Abstract
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by the production of autoantibodies against self-antigens, which occurs most often in women between 15 and 40 years of age. The innate immunity is involved in the pathogenesis of SLE through TLR- 7. Genetic factors such as copy number variation (CNV) of target genes may contribute to disease development, but this possible risk has not yet been studied in SLE patients from Yucatan, Mexico. The CNV of TLR-7 gene was determined by quantitative polymerase chain reaction assay using TaqMan probes in 80 SLE women and 150 control subjects. The results showed that 10% of SLE patients exhibited more than two copies of TLR-7 gene, whereas no mRNA overexpression was detected. These data suggested that increased CNV of the TLR-7 gene in Yucatan SLE women can be a risk factor for this disease.
Keywords: autoimmunity, toll-like receptors, innate immunity, immunogenetic mechanism
Introduction
Systemic lupus erythematosus (SLE) is a prototype autoimmune disease with diverse clinical manifestations and is characterized by the presence of various autoantibodies against self-components, especially double-strand DNA (dsDNA) and RNA-binding nuclear proteins.1 A female to male ratio of 9:1 in 80–90% of cases suggests that sex-related factors are important in the development of disease.2,3 The etiology of SLE is considered multifactorial involving multiple genes and environmental factors such as infections, hormones, UV light, and drugs.4–8 Immune system abnormalities also contribute to the pathogenesis of SLE; including abnormal clearance of apoptotic cells and immune complexes, the over production of type I interferon (IFN), the reduced B- and T-cell activation, and the production of autoantibodies against self-antigens.9,10
The incidence of female patients with SLE varies according to the population studied; in Europe, the variation is between 3.6 (Spain, north countries) and 5.8 (Iceland), and in United States, between 2.5 (Rochester, Minnesota) and 8.2 (rural Wisconsin area) per 100,000 inhabitants.11,12 In Mexico, SLE affects between 50,000 and 100,000 inhabitants, 90% are women between 20 and 45 years of age, with a prevalence of 0.06% and an incidence of 1.8–7.6/100.000 inhabitants, but no epidemiological data on SLE in Yucatan are available.13–15
The involvement of innate immunity through TLRs has been implicated in the pathogenesis of SLE.16 The TLRs recognize pathogen-associated molecular patterns (PAMPs) and endogenous ligands known as damage-associated molecular patterns (DAMPs) released by cells undergoing either apoptosis or necrosis.17,18 Genomic DNA is not accessible to the immune system under standard conditions; however, when cells die through apoptosis, apoptotic bodies containing fragmented material and abnormal antigens circulate in the body enabling the immune systems to access new epitopes.19 Alterations in apoptosis and defective removal of apoptotic cells by phagocytes induce the release and exposure of self-antigens and neoepitopes that activate antigen-presenting cells (APCs), T and B cells, generating both autoantibodies and immune complexes.20–22
SLE patients show an increase in apoptosis releasing genetic material (DNA and RNA) that can be recognized by TLRs. Moreover, most patients show an increase in the expression of regulatory genes of interferon (IFN) type I, mediated by the participation of TLR-7 and TLR-9, which recognize RNA and DNA, respectively.23,24 Dendritic cells, activated by immune complexes containing nucleic acids, are internalized by the FcγRIIA (Fc-gamma receptor IIA) to reach the endosome and activating transcription factors (IRF-5/7) inducing a massive production of IFN-α.25,26
Located at Xp22.2, TLR-7 (OMIM no. 300365) encodes proteins that play critical roles in pathogen recognition and activation of innate immunity.27 The TLR-7 recognizes endogenous RNA-containing self antigens and induces the expression of type I IFN, a pivotal cytokine in the pathogenesis of SLE.28 Studies in BXSB mice, which develop an autoimmune phenotype similar to human SLE, led to the discovery of a locus Y-linked accelerator called Yaa (autoimmune acceleration linked to Y). This factor makes male mice show a severe condition due to a species-dependent translocation of the telomeric end of the X chromosome containing the TLR-7 gene to chromosome Y.29 The increased CNV of TLR-7 gene probably intensifies the autoimmune response to nuclear material.
