Abstract
The secreted frizzled-related protein 1 (SFRP1) gene plays an important role in carcinogenesis of digestive system cancer. Previous studies proved that circulating DNA promoter methylation may be a suitable biomarker for cancer patients. The aim of the present study was to investigate whether the promoter methylation status of serum SFRP1 is a potential biomarker for gastric adenocarcinoma (GAC) and esophageal square cell carcinoma (ESCC). The blood samples obtained from 42 GAC and 36 ESCC patients were detected for the promoter methylation status of SFRP1 by methylation-specific polymerase chain reaction. The control group included 42 benign gastrointestinal disease volunteers (24 benign gastric disease and 18 benign esophageal disease) and 20 healthy volunteers. Serum SFRP1 methylation was evident in 30.95% (13/42) GAC patients and 38.89% (14/36) ESCC patients, which is clearly higher compared to 8.33% (2/24) in benign gastric disease, 11.11% (2/18) in benign esophageal disease and 5% (1/20) in healthy volunteers (P<0.05). The association between the serum SFRP1 promoter methylation status and the clinical pathological features were further analyzed and methylation of the SFRP1 gene was significantly associated with age >60 years in GAC patients (P=0.027). However, no correlations between the SFRP1 methylation status and other clinicopathological parameters were found. In conclusion, the SFRP1 promoter was detected frequently in the serum of GAC and ESCC patients. The detection of circulating methylated SFRP1 in the serum may be a useful biomarker for upper gastrointestinal cancer patients.
Keywords: methylation, SFRP1 gene, gastric adenocarcinoma, methylation-specific polymerase chain reaction, esophageal square cell carcinoma
Introduction
Gastric adenocarcinoma (GAC) and esophageal square cell carcinoma (ESCC) are two major causes of cancer-related fatalities in China. Although gastroscopy is considered the most sensitive screening tool for upper gastrointestinal cancers, it remains limited in attributing to its potential risk, high cost and intolerance of the invasive procedure in patients (1). Therefore, reliable noninvasive tests for screening and diagnostic purposes are required.
Accumulating evidence indicates that carcinogenesis is regulated and controlled not only by genetic but also by epigenetic changes (2,3). There is a marked acceleration observed in the past decade validating the concept that cancer is a disease of epigenetic alterations that are leading candidates for the development of specific markers for cancer detection, diagnosis and prognosis (4). As the most common molecular epigenetic change in human cancer, DNA methylation was proved to silence the tumor suppressor genes in GAC (5,6) and ESCC (7). Due to the relatively significant circulating DNA quantity in peripheral blood, analyses of methylation in serum DNA can potentially serve as an excellent tumor marker for diagnosis and prognosis of cancer (8). The secreted frizzled-related protein 1 (SFRP1) gene belongs to the family of five secreted glycoproteins that have been identified as modulators of the Wnt signaling pathway involved in the development of cancer (9). Previous studies have shown that SFRP1 is downregulated by promoter methylation in numerous types of digestive system cancer, which makes it a candidate for tumor gene suppression (10–12).
The present study attempted to identify the SFRP1 promoter methylation status in serum from patients with GAC and ESCC by methylation-specific polymerase chain reaction (MSPCR), compared to benign gastrointestinal disease and healthy volunteers. The correlation between serum SFRP1 gene promoter methylation and patient clinical pathological parameters were also analyzed to further evaluate the clinical significance of this epigenetic molecular change.
Materials and methods
Study population
All the samples from human subjects were collected following the approval of the study by the Ethical Committee of Jinling Hospital (Nanjing, Jiangsu, China) and informed consent was obtained from all the patients. The 140 blood samples were obtained from 42 GAC patients, 36 ESCC patients, 42 benign upper gastrointestinal disease patients (24 benign gastric disease and 18 benign esophageal disease, such as chronic gastritis, gastric ulcer, benign polyp, nonmalignant adenoma and gastroesophageal reflux disease; data not shown) and 20 healthy volunteers. Based on pathologic evidence, which were from gastroscopy, all the patients were diagnosed at the Department of Gastroenterology and Hepatology of Jinling Hospital between August 1, 2011 and November 30, 2012. All the patients and healthy volunteers were genetically unrelated and were Han Chinese.
