Abstract
Objectives
Osteoarthritis (OA) is the most common form of arthritis with a clear genetic component. To identify novel loci associated with hip OA we performed a meta-analysis of genome-wide association studies (GWAS) on European subjects.
Methods
We performed a two-stage meta-analysis on more than 78 000 participants. In stage 1, we synthesised data from eight GWAS whereas data from 10 centres were used for ‘in silico’ or ‘de novo’ replication. Besides the main analysis, a stratified by sex analysis was performed to detect possible sex-specific signals. Meta-analysis was performed using inverse-variance fixed effects models. A random effects approach was also used.
Results
We accumulated 11 277 cases of radiographic and symptomatic hip OA. We prioritised eight single nucleotide polymorphism (SNPs) for follow-up in the discovery stage (4349 OA cases); five from the combined analysis, two male specific and one female specific. One locus, at 20q13, represented by rs6094710 (minor allele frequency (MAF) 4%) near the NCOA3 (nuclear receptor coactivator 3) gene, reached genome-wide significance level with p=7.9×10−9 and OR=1.28 (95% CI 1.18 to 1.39) in the combined analysis of discovery (p=5.6×10−8) and follow-up studies (p=7.3×10−4). We showed that this gene is expressed in articular cartilage and its expression was significantly reduced in OA-affected cartilage. Moreover, two loci remained suggestive associated; rs5009270 at 7q31 (MAF 30%, p=9.9×10−7, OR=1.10) and rs3757837 at 7p13 (MAF 6%, p=2.2×10−6, OR=1.27 in male specific analysis).
Conclusions
Novel genetic loci for hip OA were found in this meta-analysis of GWAS.
Keywords: Epidemiology, Gene Polymorphism, Osteoarthritis
Introduction
Osteoarthritis (OA) is the most common form of arthritis affecting 40% of people over the age of 70 years and is one of the most common disabling diseases observed worldwide.1 2 Its aetiology is multifactorial with a clear genetic component. Inheritance studies in twins and other family-based studies have assessed the estimated heritability for OA in the range of 40–65% depending on the joint site.3–5 The established OA loci have small ORs (range 1.10–1.20)6 7 and the genetic architecture of OA is likely to consist of multiple variants of similar magnitude.
During the last few years, extensive efforts have led to the identification of a number of OA susceptibility signals in European populations that have surpassed the genome-wide significance (GWS) level (p<5×10−8). A locus on chr7q22 near the orphan receptor GPR22 derived from genome-wide association studies (GWAS)8 9 and a variant in the GDF5 gene, originating from a candidate gene approach reached GWS for knee OA.10 An analysis using a 1000-genomes-project-based imputations identified a variant on chromosome 13q34 near the MCF2L gene.7 A recent GWAS in UK subjects revealed eight more loci that increase risk for OA. From those eight loci, four signals in 9q33 (ASTN2), 6q14 (FILIP1/SENP6), 12p11 (KLHDC5/PTHLH) and 12q23 (CHST11) were found to be associated with total hip replacement (THR) or hip OA in European populations.6 Finally, GWAS and functional studies revealed that the DOT1L gene on 19q13 is also associated with hip OA and cartilage thickness.11 12
In this study a large-scale GWAS meta-analysis for hip OA was performed under the auspices of the Translational Research in Europe Applied Technologies in Osteoarthritis (TreatOA) consortium including eight sample sets, in the discovery stage. With a total of 4349 hip OA cases and 46 903 controls in the discovery stage, and a total of 11 277 cases and 67 473 controls, this is the largest study of hip OA to date.
Methods
Study design and analysis plan
A two-stage design was used for the identification of potential associations. In the discovery stage imputed and directly genotyped autosomal single nucleotide polymorphism (SNPs) were assessed using a quality control (QC) procedure that is described in the online supplementary material (section 1.a). Briefly, we excluded SNPs based on low minor allele frequency (MAF <1%), low imputation quality, low call rate and deviation from the Hardy-Weinberg equilibrium. Genomic control was applied to each study before meta-analysis. The effect estimates of each study where synthesised using an additive model. The variants that surpassed the p<1×10−6 threshold in the meta-analysis were selected for further follow-up. Besides the main analysis including all participants a separate analysis stratified by sex was performed to detect possible sex-specific signals. In silico and de novo replication was sought for the discovery signals in 10 additional studies. All the derived effects from the discovery and the replication stage were finally synthesised using inverse variance fixed-effect models and the between-study heterogeneity was assessed using the I2metric.13 Moreover, a random-effects (RE) model was applied.14 A p-value of <5×10−8 was considered GWS. The associations of the top findings at the discovery stage were also assessed when adjusted for other risk factors such as age, height and body mass index (BMI). We also examined the association of these markers with height and BMI in the large publicly available sample set of the Genetic Investigation of Anthropometric Traits (GIANT) consortium (133 000 individuals in height analysis and 123 000 individuals in BMI analysis.15 16 The detailed analysis plan is presented in the online supplementary material (section 1.b).