Several studies to identify genetic factors involved in the development of SLE.30–35 The role of CNV of genes is currently the topic of intense research in the genetics of SLE. The CNVs can arise either when a complete gene or gene segment had been duplicated or when a gene is abnormally absent. Additional copies of genes may contribute to the overexpression of the proteins and the suppression of a gene can lead to a deficiency, both results have functional consequences.34,35
The CNV of TLR-7 gene have been examined in SLE. Kelly et al studied whether an increase in the copy number of TLR-7 gene in SLE patients could influence the autoantibody profiles. They examined the relative copy number of TLR-7 gene from 50 Caucasian and 49 African-American SLE patients (55 women and 44 men) and 91 sex and ethnicity-matched healthy control subjects. Their results found a variation of copy number of TLR-7 gene in both SLE patients and healthy controls, but no significant association was detected related to the autoantibody profile in SLE patients.36
In the northern region of Mexico, the CNV of TLR-7 gene was associated with the childhood onset of the disease.37 The results indicated a significant increase in the copy number of TLR-7 gene in SLE women when compared with control ones, and showed increased association (OR = 6.61) in female than in male patients (OR = 3.07). These data suggested that increasing the copy number of TLR-7 gene can be a risk factor for the disease in the Mexican population. These results also support the role of TLR-7 and the X-linked susceptibility in SLE pathogenesis. However, the Maya population from the state of Yucatan has not yet been studied. Our objective was to determine the copy number of TLR-7 gene in SLE women and its association with the development of disease in a Mayan population in Yucatan Mexico.
Materials and Methods
Selection of the Mayan and Mayan Mestizo ethnics
Both SLE and control group were selected using the anthropologic and demographic parameters such as language, birth place, surnames, genealogy, and history of lifestyle. The ethnic group of Mayan Mestizo, defined as individuals born in the country having a Spanish-derived last name, with Mexican ancestors back at least to the third generation was included in the research. The second criterion for selection was that at least one parent was born in Yucatan for two generations including their own. In addition, we determined the absence of substructure or population stratification within the population of Yucatan by using 16 autosomal short tandem reagents (STR) markers, which may represent a confounder.38
Eighty Yucatan Mayan SLE women were recruited at the Rheumatology outpatient of the Agustin O’Horán Hospital, Mérida. Diagnosis of SLE was established according the American College of Rheumatology criteria and disease activity was evaluated by SLEDAI score.39–41 The control group was formed by 150 healthy Yucatan women volunteers. Subjects with a chronic or degenerative disease, such as diabetes mellitus type 2, obesity, or hypertension, were excluded. Informed consent was obtained from all the participants according to the Helsinki Declaration recommendations. The study was approved by the Ethics Committee of Agustin O’Horán Hospital. Confidentiality of participants was strictly maintained.
DNA and RNA extraction
Venous peripheral blood samples (5 mL) were collected in EDTA tubes from each patient and control subject. Genomic DNA was extracted by the method of Bounce42 and the RNA extraction was performed using the PAXgene kit. The DNA and RNA obtained were quantified at 260 and 280 nm using the computer-Nano BioSpec (Shimadzu).