Sample collection
Each patient donated 5 ml of peripheral venous blood collected from 1 day after the patients were admitted to the hospital. At this time point, the patients did not start their treatment (surgery and/or chemo-radiation therapy). All the blood samples were kept in tubes containing clot activator at 4°C for 2 h and samples were centrifuged at ~2,000 x g for 10 min to isolate sera. Twenty serum samples from healthy volunteers were obtained from the Blood Center of Jinling Hospital and were used as normal controls. All the serum samples were stored at −80°C until use.
DNA extraction and MSPCR
Serum genomic DNA, extracted by the Axygen blood mini kit (Axygen, Union City, CA, USA) according to the manufacturer's instructions, was stored at −80°C until use. The serum DNA was modified with sodium bisulfite using the DNA methylation kit (Sigma, St. Louis, MO, USA), also according to the manufacturer's instructions. The methylation status of the CpG islands in the promoter region of SFRP1 was determined by MSPCR in GAC, ESCC, benign gastrointestinal disease and healthy control samples. Two sets of SFRP1 primers, described elsewhere (13), were used to discriminate between the methylated and unmethylated alleles. MSPCR was determined with two-step amplification/detection MSP instructions, as in previous studies (8,10,13,14). Briefly, the PCR mixture contained 6.25 µl AmpliTaq Gold 360 Master mix (Applied Biosystems, Foster City, CA, USA), 0.5 µmol of each primer (Invitrogen, Shanghai, China), 3.25 µl modified DNA and was adjusted by double-distilled H2O to a final volume of 25 µl. The cycling conditions consisted of one incubation for 2 min at 94°C, followed by 38 cycles of a 30-sec denaturation at 94°C, a 30-sec anneal (unmethylation at 56°C, methylation at 58°C), a 45-sec extension at 72°C and a final extension at 72°C for 7 min. The PCR products were electrophoresed through 2% agarose gels, stained with ethidium bromide and visualized with ultraviolet illumination. Table I lists the sequences of the PCR primers, products size and annealing temperature. All the experiments were performed in duplicate.
Table I.
SFRP1 sequences of the primers used in MSPCR.
| Gene | Primer sequence (5′-3′) | Product size, bp | Annealing temperature, °C |
|---|---|---|---|
| U | F: 5′-GTAGTTTTTGGAGTTAGTGTTGTGT-3′ | 126 | 56 |
| R: 5′-ACCTACAATCAAAAACAACACAAACA-3′ | |||
| M | F: 5′-GTTTTCGGAGTTAGTGTCGCGC-3′ | 119 | 58 |
| R: 5′-ACGATCGAAAACGACGCGAACG-3′ |
SFRP1, secreted frizzled-related protein 1; MSPCR, methylation-specific polymerase chain reaction; bp, basepair; U, unmethylated; M, methylated; F, forward; R, reverse.
Statistical analysis
All the data were analyzed by the SPSS 17 software (SPSS, Inc., Chicago, IL, USA). Statistical analysis was performed using the χ2 and Fisher's exact tests. Two-sided tests were used to determine significance and P<0.05 was considered to indicate a statistically significant difference.