Study populations and phenotype definition
The studies included in the discovery and replication efforts are described in table 1 and more details are given in the online supplementary material (section 1.c). All studies had standardised definitions of the phenotypes. Specifically, the definition of the hip OA in the studies was either a radiographic Kellgren and Lawrence (K/L) grade of ≥2 or history of a THR surgery because of OA. THR subjects were excluded from the study if they had: other major arthropathy (eg, rheumatoid arthritis, ankylosing spondylitis); Paget's disease affecting the pelvis or femur; THR due to hip trauma or avascular necrosis of the femoral head; or terminal illness. The control groups consisted of subjects who had no known affected joints. Population-based controls were used by the arcOGEN study.
Table 1.
Studies included in the TreatOA GWAS meta-analysis
| Study | N cases | N controls | λ | N SNPs | ROA/SOA | Controls | Genotyping platform | Imputation method |
|---|---|---|---|---|---|---|---|---|
| Discovery stage | ||||||||
| arcOGEN stage 1 | 1728 | 4896 | 1.058 | 2.454.242 | ROA/SOA | Population | Illumina Human610 (cases)+Illumina 1.2M Duo (controls) | Impute |
| deCODE | 1423 | 31 385 | 1.182 | 2.399.690 | SOA | Osteoarthritis-free | Infinium HumanHap 300+humanCNV370 | Impute |
| EGCUT | 64 | 2531 | 0.994 | 2.242.156 | ROA | Population | Illumina HumanCNV370 or HumanOmniExpress | Impute |
| GARP | 106 | 1671 | 1.294 | 2.406.007 | SOA/ROA | KL<2 | Illumina Infinium HD Human660W-Quad | Impute |
| RSI | 760 | 3233 | 1.009 | 2.450.385 | ROA | KL<2 | Illumina HumanHap550v3 | MACH |
| RSII | 159 | 1472 | 0.993 | 2.442.419 | ROA | KL<2 | Illumina HumanHap550-Duo | MACH |
| RSIII | 41 | 1487 | 0.962 | 2.397.764 | ROA | KL<2 | Illumina Human660W-Quad | MACH |
| TwinsUK | 68 | 228 | 0.993 | 2.358151 | ROA | KL<2 | Infinium HumanHap300 | Impute |
| Total discovery | 4349 | 46 903 | 1.028 | 2.567.279 | – | – | ||
| Replication stage | ||||||||
| arcOGEN stage 2 | 1763 | 6157 | 1.058 | – | ROA/SOA | – | Illumina Human610 (cases)+Illumina 1.2M Duo (controls) | Impute |
| SOF* | 761 | 2376 | 1.008 | – | ROA | – | Illumina Omni 1 array (1.1 million probes) | MACH |
| MrOS* | 446 | 2837 | 1.002 | – | ROA | – | Illumina Omni 1 array (1.1 million probes) | MACH |
| arcOGEN plus | 223 | 1828 | – | – | ROA/SOA | – | Illumina Human610 (cases)+Illumina 1.2M Duo (controls) | Impute |
| EGCUT | 977 | 1131 | – | – | ROA | – | Illumina HumanCNV370 or HumanOmniExpress | Impute |
| Greek TJR cases | 93 | 361 | – | – | SOA | – | Single base extension using SNaPshot Multiplex Kit (Applied Biosystems, Foster City, USA)) | – |
| Paprika Study | 602 | 2321 | – | – | SOA | – | Sequenom (MassARRAY iPLEX Gold) | – |
| Spanish TJR cases | 697 | 783 | – | – | SOA | – | Single base extension using SNaPshot Multiplex Kit (Applied Biosystems, Foster City, USA)) | – |
| Icelandic cases | 857 | 1857 | – | – | SOA | – | Centaurus (Nanogen)37 | |
| Swedish MDC study | 509 | 919 | – | – | SOA | – | Centaurus (Nanogen)37 | – |
| Total replication | 6928 | 20 570 | – | – | ||||
| Total | 11 277 | 67 473 | – | – | ||||
EGCUT, Estonian Genome Center, University of Tartu; GWAS, genome-wide association studies; HD, high density; KL, Kellgren-Lawrence score; MDC, Malmo Diet Cancer study; MrOS, Osteroporotic Fractures in Men study; ROA, radiographic osteoarthritis; SOA, symptomatic osteoarthritis; SOF, Study of Osteoporotic Fractures; TJR, total joint replacement.