Determination of the copy number of TLR-7 gene by real-time quantitative polymerase chain reaction assay
The copy number of TLR-7 gene was carried out using the TaqMan probes (Assay-by-Design, ID: Hs00226289_cn) for TLR-7 (Applied Biosystems, Foster City, CA), according to the protocol described by Kelly et al.36 Thermal cycling conditions consisted of initial denaturation at 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds each and at 60°C for one minute each. The differences in DNA concentration between samples were normalized against RNase P as housekeeping gene. The reactions were conducted in triplicate using 10 ng of DNA on the OneStep Applied Biosystems thermocycler and calculations were performed using the 2−^^Ct method.43 This method calculates the difference in cycle thresholds (the number of polymerase chain reaction (PCR) cycles required to produce a set of fixed thresholds) between the gene of interest and the housekeeping gene (^Ct). Subsequent calculations normalized the ^Ct of each sample to a calibrator (DNA with 2 copies of TLR-7 gene) that was assigned a relative expression value of 1.00 (^^Ct). Assuming that the amount of PCR product doubled with each successive PCR cycle, calculating the 2−^^Ct value will provide the relative amount of DNA initially available for amplification in each quantitative PCR run. Therefore, the 2−^^Ct method revealed differences in the relative gene copy numbers between the samples tested.43 A range for each expression value was calculated based on the standard deviations of the ^^Ct value, where 2−(^^Ct + s) was the lower limit and 2−(^^Ct − s) was the upper one.
mRNA expression of TLR-7
The reverse transcription (RT) reaction was performed to obtain cDNA from 30 ng of RNA using the QuantiTect® Whole Transcriptome kit. Reactions by real-time quantitative of RT-PCR were conducted in triplicate using 100 ng of cDNA, using Taq-Man probes (Hs01933259_s1) (Applied Biosystems, Foster City, CA) in the OneStep Applied Biosystems thermocycler. The estimation of the expression was performed by the 2−^^Ct method previously described, using a calibrator cDNA (mRNA expression constant of TLR-7), and the 18S rRNA as housekeeping gene.
Statistical analysis
The non-parametric Mann–Whitney U-test was used to compare the distribution of the relative TLR-7 copy number between the cases and controls group. ORs with 95% CI were calculated by logistic regression models using STATA 10.2 software, using absolute TLR-7 gene copy number. Paired t-test was used to assess the difference of expression of mRNA between SLE patients and control subjects. The Spearman correlation coefficient rank test using the GraphPad Prism 5.0 software (GraphPad, San Diego, CA, USA) was used to determine the correlation between TLR-7 relative copy number with the mRNA expression of TLR-7 (p < 0.05).
Results
The study is the first record of the determination of the variability of copy number of TLR-7 gene in women with SLE of the Mayan and Mestizo people from Mexico. The location, age, and treatment characteristics of the patients and control group are listed in Table 1. The copy numbers of TLR-7 gene predominantly found in SLE cases were 2 (89%) and 3 (10%), which was significantly (p = 0.0001) different from the homogeneous 2 copies found in controls (Table 2). One subject of each group showed only one copy. The mean copy TLR-7 number obtained for SLE patients was significantly higher than controls (p < 0.0001) (Table 2 and Fig. 1). The relative risk was estimated with “2 copies” as reference (OR = 16.5, IC: 2.03–134.04, p = 0.001), which suggested that 3 copies of TLR-7 gene is associated with a higher genetic risk for developing SLE in the studied population.
Table 1.
Geographic distribution of both SLE patients and control subjects, in relation with age and treatment.
| SLE FEMALE PATIENT | CONTROL SUBJECT | |
|---|---|---|
| Number | 80 | 150 |
| Age, year, mean ± s.d. | 34.01 ± 11.27 | 39.62 ± 15.28 |
| Locality | ||
| Mérida | 32/80 (40%) | 48/150 (32%) |
| Motul | 5/80 (6.25%) | – |
| Progreso | 4/80 (5%) | – |
| Acanceh | 2/80 (2.25%) | – |
| Buczot | 2/80 (2.25%) | – |
| Sacalum | 2/80 (2.25%) | – |
| Teabo | 2/80 (2.25%) | – |
| Ticul | 2/80 (2.25%) | – |
| Caucel | – | 28/150 (19%) |
| Tekal de Venegas | – | 74/150 (49%) |
| Other | 29/80 (36%) | – |
| Treatment | ||
| Prednisone | 44/80 (55%)* | |
| Azathioprine | 24/80 (30%)* | |
| Hydroxycloroquine | 4/80 (11.25%)* | |
| Metrotexate | 7/80 (8.75%)* | |
| Deflazacort | 5/80 (6.25%)* | |
Note:
Percentage of patients receiving the drug in combination with other one.