Results
SFRP1 promoter methylation status in all the patients and controls
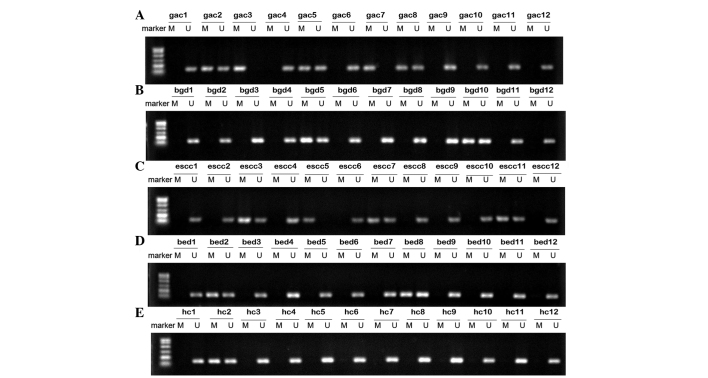
The methylation status of the SFRP1 promoter in serum DNA from 42 GAC, 36 ESCC, 24 benign gastric disease, 18 benign esophageal disease patients and 20 healthy volunteers was successfully performed. The representative agarose gel electrophoresis results are shown in Fig. 1. As shown, there were three statuses of the methylation analysis result: Complete methylation (only the methylated SFRP1 promoter was amplified), incomplete methylation (methylated and unmethylated SFRP1 promoter were amplified) and unmethylation (only the unmethylated SFRP1 promoter was amplified). Complete and incomplete methylation were defined as the methylation status. As a result, serum SFRP1 promoter methylation was detected in 13 GAC, 14 ESCC, two benign gastric disease, two benign esophageal disease patients and one healthy volunteer. The detection frequencies of serum SFRP1 promoter methylation was 30.95% in GAC and 38.89% in ESCC, which were significantly higher than those in the benign gastric disease (8.33%, P<0.05), benign esophageal disease (11.11%, P<0.05) and healthy volunteers (5.00%, P<0.05), respectively.
Figure 1.
Representative results showing the SFRP1 promoter methylation status identified by MSPCR. Identification of the SFRP1 promoter methylation status in serum samples from (A) gastric adenocarcinoma patients, (B) benign gastric disease patients, (C) esophageal square cell carcinoma patients, (D) benign esophageal disease patients and (E) healthy control. Lanes M and U indicate the amplified products with primers recognizing specific methylated and unmethylated sequences, respectively. SFRP1, secreted frizzled-related protein 1; MSPCR, methylation-specific polymerase chain reaction; M, methylated; U, unmethylated; GAC, gastric adenocarcinoma; BGD, benign gastric disease; ESCC, esophageal square cell carcinoma; BED, benign esophageal disease; HC, healthy control.
Correlation between the serum RASSF1A promoter methylation and clinicopathological parameters in patients with GAC and ESCC
The association between the serum SFRP1 promoter methylation status and clinicopathological parameters were analyzed in the GAC and ESCC patients. The results are listed in Table II. As indicated, methylation of the SFRP1 gene was significantly associated with age >60 years in the GAC patients (P=0.027). However, in the ESCC patients the difference was not significant (P=0.145). There was no other correlation between the SFPR1 promoter methylation status and patient gender, clinical stage, tumor differentiation grade, metastasis or serum carcinoembryonic antigen levels.
Table II.
Correlation between the serum SFRP1 methylation gene promoter methylation status and clinicopathological parameters in gastric adenocarcinoma and esophageal square cell carcinoma patients.
| Gastric adenocarcinoma | Esophageal square cell carcinoma | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characters | No. | M | U | χ2 | P-value | No. | M | U | χ2 | P-value |
| Total no. | 42 | 13 | 29 | 36 | 14 | 22 | ||||
| Age, years | ||||||||||
| <60 | 17 | 2 | 15 | 4.92a | 0.027a,b | 23 | 8 | 18 | 7.882a | 0.140c |
| >60 | 25 | 11 | 14 | 13 | 6 | 4 | ||||
| Gender | ||||||||||
| Male | 30 | 8 | 22 | 0.902a | 0.463c | 25 | 8 | 17 | 1.634a | 0.273c |
| Female | 12 | 5 | 7 | 11 | 6 | 5 | ||||
| Differentiation | ||||||||||
| Well/moderate | 16 | 4 | 12 | 0.428c | 0.733a | 21 | 9 | 12 | 0.334a | 0.563a |
| Poor/UN | 26 | 9 | 17 | 15 | 5 | 10 | ||||
| Stage | ||||||||||
| I/II | 9 | 2 | 7 | 0.408c | 0.695a | 24 | 6 | 15 | 2.258a | 0.133a |
| III/IV | 33 | 11 | 22 | 12 | 8 | 7 | ||||
| Metastasis | ||||||||||
| Yes | 15 | 4 | 11 | 0.278a | 0.734c | 13 | 3 | 10 | 2.141a | 0.143a |
| No | 27 | 9 | 17 | 23 | 11 | 12 | ||||
| CEA (ng/ml) | ||||||||||
| >9.7 | 18 | 7 | 11 | 0.928a | 0.335a | 6 | 4 | 2 | 2.338a | 0.181c |
| <9.7 | 24 | 6 | 18 | 30 | 10 | 20 | ||||
χ2test;
statistically significant (P<0.05);
Fisher's exact test. SFRP1, secreted frizzled-related protein 1; M, methylated; U, unmethylated; UN, undifferentiated; CEA, carcinoembryonic antigen.