Genotyping and imputation
Genotyping of GWAS genotyping was performed by each study following standard protocols and imputation was then carried out at the individual study level on the ∼2.5 million SNPs from HapMap Phase 2 release 22 using genome build 36 (Utah residents with ancestry from northern and western Europe (CEPH))17 on MACH or IMPUTE18 software. Imputation quality scores for each SNP were obtained from IMPUTE and MACH statistics, as appropriate. An overview of all studies and the genotyping platforms and imputation method used is given in table 1. Ten studies of European ancestral origin provided data for independent replication. Four datasets (arcOGEN stage 2, arcOGEN plus, Osteroporotic Fractures in Men study and the Study of Osteoporotic Fractures) provided ‘in silico’ replication whereas ‘de novo’ replication was performed in six other study groups (Icelandic, Swedish, Estonian Genome Center, University of Tartu (EGCUT), Paprika study, Greek, Spanish). QC criteria for deviation from Hardy-Weinberg equilibrium, MAF inconsistencies with the discovery data and outliers were applied in the replication data before including all the available data in the final analysis.
Gene expression
Expression was determined by Illumina HT-12 V3 microarrays using standard methods using 47 000 probes corresponding to over 25 000 well-characterised genes. DNA was available from blood and cartilage. Using the Beadstudio software the intensity values were normalised using the ‘rsn’ option in the Lumi R-package. The corresponding signals increase exponentially with relative levels and units are light intensity (Illumina provided values). The obtained raw probe-level data (overall mean normalised probe level value of measured genes in cartilage) were exported for analyses using Limma.19 As implemented in Limma, a paired t test was used on all samples. There were two probes, approximately 2.2 kb apart, on the array used for NCOA3.
Heritability of hip OA explained by genetic variants
We calculated the sibling recurrence risk and the expected genetic variance explained for hip OA hits that were identified previously and in this study as described in online supplementary material (section 1.e)
Results
The final analysis included a total of 11 277 radiographic and symptomatic hip OA cases and 67 473 controls of European ancestry, with 4349 cases and 46 903 controls included in the discovery stage and 6928 cases and 20 570 controls in the follow-up effort (table 1). In the sex-specific analyses 2045/20 823 male cases and controls and 2689/25 384 female cases and controls were analysed in the discovery. After QC, 2 567 279 SNPs were analysed. Low genomic inflation factor was observed for the first stage with λ=1.028. The results of the discovery stage were uncorrected for the overall inflation factor. Quantile-Quantile plots showed an excess of signals compared with what was expected by chance, indicating the presence of true association signals that could confer susceptibility to hip OA (see online supplementary figures S1–S3). For male-specific analysis (see online supplementary figure S3) there is an early deviation from the neutrality line, therefore positive signals should be treated with caution.
Following analysis of the discovery stage it was found that eight independent loci reached the prespecified threshold of p<1.0×10−6 required for further replication; five from the combined analysis of sexes, two male-specific and one female-specific loci. The number of independent SNPs is larger compared with three estimated independent SNPs expected under the null for the main and the sex-specific analyses (binomial test p=0.012). Two of these signals (rs6094710 at 20q13 and rs640070 at 11q25) were imputed, had MAF <5% and moderate or large effect sizes were observed (OR >1.2). Therefore, we examined for possible imputation errors by comparing with de novo genotyping in a random sample of the RS-I and the Twins UK studies to exclude any chance of false positive findings. The obtained MAF estimates between the imputation and the genotyping efforts were not consistent for rs640070, which was therefore excluded from further consideration, minimising the chance of any false positive signals (see online supplementary material; section 1.d). Moreover, we included in the replication stage rs17610181 at 17q23, a SNP that was just below the desired threshold, but was previously shown to be associated with height.15 Therefore, eight independent SNPs were included in the replication phase. The combined effect sizes and p values for the eight signals are presented in table 2 and in online supplementary figures S4–S11. None of these variants associate with height or BMI in the large publicly available GIANT databases (see online supplementary table S2) nor did including these covariates, or age, in the analysis change the association of OA with these genetic variants (see online supplementary table S3).
Table 2.