Table 2.
Distribution of the numbers of TLR-7 gene copy and their means between SLE female patients and control subjects and their statistical significances.
| NUMBER OF TLR-7 GENE COPY | SLE FEMALES PATIENT N = 80 | CONTROL SUBJECT N = 150 | P |
|---|---|---|---|
| 2 copies | 71 (88.75%) | 149 (99.33%) | 0.0001 |
| 3 copies | 8 (10.00%) | 0 (0.00%) | |
| 1 copy | 1 (1.25%) | 1 (1.25%) | |
| Mean copy number of TLR-7 gene | 2.195 ± 0.241 | 2.04 ± 0.158 | <0.0001 |
Figure 1.
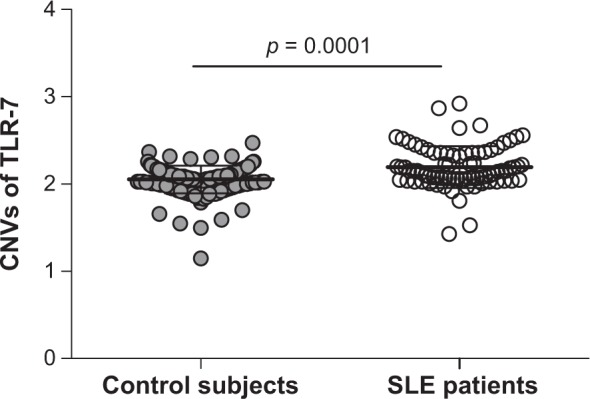
Scatter plot, means and standard deviations of numbers of TLR-7 copy in SLE patients (n = 80) and control subjects (n = 150) analyzed by Mann–Whitney U-test.
To perform a paired t-test and document the increasing copy number and mRNA expression of TLR-7 gene, 30 samples of SLE patients with the highest relative number of copies (3) and 30 control subjects with 2 copies were randomly selected; however, no statistical difference was detected (p = 0.1763) (Fig. 2). Moreover, no correlation was found between the copy number and mRNA expression of TLR-7 (r = 0.125, p = 0.5243) in SLE patients.
Figure 2.
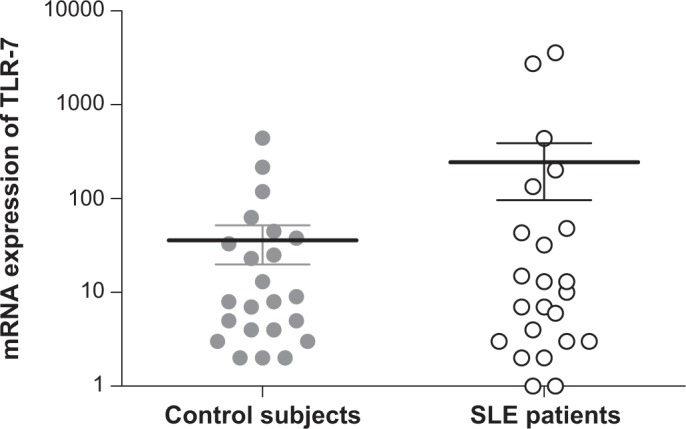
Paired t-test of the scale log mRNA expression of TLR-7 in SLE patients (n = 30) and control subjects (n = 30) (p = 0.1763).
Discussion
SLE is a complex systemic autoimmune disease that occurs most often in women. Several studies have identified genetic factors in the development of SLE in various ethnic groups.44 Despite the importance of CNV of genes involved in the pathogenesis of SLE, studies of copy number of TLR-7 gene are scarce, especially in Mexican women. Association was found between copy numbers of TLR-7 gene and the disease in northern Mexico, but no information is available in the Yucatan women population. In this study of Mexican women of Mayan ancestry, we demonstrated that the increased copy number of TLR-7 gene to 3 is associated with the genetic risk of SLE.