Discussion
Indentation
Patients with early stage upper gastrointestinal cancer have no typical disease-related symptoms, which is why numerous gastric and esophageal cancers are detected at an advanced stage, even with incurable distant metastases (15). Thus, there is an urgent requirement to identify the valuable markers for early diagnosis evaluation of these types of cancer.
Aberrant methylation of promoter DNA regions that are rich in CpG islands is the key step in epigenetic gene silencing (16). Molecular alterations of the epigenome, particularly DNA methylation, have emerged as alternative targets of biomarker research and exhibit potentially great clinical significance (3). In numerous types of cancers, SFRP1 defects at the genetic (mutations) and epigenetic (transcriptional inactivation) levels have been reported (17). Epigenetically, SFRP1 is inactivated via promoter methylation, and tumor suppressor genes are silenced in cancers (18).
In the present study, the arbitrary promoter region methylation in SFRP1 was frequently observed in the serum of GAC and ESCC patients. The frequency of SFRP1 methylation was 30.92% in the serum of GAC patients, clearly higher compared to 8.33% in benign gastric disease (P=0.035) and 5% in healthy volunteers (P=0.018). The frequency is higher when compared to 11.3%, reported in the study by Tahara et al (5) that used quantitative MSPCR. However, Guo et al (19) detected the methylation of SFRP1 in gastric cancer tissue and obtained the frequency of 78.7% (74/94). The difference may result from the different methods and sample sizes. Another possibility is that in certain clinical cases, the methylation of tumor suppressor genes may be exhibited in tumor tissue, but have not been detected in serum (14). In 2005, the study by Zou et al (11) detected the mRNA expression of the SFRP genes and quantified by quantitative reverse transcription PCR in esophageal adenocarcinoma cell lines, and found that SFRP1 was detected in 93% of 40 cancers tissue and 81% of 37 Barrett's epithelia. This indicates that aberrant promoter methylation results in downregulation of the SFRP gene expression and occurs commonly in Barrett's esophagus and the early stage of esophageal cancer. Subsequently, Liu et al (20) detected hypermethylation of the promoter of SFRP1 using plasma DNA from 81 ESCC patients, with a percentage of 29.6%, but the study did not compare the result with benign esophageal disease. The present study had a similar, but higher, result of 38.89% in 36 ESCC patients. SFRP1 methylation was also detected in the benign esophageal disease with a frequency of 11.11%, which is markedly lower compared to in ESCC patients (P=0.035). These above findings confirmed that aberrant promoter methylation of SFRP1 may be an early event in the development of gastric and esophageal cancer and it can be a candidate biomarker for population screening. When it is detected in benign upper gastrointestinal disease patients and the healthy population, this may be a warning to perform gastroscopy and provide attention to this.
In the present study, the serum SFRP1 promoter methylation status was further compared to the clinicopathological parameters. As with a number of previous studies, no association was found between serum SFRP1 methylation and patient gender, tumor differentiation grade and distal metastasis in these two types of cancers. However, in GAC, methylation of the SFRP1 gene was significant associated with age >60 years in GAC patients (P=0.027). The similar result was reported by Wang et al (21) in which the age was observed to be significantly associated with the methylation status of the SFRP1 gene in breast cancer tissue. Age is an important risk factor and the association between DNA methylation and aging has been reported constantly (2,5,22,23). As early as 1987, Wilson et al (22) found that a significant loss of DNA 5-methyldeoxycytidine residues in old age disrupt the cellular gene expression and contribute to the physiological decline of the animal. Teschendorff et al (23) also found that the age-associated methylation signature is present in preneoplastic conditions and may drive gene expression changes associated with carcinogenesis. To verify the association of the SFRP1 gene methylation and age, this requires a large group of clinic observation and follow-up. The research regarding the biological mechanism of SFRP1 promoter methylation and age is also noteworthy to GAC.