Association results for hip OA meta-analysis of the discovery and the replication studies
| SNP | Chr | Gene | A1 | EAF (%) | Discovery stage | Replication stage | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | I2 (%) | OR (95% CI) | p Value | OR (95% CI) | p Value | I2 (%) | H&E RE p value | |||||
| Combined analysis of both genders: | |||||||||||||
| rs6094710 | 20q13 | NCOA3 | A | 4 | 1.40 (1.24 to 1.58) | 5.6×10−8 | 0 | 1.20 (1.08 to 1.34) | 7.3×10−4 | 1.28 (1.18 to 1.39) | 7.9×10−9 | 64 | 2.0×10−10 |
| rs1577792 | 6q14 | HMGN3 | A | 39 | 1.14 (1.09 to 1.20) | 7.3×10−8 | 0 | 1.03 (0.99 to 1.07) | 0.17 | 1.07 (1.04 to 1.11) | 7.8×10−5 | 50 | 9.6×10−6 |
| rs5009270 | 7q31 | IFRD1 | A | 30 | 1.15 (1.09 to 1.21) | 5.7×10−7 | 21 | 1.05 (1.00 to 1.11) | 0.061 | 1.10 (1.06 to 1.14) | 9.0×10−7 | 46 | 3.1×10−6 |
| rs10773046 | 12q24 | DNAH10 | G | 45 | 1.13 (1.08 to 1.19) | 9.7×10−7 | 10 | 1.01 (0.96 to 1.06) | 0.72 | 1.07 (1.03 to 1.11) | 2.0×10−4 | 64 | 1.9×10−6 |
| rs17610181 | 17q23 | NACA2 | A | 14 | 1.20 (1.11 to 1.29) | 1.9×10−6 | 32 | 1.05 (0.99 to 1.12 | 0.12 | 1.12 (1.06 to 1.18) | 1.3×10−5 | 42 | 7.5×10−6 |
| Female-specific analysis: | |||||||||||||
| rs10878630 | 12q15 | DYRK2 | A | 41 | 1.19 (1.11 to 1.26) | 1.2×10−7 | 25 | 0.98 (0.91 to 1.05) | 0.52 | 1.09 (1.04 to 1.14) | 3.9×10−4 | 65 | 1.1×10−5 |
| Male-specific analysis: | |||||||||||||
| rs12551314 | 9q22 | PHF2 | A | 12 | 1.30 (1.18 to 1.44) | 6.3×10−7 | 28 | 0.98 (0.86 to 1.11) | 0.72 | 1.16 (1.07 to 1.25) | 2.9×10−4 | 66 | 4.3×10−5 |
| rs3757837 | 7p13 | CAMK2B | C | 6 | 1.46 (1.26 to 1.69) | 8.3×10−7 | 46 | 1.15 (1.00 to 1.32) | 0.044 | 1.27 (1.15 to 1.41) | 2.2×10−6 | 78 | 7.7×10−10 |
The discovery sample set comprised 4349 hip OA cases and 46 903 controls, and the replication samples were 6928 cases and 20 570 controls in the analysis of both genders. The female-specific analysis comprised 2689 hip OA cases and 25 384 controls in the discovery set and 2398 cases and 8787 controls in the replication stage. For the male-specific analysis there were 2045 hip OA cases and 20 823 controls in the discovery set and 1386/7087 and 1451/4956 cases/controls in the replication stage for rs12551314 and rs3757837 respectively.
A1, coded/effect allele; Chr, chromosome; EAF, effect allele frequency; Gene, nearest gene; HE RE P, Han & Eskin Random Effects P; I2, measure of heterogeneity; OA, osteoarthritis.
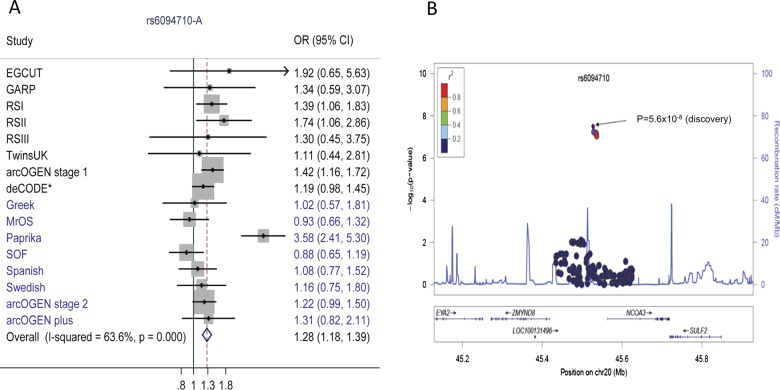
Two SNPs, rs6094710 at 20q13 and rs1577792 at 6q14, were borderline GWS in the discovery stage with p=5.8×10−8 and 7.3×10−8, respectively, with no observed heterogeneity (I2=0). The SNP rs6094710 replicated with p=7.3×10−4 in the follow-up samples and reached GWS level with p=9.3×10−9 and OR=1.28 (95% CI 1.17 to 1.39) when we combined the discovery and the replication data, although large heterogeneity was observed I2=64% (figure 1). The SNP remained GWS after the second genomic control. rs6094710 is annotated near NCOA3 (nuclear receptor coactivator 3) gene. The OR of rs1577792 at 6q14 was close to unity in the replication effort with one study being significant in the opposite direction.