In Caucasian and African-Americans, the relative gene copy number of TLR-7 varied both in SLE patients and the control groups thus suggesting that CNV is neither a genetic risk factor for SLE nor is associated with the autoantibody profile.36 Opposite to the report of Kelly et al, in the Mayan population of Yucatan, significant differences in copy numbers of TLR-7 gene between SLE cases and controls were observed suggesting ethnical variations. The ethnic genetic makeup has often been related to variations in susceptibility to specific diseases.45
However, this study was consistent with the northern Mexican population, where SLE women had higher number of copies of TLR-7 gene (p < 0.0001) compared with controls.37 Both studies might suggest that the positive association of increased copy number of TLR-7 with the risk for developing SLE might be in the Mexican women population. Additionally, stratification by gender in Mexican population has showed a higher risk for SLE in male patients with more than one copy than in female patients with more than two copies.37 In this study, the stratification by gender was not possible because all patients were of female gender of Mayan ancestry, whereas Garcia et al studied female and male pediatric patients of different ethnical origin.
Similarly, several single nucleotide polymorphisms (SNPs) of TLR-7 gene were analyzed in 7,107 SLE patients of different ethnicities (European- American, African- American, and Amerindian-Hispanic), and the highest frequency of the risk allele (G) was observed in Amerindian-Hispanic (44.8%). However, the Asian population has an even higher rate (81%).46 In this respect, the results obtained in the study of SLE patients from northern Mexico may be influenced by the Amerindian-Hispanic descent in the central and northern region of Mexico. Significant mRNA expression of TLR-7 was observed in SLE patients who showed the highest relative copy number (p = 0.0001, r = 0.796).37 In this study, all the SLE patients were residents of the state of Yucatan (Table 1), a more homogeneous Maya population with a limited genetic variability. The increase of TLR-7 copies represented a risk factor in the Yucatan women. However, we observed no significant difference in mRNA expression of TLR-7 among either patients or the controls (Fig. 2). These differences could be due to more specific ethnic variations within the Mexican population.
However, the impact of previous treatments should not be ignored (Table 1). Immunosuppressive agents such as azathioprine, a purine analog, can inhibit the production of nucleic acids and other enzymes involved in the synthesis of DNA, RNA, and proteins. Azathioprine may block the functions of T cells, inhibiting the synthesis of antibodies and reduce the number of circulating monocytes and granulocytes, but can also inhibit the synthesis of the mRNA of TLR-7.47 In this study, 30% of SLE patients were receiving azathioprine in combination with other immunosuppressive agents (Table 1); thus, further studies are needed to determine the inhibitory effect of drugs on RNA synthesis.
No association between the increase in copy number and mRNA expression of TLR-7 was observed, probably due to drug administration and the role of DNA methylation and some genetic regulators. The DNA methylation was reported as an epigenetic process that directly regulates gene expression preventing the binding of transcription factors and indirectly promoting closed chromatin structure.48,49 Further epigenetic studies are needed to determine its effect on mRNA synthesis and its role in the immunogenetic mechanisms related to the development of the disease in our patients.
Copy number of TLR-7 may contribute to overproduction of IFN-α, and this association was found in the SLE murine model.50 To determine the possible association between the TLR-7 CNV and the serum levels of IFN-α in the Mayan SLE women, an analysis was performed but no correlation was observed (data not shown) probably due to drug administration or disease activity.
Our results suggested that the variation in the copy numbers of TLR-7 is important in the development of SLE in the Mayan women population; however, presenting more than 2 TLR-7 copies does not mean that the person will develop the disease, as it depends on other factors which together can onset the disease. Early detection of genetic factors in SLE as CNV and SNPs of TLR-7 gene in the Mayans of the Yucatan state can help to establish preventive measures contributing to increased women’ quality of life.
Acknowledgments
We are grateful to Nicole R. Van Wynsberghe for editing.
