Bessler M. The genetics of dyskeratosis congenita. Cancer Genet. 2011;204:635–645. doi: 10.1016/j.cancergen.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kong CM, Lee XW, Wang X. Telomere shortening in human diseases. FEBS J. 2013;280:3180–3193. doi: 10.1111/febs.12326. [DOI] [PubMed] [Google Scholar]
- 137.Batista LF, Artandi SE. Understanding telomere diseases through analysis of patient-derived iPS cells. Curr Opin Genet Dev. 2013;23:526–533. doi: 10.1016/j.gde.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng Z, Glousker G, Molczan A, et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal–Hreidarsson syndrome. Proc Natl Acad Sci U S A. 2013;110:E3408–E3416. doi: 10.1073/pnas.1300600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ballew BJ, Joseph V, De S, et al. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson syndrome. PLoS Genet. 2013;9:e1003695. doi: 10.1371/journal.pgen.1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kirwan M, Beswick R, Walne AJ, et al. Dyskeratosis congenita and the DNA damage response. Br J Haematol. 2011;153:634–643. doi: 10.1111/j.1365-2141.2011.08679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Montanaro L, Brigotti M, Clohessy J, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- 142.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 143.Jobert L, Skjeldam HK, Dalhus B, et al. The human base excision repair enzyme SMUG1 directly interacts with DKC1 and contributes to RNA quality control. Mol Cell. 2013;49:339–345. doi: 10.1016/j.molcel.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 144.Montanaro L, Calienni M, Bertoni S, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 145.Rocchi L, Pacilli A, Sethi R, et al. Dyskerin depletion increases VEGF mRNA internal ribosome entry site-mediated translation. Nucleic Acids Res. 2013;41:8308–8318. doi: 10.1093/nar/gkt587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ruggero D, Grisendi S, Piazza F, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 147.Montanaro L. Dyskerin and cancer: more than telomerase. The defect in mRNA translation helps in explaining how a proliferative defect leads to cancer. J Pathol. 2010;222:345–349. doi: 10.1002/path.2777. [DOI] [PubMed] [Google Scholar]
- 148.Alawi F, Lin P. Dyskerin is required for tumor cell growth through mechanisms that are independent of its role in telomerase and only partially related to its function in precursor rRNA processing. Mol Carcinog. 2011;50:334–345. doi: 10.1002/mc.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thiel CT. Cartilage-hair hypoplasia—anauxetic dysplasia spectrum disorders [Internet] In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong C-T, Stephens K, editors. GeneReviews™. Seattle, WA: University of Washington; 2012. [cited October 4, 2013]. Available at http://www.ncbi.nlm.nih.gov/books/NBK84550/ [Google Scholar]
- 150.Taskinen M, Ranki A, Pukkala E, Jeskanen L, Kaitila I, Makitie O. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am J Med Genet A. 2008;146A:2370–2375. doi: 10.1002/ajmg.a.32478. [DOI] [PubMed] [Google Scholar]
- 151.Ridanpaa M, van Eenennaam H, Pelin K, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 152.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci U S A. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maida Y, Yasukawa M, Furuuchi M, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–U104. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rogler LE, Kosmyna B, Moskowitz D, et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet. 2014;23:368–382. doi: 10.1093/hmg/ddt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schlump J-U, Stein A, Hehr U, et al. Treacher Collins syndrome: clinical implications for the paediatrician—a new mutation in a severely affected newborn and comparison with three further patients with the same mutation, and review of the literature. Eur J Pediatr. 2012;171:1611–1618. doi: 10.1007/s00431-012-1776-7. [DOI] [PubMed] [Google Scholar]
- 157.Gonzales B, Henning D, So RB, Dixon J, Dixon MJ, Valdez BC. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum Mol Genet. 2005;14:2035–2043. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- 158.Dixon J, Jones NC, Sandell LL, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci U S A. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Masotti C, Ornelas CC, Splendore-Gordonos A, et al. Reduced transcription of TCOF1 in adult cells of Treacher Collins syndrome patients. BMC Med Genet. 2009;10:136. doi: 10.1186/1471-2350-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jones NC, Lynn ML, Gaudenz K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dauwerse JG, Dixon J, Seland S, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 162.Armistead J, Khatkar S, Meyer B, et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am J Hum Genet. 2009;84:728–739. doi: 10.1016/j.ajhg.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wurm JP, Meyer B, Bahr U, et al. The ribosome assembly factor Nep1 responsible for Bowen–Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 2010;38:2387–2398. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meyer B, Wurm JP, Kötter P, et al. The Bowen–Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Ψ1191 in yeast 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Thomas SR, Keller CA, Szyk A, Cannon JR, LaRonde-LeBlanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chagnon P, Michaud J, Mitchell G, et al. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet. 2002;71:1443–1449. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010;38:4798–4806. doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Freed EF, Prieto J-L, McCann KL, McStay B, Baserga SJ. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012;8:e1002892. doi: 10.1371/journal.pgen.1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bolze A, Mahlaoui N, Byun M, et al. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Khan S, Pereira J, Darbyshire PJ, et al. Do ribosomopathies explain some cases of common variable immunodeficiency? Clin Exp Immunol. 2011;163:96–103. doi: 10.1111/j.1365-2249.2010.04280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Allenspach EJ, Bellodi C, Jeong D, et al. Common variable immunodeficiency as the initial presentation of dyskeratosis congenita. J Allergy Clin Immunol. 2013;132:223–226. doi: 10.1016/j.jaci.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Leegwater PAJ, Vermeulen G, Könst AAM, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–388. doi: 10.1038/ng764. [DOI] [PubMed] [Google Scholar]
- 173.Van der Knaap MS, Pronk JC, Scheper GC. Vanishing white matter disease. Lancet Neurol. 2006;5:413–423. doi: 10.1016/S1474-4422(06)70440-9. [DOI] [PubMed] [Google Scholar]
- 174.Perlman SJ, Mar S. Leukodystrophies [Internet] In: MSc SIABs, editor. Neurodegenerative Diseases. US: Springer; 2012. pp. 154–171. [cited October 7, 2013] Available from: http://link.springer.com/chapter/10.1007/978-1-4614-0653-2_13. [Google Scholar]
- 175.Neves-Pereira M, Müller B, Massie D, et al. Deregulation of EIF4E: a novel mechanism for autism. J Med Genet. 2009;46:759–765. doi: 10.1136/jmg.2009.066852. [DOI] [PubMed] [Google Scholar]
- 176.Santini E, Huynh TN, MacAskill AF, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gkogkas CG, Khoutorsky A, Ran I, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 2013;493:371–377. doi: 10.1038/nature11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5:pii:a012336. doi: 10.1101/cshperspect.a012336. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 180.Macias E, Jin A, Deisenroth C, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Challagundla KB, Sun X-X, Zhang X, et al. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31:4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 183.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116:3715–3723. doi: 10.1182/blood-2010-02-251090. [DOI] [PMC free article] [PubMed] [Google Scholar]