Figure 1.
(A) Forest plot for meta-analysis of rs6094710. The blue diamond in the forest plot denotes the summary effect size and its edges the respective 95% CIs. Studies shaded in blue were included in the replication stage. *Discovery and replication estimate combined; and (B) regional plot for rs6094710 comprising directly genotyped and imputed SNPs. Case-control association results (−log10 P) in the discovery set are plotted against genomic position (National Center for Biotechnology Information build 36) for the stratum where the most significant meta-analysis p was observed. The colour reflects the correlation coefficients (r2) of each genotyped SNP with the index SNP estimated using the CEU HapMap II panel.
The rs3757837 SNP at 7p13 from the male-specific analysis replicated nominally (p=0.044, OR=1.15) in the follow-up samples, but did not reach GWS in the overall analysis with p=2.2×10−6 and OR=1.27. Moderate heterogeneity was observed for this SNP in the discovery analysis (I2=46%). Heterogeneity was increased, in the overall analysis with all the studies combined (I2=78%), reflecting the further heterogeneity introduced by the replication data. We, therefore, also applied the Han and Eskin (RE) model, an approach that allows more heterogeneity in the data compared with traditional models.14 Using this model rs3757837 showed a stronger association with p=7.7×10−10 in this analysis. The strength of the association of this approach was not substantially different from the fixed-effect model for any other SNP in our study (table 2). rs3757837 resides in intron 8 of the CAMK2B (calcium/calmodulin-dependent protein kinase II β) gene.
The rs5009270 SNP at 7q31 remained suggestively associated in the final analysis (p<5×10−6) with combined two-stage p=9.9×10−7 and summary OR 1.10. rs5009270 is located near the IFRD1 (interferon-related) gene. The p with the RE approach was 3.1×10−6.
Gene expression data
To further investigate our findings for the top GWS hit at 20q13 we explored mRNA expression profile of the NCOA3 gene in articular cartilage, a highly relevant tissue for OA. Expression of the gene was examined by exploring a micro array mRNA expression dataset generated on Illumina V3 Human-12 chips in cartilage samples of 33 patients (13 men and 20 women of European descent aged 54 years to 80 years) that underwent joint replacement due to end-stage OA disease. Expression levels of NCOA3 in cartilage displaying symptoms of OA were compared with expression levels in cartilage that appeared macroscopically normal but isolated from the same joint (preserved cartilage). A moderate level of expression, as determined by the mean normalised probe level value, was observed for NCOA3 (mean level 8.49), which was above the observed average expression of genes in the articular cartilage (mean normalised probe level value of measured genes in cartilage was 7.4; range 6.6–14.9). Expression was also high in blood (6.96; range 6.3–14.7). When we tested for differential expression of NCOA3 among the pairs of preserved and OA-affected cartilage, we observed a significantly lower expression of NCOA3 in the OA-affected cartilage (p=0.0064).
Heritability of hip OA explained by genetic variants
Table 3 summarises genome-wide significant and suggestive signals of hip OA including this study. Based on these findings and if we consider a sibling recurrence ratio λs=520 then the discovered signals of OA contribute 3% of the heritability in OA when all hits are considered and 2.1% if only GWS are evaluated.
Table 3.