Footnotes
Author Contributions
Conceived and designed the experiments: GVP, LJGH. Analyzed the data: DCC, GIAA, GPM, and YENU. Contributed to the writing of the manuscript: AVAR. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Christian Bronner, Editor in Chief
FUNDING: This work was supported by CONACYT (National Council of Science and Technology), grant FONSEC SALUD 2010-1-139788.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.Mills JA. Systemic lupus erythematosus. N Engl J Med. 1994;330:1871–1879. doi: 10.1056/NEJM199406303302608. [DOI] [PubMed] [Google Scholar]
- 2.Schwartzman MJ, Putterman C. Gender differences in the pathogenesis and outcome of lupus and of lupus nephritis. Clin Dev Immunol. 20122012:ID 604892. doi: 10.1155/2012/604892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40(1):42–49. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vyse TJ, Kotzin BL. Genetic susceptibility to systemic lupus erythematosus. Annu Rev Immunol. 1998;16:261–292. doi: 10.1146/annurev.immunol.16.1.261. [DOI] [PubMed] [Google Scholar]
- 5.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155(3):109–117. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doria A, Canova M, Tonon M, et al. Infections as triggers and complications of systemic lupus erythematosus. Autoimmun Rev. 2008;8(1):24–28. doi: 10.1016/j.autrev.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Zandman-Goddard G, Shoenfeld Y. Infections and SLE. Autoimmunity. 2005;38(7):473–485. doi: 10.1080/08916930500285352. [DOI] [PubMed] [Google Scholar]
- 9.Lispky PE. Systemic lupus erythematosus: an autoimmune disease of B cell hyperactivity. Nat Immunol. 2001;2:764–766. doi: 10.1038/ni0901-764. [DOI] [PubMed] [Google Scholar]
- 10.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31(12):887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty DJ, Manzi S, Medsger TA, Jr, Ramsey-Goldman R, LaPorte RE, Kwoh CK. Incidence of systemic lupus erythematosus. Race and gender differences. Arthritis Rheum. 1995;38(9):1260–1270. doi: 10.1002/art.1780380914. [DOI] [PubMed] [Google Scholar]
- 12.Danchencho N, Satia JA, Anthony SM. Lupus around the world. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15:308–318. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 13.Gómez Puerta J, Cervera R. Lupus Eritematoso Sistémico. Med Lab. 2008;14:211–223. [Google Scholar]
- 14.Secretaría de Salud . Comunicado de prensa No. 165. México: Secretaria de Salud; 2012. [Google Scholar]
- 15.Guías de diagnóstico y tratamiento; servicio de reumatología. México: hospital General de México; 2011. [Google Scholar]
- 16.Marshak Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 19.Casciola Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179(4):1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunh A, Bijl M. Pathogenesis of cutaneous lupus erythematosus. Lupus. 2008;17:389–393. doi: 10.1177/0961203308090019. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz L, Van Bavel C, Franz S, Berden J. Apoptosis in the pathogenesis of systemic lupus erythematosus. Lupus. 2008;17:371–375. doi: 10.1177/0961203308089990. [DOI] [PubMed] [Google Scholar]
- 22.White S, Rosen A. Apoptosis in systemic lupus erythematosus. Curr Opin Rheumatol. 2003;15:557–562. doi: 10.1097/00002281-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Rahman A, Isenberg D. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 24.Rönnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rönnblom L, Eloranta M, Alm G. The type interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54:408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- 26.Tian J, Avalos A, Mao S. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 27.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 28.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian S, Tus K, Li QZ, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croker J, Kimberly R. Genetics of susceptibility and severity in systemic lupus erythematosus. Curr Opin Rheumatol. 2005;17:529–537. doi: 10.1097/01.bor.0000169360.15701.27. [DOI] [PubMed] [Google Scholar]