The present study had limitations, such as the small sample size and lack of follow-up due to the limited time constraint. Thus, whether SFRP1 is a diagnostic, progressive, predictive or prognostic biomarker remains unknown. However, the sensitive and specific detection of the SFRP1 methylation patterns makes it a choice of biomarker for the clinical management of cancer patients. These investigations will continue in our future study.
In conclusion, the serum promoter methylation of the SFRP1 gene can be frequently detected in GAC and esophageal carcinoma patients and it may be a promising biomarker for such cancers. In general, more prospective studies are required to prove the use as a biomarker and to use it to aid in the clinical decision-making. Therefore, investigating a larger study population may reveal more significant results.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81270453) to Fangyu Wang, the National Natural Science Foundation of China (grant no. 81302162) to Nan Li and Jinling Hospital Foundation (grant no. 2011052) to Chang Liu.
References
- 1.Tanzer M, Liebl M, Quante M. Molecular biomarkers in esophageal, gastric, and colorectal adenocarcinoma. Pharmacol Ther. 2013;140:133–147. doi: 10.1016/j.pharmthera.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 3.How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications. Biochimie. 2012;94:2314–2337. doi: 10.1016/j.biochi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahara T, Maegawa S, Chung W, et al. Examination of whole blood DNA methylation as a potential risk marker for gastric cancer. Cancer Prev Res (Phila) 2013;6:1093–1100. doi: 10.1158/1940-6207.CAPR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao D, Shi J, Shi B, et al. Quantitative assessment of gene methylation and their impact on clinical outcome in gastric cancer. Clin Chim Acta. 2012;413:787–794. doi: 10.1016/j.cca.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Ishii T, Murakami J, Notohara K, et al. Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa. Gut. 2007;56:13–19. doi: 10.1136/gut.2005.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, Zhu RM, Li GL, Xia XY, Wei XW, et al. Detection of serum RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol. 2008;14:3074–3080. doi: 10.3748/wjg.14.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 10.Nojima M, Suzuki H, Toyota M, et al. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–4713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 11.Zou H, Molina JR, Harrington JJ, et al. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett's esophagus. Int J Cancer. 2005;116:584–591. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 12.Bu XM, Zhao CH, Dai XW. Aberrant expression of Wnt antagonist SFRP1 in pancreatic cancer. Chin Med J (Engl) 2008;121:952–955. [PubMed] [Google Scholar]
- 13.Urakami S, Shiina H, Enokida H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109–2116. doi: 10.1158/1078-0432.CCR-05-2468. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Yu Z, Wang T, Zhang J, Hong L, Chen L. Identification of epigenetic aberrant promoter methylation of RASSF1A in serum DNA and its clinicopathological significance in lung cancer. Lung Cancer. 2007;56:289–294. doi: 10.1016/j.lungcan.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Tani N, Ichikawa D, Ikoma D, et al. Circulating cell-free mRNA in plasma as a tumor marker for patients with primary and recurrent gastric cancer. Anticancer Res. 2007;27:1207–1212. [PubMed] [Google Scholar]
- 16.Liu JB, Wu XM, Cai J, et al. CpG island methylator phenotype and Helicobacter pylori infection associated with gastric cancer. World J Gastroenterol. 2012;18:5129–5134. doi: 10.3748/wjg.v18.i36.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urakami S, Shiina H, Enokida H, et al. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–6997. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 18.Mao W, Wordinger RJ, Clark AF. Focus on molecules: SFRP1. Exp Eye Res. 2010;91:552–553. doi: 10.1016/j.exer.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Guo Y, Guo W, Chen Z, Kuang G, Yang Z, Dong Z. Hypermethylation and aberrant expression of Wnt-antagonist family genes in gastric cardia adenocarcinoma. Neoplasma. 2011;58:110–117. doi: 10.4149/neo_2011_02_110. [DOI] [PubMed] [Google Scholar]
- 20.Liu JB, Qiang FL, Dong J, et al. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17:4917–4921. doi: 10.3748/wjg.v17.i44.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Dorsey TH, Terunuma A, et al. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7:e37928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 23.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]