Summary of the genome-wide significant and suggestive SNPs for hip OA and their estimated heritability
| Reference | SNP | Gene | OR | EAF | λs | Expected GV explained (%) |
|---|---|---|---|---|---|---|
| Genome wide significant findings (p<5×10−8) | ||||||
| This study | rs6094710 | NCOA3 | 1.28 | 0.04 | 1.0029 | 0.46 |
| arcOGEN6 | rs6976 | GLT8D1 | 1.12 | 0.37 | 1.0031 | 0.60 |
| arcOGEN6 | rs11177 | GNL3 | 1.12 | 0.38 | 1.0031 | 0.60 |
| arcOGEN6 | rs4836732 | ASTN2 | 1.20 | 0.47 | 1.0083 | 1.66 |
| arcOGEN6 | rs9350591 | FILIP1;SENP6 | 1.18 | 0.11 | 1.0030 | 0.54 |
| arcOGEN6 | rs10492367 | KLHDC5;PTHLH | 1.14 | 0.19 | 1.0029 | 0.53 |
| 3arcOGEN6 | rs835487 | CHST11 | 1.13 | 0.34 | 1.0034 | 0.67 |
| TreatOA12 | rs12982744 | DOT1L | 1.17 | 0.38 | 1.0060 | 1.16 |
| Suggestive findings (5×10−6<p<5×10−8) | ||||||
| This study | rs5009270 | IFRD1 | 1.10 | 0.30 | 1.0019 | 0.38 |
| This study | rs3757837 | CAMK2B | 1.27 | 0.06 | 1.0040 | 0.64 |
| arcOGEN6 | rs12107036 | TP63 | 1.21 | 0.52 | 1.0090 | 1.81 |
| arcOGEN6 | rs8044769 | FTO | 1.11 | 0.50 | 1.0027 | 0.54 |
| arcOGEN6 | rs10948172 | SUPT3H;CDC5L | 1.14 | 0.29 | 1.0037 | 0.71 |
EAF, effect allele frequency; GV, genetic variance; OA, osteoarthritis.
Discussion
In this report we attempted to further clarify the genetic architecture of the genetic background of hip OA by using the largest sample-size for hip OA to-date of more than 78 000 genotyped individuals under a GWAS framework. During the discovery phase, we identified eight signals that qualified for further independent replication. Of these, the signal at chromosome 20q13 near NCOA3 gene was found to be GWS and two other loci were suggestively significant at a p<5×10−6 level in the joint analysis of the discovery and replication stages. Adjusted analyses of the prioritised signals revealed that these markers were not associated with body size. These genetic risk factors contribute to our knowledge base in the field of the susceptibility for hip OA by conferring a medium OA risk.
The top signal identified in this meta-analysis was rs6094710, a variant that is annotated on chromosome 20q13 near the NCOA3 gene, increasing the risk for hip OA for the carriers of the A allele by almost 30%. Furthermore, we showed that the identified NCOA3 gene was expressed in articular cartilage and its expression was significantly reduced in OA affected cartilage, further supporting a role of the NCOA3 gene signal in OA disease process. Interestingly, rs6094710 is in complete linkage disequilibrium with rs6094752 (r2=1) which is a missense SNP leading to an amino acid change at position 218 in the protein (Arg>Cys). This amino acid change is predicted to have a benign and damaging effect on the protein by PolyPhen-2,21 dependent on the variant protein. The functional consequences of SNP rs6094752 are unknown, and further research is therefore needed to unravel the biological mechanism of this amino acid change in relation to OA.
The NCOA3 gene is a nuclear receptor coactivator that directly binds nuclear receptors and stimulates the transcriptional activities in a hormone-dependent fashion. In this signalling process, NCOA3 recruits histone acetyltransferases and methyltransferases for chromatin remodelling and facilitating downstream gene transcription. NCOA3 is involved in the coactivation of different nuclear receptors, such as for steroids, retinoids, thyroid hormone, vitamin D3 and prostanoids. Many of these hormones have been implicated in skeletal metabolism and OA, which makes NCOA3 a compelling causal candidate gene. Previously, NCOA3 knockout mice were generated through homologous recombination in embryonic stem cells.22 These mice showed growth retardation and reduced adult body size, but the molecular mechanism responsible for this growth retardation remains largely unknown. In addition, female mice exhibited abnormal development and function of their reproductive system and oestrogen levels were significantly lower in the knockout mice compared with the wild type,22 possibly indicating involvement of NCOA3 in steroid regulation.
NCOA3 could be also implicated through regulation of the target tissue responses to thyroid hormone (T3).23 Since intracellular T3 is tightly regulated by deiodinase, iodothyronine, type-2 and deiodinase, iodothyronine, type-3 encoded by the DIO2 and DIO3 genes, respectively, that were previously recognised as OA susceptibility genes,24 25 the current NCOA3 findings complement the previous outlined hypothesis that local T3 signalling may affect OA susceptibility.26
Another possible mechanism by which NCOA3 might be involved in cartilage homoeostasis is through transcriptional regulation in mechanotransduction. NCOA3 is upregulated by the signal transducer and activator of transcription 6 (STAT6) in naïve splenic B cells from BALB/c and serves as a positive regulator of transcriptional activation by STAT6.27 STAT6 is known to be the common signal transducer of interleukin (IL)-4 receptor α chain and mediates IL-4- and IL-13-induced responses.28 Chondrocytes from normal and OA cartilage signal through a type II IL-4R in in human articular chondrocyte mechanotransduction. This signalling is via a STAT6-independent pathway. Differences in IL-4 signalling are likely due to crosstalk between integrin and cytokine signalling pathways.29 Therefore, NCOA3 may be related to cartilage function and molecular signalling and transcriptional regulation in mechanotransduction.