- 31.Gregersen P, Behrens T. Genetics of autoimmune diseases-disorders of immune homeostasis. Nat Rev Genet. 2006;7:917–928. doi: 10.1038/nrg1944. [DOI] [PubMed] [Google Scholar]
- 32.Brown E, Edberg J, Kimberly R. Fc receptor genes and the systemic lupus erythematosus diathesis. Autoimmunity. 2007;40:567–581. doi: 10.1080/08916930701763710. [DOI] [PubMed] [Google Scholar]
- 33.International Consortium for Systemic Lupus Erythematosus Genetics (SLE-GEN), Harley JB, Alarcón-Riquelme ME, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ptacek T, Li X, Kelley J, Edberg J. Copy number variants in genetic susceptibility and severity of systemic lupus erythematosus. Cytogenet Genome Res. 2008;123:142–147. doi: 10.1159/000184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichler E, Nickerson D, Altshuler D, Bowcock A, et al. Completing the map of human genetics variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley J, Johnson M, Alarcón G, Kimberly R, Edberg J. Variation in the relative copy number of the TLR-7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56(10):3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
- 37.García-Ortiz H, Velázquez-Cruz R, Espinosa-Rosales F, Jiménez-Morales S, Baca V, Orozco L. Association of TLR7 copy number variation with susceptibility to childhood-onset systemic lupus erythematosus in Mexican population. Ann Rheum Dis. 2010;69:1861–1865. doi: 10.1136/ard.2009.124313. [DOI] [PubMed] [Google Scholar]
- 38.Herrera Lizbeth González, Navarrete Lourdes Vega, Canto Cinthia Roche, et al. Forensic parameters and genetic variation of 15 autosomal STR loci in Mexican Mestizo populations from the States of Yucatan and Nayarit. Open Forensic Sci J. 2010;3(1):57–63. [Google Scholar]
- 39.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 40.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [Letter] [DOI] [PubMed] [Google Scholar]
- 41.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI: a disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum. 1992;35:630–640. doi: 10.1002/art.1780350606. [DOI] [PubMed] [Google Scholar]
- 42.Graham D. The isolation of high molecular weight DNA from whole organisms of large tissues masses. Anal Biochem. 1978;85:609–613. doi: 10.1016/0003-2697(78)90262-2. [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−^^Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Johnson MR, Wang K, Smith JB, Heslin MJ, Diasio RB. Quantitation of dihydropyrimidine dehydrogenase expression by real time reverse transcription polymerase chain reaction. Anal Biochem. 2000;278:175–184. doi: 10.1006/abio.1999.4461. [DOI] [PubMed] [Google Scholar]
- 45.Merino J, Merino R. Aportación de los modelos animales al estudio y el tratamiento de las enfermedades autoinmunitarias sistémicas. Reumatol Clin. 2008;4(Supl 1):S5–S10. doi: 10.1016/S1699-258X(08)76132-1. [DOI] [PubMed] [Google Scholar]
- 46.Sosa P, Batista F, González M, Bouza N. In: La conservación genética de las especies amenazadas en: Biología de la conservación de plantas amenazadas: Técnicas de diagnostico del estado de conservación. Bañares A, editor. Organismo Autónomo de Parques Nacionales; España: 2002. pp. 133–160. [Google Scholar]
- 47.Deng Y, Zhao J, Sakurai D, et al. MicroRNA-3148 Mudulates allelic expresion of Toll-Like receptor 7 variant associated with systemic lupus erythematosus. PLoS Genet. 2013;9(2):e1003336. doi: 10.1371/journal.pgen.1003336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberman A, Druker J, Refojo D, Arzt E. Mecanismos moleculares de acción de algunas drogas inmunosupresoras. Medicina. 2008;68:455–464. [PubMed] [Google Scholar]
- 49.Mesa Cornejo V, Barros Núñez P, Medina Lozano L. Metilación del ADN: marcador diagnóstico y pronóstico de cáncer. Gac Med Méx. 2006;142:1. [PubMed] [Google Scholar]
- 50.Machado LR, Hardwick RJ, Bowdrey J, et al. Evolutionary history of copy-number-variable locus for the low-affinity Fcγ receptor: mutation rate, autoimmune disease, and the legacy of helminth infection. Am J Hum Genet. 2012;90(6):973–985. doi: 10.1016/j.ajhg.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