The male-specific locus on 7p13, represented by rs3757837, showed considerable heterogeneity between studies. The signal was strongly supported by the RE model. Unlike the conservative traditional RE methods this new method has been shown to achieve higher statistical power when heterogeneity exists, allowing for new discoveries in the field of genetic epidemiology.14 rs3757837 is located in CAMK2B gene, which belongs to the calcium/calmodulin-regulated kinase (CaMKII) subfamily. There is evidence that CaMKII-signalling may be important in onset and progression of OA.30 31 This pathway has been described as central to the molecular events that regulate chondrocyte responses to mechanical stimulation and in particular, to the upstream effect of IL-4. This would be linked to the STAT6 pathway, which in turn is regulated by NCOA3. Thus these two genetic signals, NCOA3 and CAMK2B, both point to pathways related to cartilage mechanotransduction, suggesting that genetic defects in this pathway may be central to the degeneration of cartilage that takes place in hip OA. OA is a disease affecting articular cartilage and the underlying bone, resulting from many biological and mechanical interacting factors which change the extracellular matrix and cells and lead to increasing levels of cartilage degeneration. Joint tissues are exquisitely sensitive to their mechanical environment, and mechanical loading may be the most important external factor regulating the development and long-term maintenance of joint tissues.32 Finally, rs5009270 resides near IFDRD1 which codes an interferon-related developmental regulator and has been implicated in skeletal muscle regeneration.33 Reduction in muscle strength is strongly associated with functional decline, and individuals with lower quadriceps strength adjusted for body weight are more likely to develop OA.34
Our study has certain limitations. In our discovery stage, two out of the eight independent loci had no heterogeneity and in five signals the heterogeneity was moderate or low. However, when we included the replication data, large heterogeneity was introduced for all SNPs. Conflicting results in the replication data, besides chance, could be explained by inconsistent definitions of the OA phenotypes but also from the different population structure that can introduce heterogeneity. The TreatOA consortium has addressed the need of standardisation of OA phenotypes35 and this effort may have diminished the observed heterogeneity in the discovery stage. Efforts including more data using common and stringent QC criteria and standardisation methods should substantially improve the power of GWAS to identify novel findings in the near future. The main limitation of the expression study is that there are few individuals included, in particular only two carriers of the rs6094710 variant.
In conclusion, novel loci involved in hip OA were discovered through a large-scale meta-analysis of GWAS. The exact underlying mechanism leading to a higher risk of OA remains to be elucidated by functional experiments. It is evident from this work and other recent studies that deciphering the architecture of the genetics of OA requires major large-scale efforts, and in this regard calls for international worldwide collaborations are not fruitless. In the near future emphasis should be given to the enhancement of the total sample size, the adoption of stringent and standardised definitions of the phenotypes and the application of imputation-based meta-analysis using the panel of the 1000 Genomes Project.36 In addition, linking results from genetic association studies to for example genome-wide RNA expression data might further improve our understanding. Eventually, large-scale studies with whole-genome sequencing will be needed to target the heritability caused by less common variants.
Supplementary Material
Footnotes
Correction notice: This article has been corrected since it was published Online First. The affiliations for P Eline Slagboom have been corrected and affiliations 1 and 2 have been amended.
Acknowledgements: The authors are very grateful to all study participants, the staff from all studies and the participating physicians and pharmacists. Acknowledgements for each study are given in online supplementary material (section 4).
Contributors: EE, HJK, US, IM, JPAI, TDS, JBvM and AMV conceived and designed the study. EE, HJK, EEN and KKT analysed the data. EE, HJK, US, EEN, EZ, IM, JPAI, TDS, JBvM and AMV wrote the first draft of the manuscript. All authors interpreted the results, critically commented and approved the final version of the manuscript.
Funding: This project was supported through Coordination Theme 1 (Health) of the European Community's FP7 (grant 200800) TreatOA. The work was also supported by EU FP7 Small-scale focused research collaborative projects EurHEALTHAging (grant 277849).
Competing interests: US, IJ, GT, HTH, UT and KS are employed by deCODE Genetics/Amgen. HJK is currently employed by Pfizer.
Ethics approval: Meta-analysis of GWAS, different review boards approved the individual studies.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 2008;58:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valdes AM, Spector TD. The contribution of genes to osteoarthritis. Med Clin North Am 2009;93:45–66. [DOI] [PubMed] [Google Scholar]
- 3.Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage 2004;12(Suppl A):S39–44. [DOI] [PubMed] [Google Scholar]
- 4.Jonsson H, Manolescu I, Stefansson SE, et al. The inheritance of hand osteoarthritis in Iceland. Arthritis Rheum 2003;48:391–5. [DOI] [PubMed] [Google Scholar]
- 5.Kraus VB, Jordan JM, Doherty M, et al. The Genetics of Generalized Osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage 2007;15:120–7. [DOI] [PubMed] [Google Scholar]
- 6.arcOGEN consosrtium; arcOGEN Collaborators. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 2012;380:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day-Williams AG, Southam L, Panoutsopoulou K, et al. A variant in MCF2L is associated with osteoarthritis. Am J Hum Genet 2011;89:446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerkhof HJ, Lories RJ, Meulenbelt I, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum 2010;62:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis 2011;70:349–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valdes AM, Evangelou E, Kerkhof HJ, et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis 2011;70:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castano Betancourt MC, Cailotto F, Kerkhof HJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci USA 2012;109:8218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelou E, Valdes AM, Castano-Betancourt MC, et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann Rheum Dis 2013;72:1264–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 2011;88:586–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010;467:832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International HapMap C, Frazer KA, Ballinger DG, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007;449:851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13. [DOI] [PubMed] [Google Scholar]
- 19.Smyth G. Linear models and empirical bayes methods for assessing differential expression in microarry experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 20.Valdes AM, Spector TD. Genetic epidemiology of hip and knee osteoarthritis. Nature reviews. Rheumatology 2011;7:23–32. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Liao L, Ning G, et al. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci USA 2000;97:6379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholls JJ, Brassill MJ, Williams GR, et al. The skeletal consequences of thyrotoxicosis. J Endocrinol 2012;213:209–21. [DOI] [PubMed] [Google Scholar]
- 24.Meulenbelt I, Bos SD, Chapman K, et al. Meta-analyses of genes modulating intracellular T3 bio-availability reveal a possible role for the DIO3 gene in osteoarthritis susceptibility. Ann Rheum Dis 2011;70:164–7. [DOI] [PubMed] [Google Scholar]
- 25.Meulenbelt I, Min JL, Bos S, et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum Mol Genet 2008;17:1867–75. [DOI] [PubMed] [Google Scholar]
- 26.Bos SD, Slagboom PE, Meulenbelt I. New insights into osteoarthritis: early developmental features of an ageing-related disease. Curr Opin Rheumatol 2008;20:553–9. [DOI] [PubMed] [Google Scholar]
- 27.Arimura A, vn Peer M, Schroder AJ, et al. The transcriptional co-activator p/CIP (NCoA-3) is up-regulated by STAT6 and serves as a positive regulator of transcriptional activation by STAT6. J Biol Chem 2004;279:31105–12. [DOI] [PubMed] [Google Scholar]
- 28.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994;264:1415–21. [DOI] [PubMed] [Google Scholar]
- 29.Millward-Sadler SJ, Khan NS, Bracher MG, et al. Roles for the interleukin-4 receptor and associated JAK/STAT proteins in human articular chondrocyte mechanotransduction. Osteoarthritis Cartilage 2006;14:991–1001. [DOI] [PubMed] [Google Scholar]
- 30.Shimazaki A, Wright MO, Elliot K, et al. Calcium/calmodulin-dependent protein kinase II in human articular chondrocytes. Biorheology 2006;43:223–33. [PubMed] [Google Scholar]
- 31.Huser CA, Davies ME. Calcium signaling leads to mitochondrial depolarization in impact-induced chondrocyte death in equine articular cartilage explants. Arthritis Rheum 2007;56:2322–34. [DOI] [PubMed] [Google Scholar]
- 32.Sun HB. Mechanical loading, cartilage degradation, and arthritis. Ann NY Acad Sci 2010;1211:37–50. [DOI] [PubMed] [Google Scholar]
- 33.Micheli L, Leonardi L, Conti F, et al. PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kappaB. J Biol Chem 2011;286:5691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trudelle-Jackson E, Ferro E, Morrow JR., Jr Clinical implications for muscle strength differences in women of different age and racial groups: the WIN study. J Womens Health Phys Therap 2011;35:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerkhof HJ, Meulenbelt I, Akune T, et al. Recommendations for standardization and phenotype definitions in genetic studies of osteoarthritis: the TREAT-OA consortium. Osteoarthritis Cartilage 2011;19:254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutyavin IV, Milesi D, Belousov Y, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res 2006;34:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.