Abstract
Hepatits C virus (HCV) genotype 4 (GT4) shows low treatment response rates and discrepancies when compared to other genotypes. However, the reason underlying these discrepancies remains unclear due to the limited number of studies on GT4. microRNA-155 (miR-155) is a noteworthy example of a discrepancy in GT4, as it was found to be upregulated in genotypes 1, 2 and 3 HCV infection, but downregulated in GT4-HCV-infected peripheral blood mononuclear cells (PBMCs). The present study aimed to investigate the expression of miR-155 in PBMCs, serum and liver tissues of GT4-HCV-infected patients. miR-155 expression was assessed using reverse transcription-quantitative polymerase chain reaction in GT4-HCV-infected PBMCs, serum and liver tissues, as well as GT2- and GT4-infected Huh7 cells, and compared to the healthy controls. There was no difference in miR-155 expression observed between naïve GT4-HCV patients and healthy controls in the PBMCs and serum. In HCV-infected liver tissues, however, a significant downregulation was observed. The unique miR-155 expression pattern during GT4 infection was confirmed in the infected Huh7 cell lines when compared to GT2 infection. Clinical data showed a positive correlation between liver transaminases and serum miR-155 expression. In addition, serum miR-155 expression was significantly lower in naïve non-responders (NRs) than naïve sustained virological responders (SVRs), and in post-treatment NRs compared to post-treatment SVRs. In conclusion, miR-155 was not only proven to be a genotype-specific microRNA that is not induced during GT4-HCV infection, but also a good prognostic factor and predictor of response to treatment enabling a non-invasive differentiation between NRs and SVRs during GT4-HCV infection.
Keywords: hepatitis C virus, genotype 4, microRNA-155, treatment response
Introduction
Abnormal expression of numerous microRNAs (miRNAs) has been associated with disease development and progression (1). microRNA-155 (miR-155) has been shown to play a key role in cancer, inflammation and immune response to infections (2,3). miR-155 was reported to regulate the immune system and to be regulated by it (2,4–11). Increasing evidence indicates that upregulation of miR-155 is a consistent feature of the inflammatory response (5), in which several Toll-like receptor (TLR) ligands, as well as inflammatory cytokines produced by the TLR signaling pathway, were reported to induce miR-155 expression (5–11). miR-155 in turn regulates the host immune system by targeting suppressor of cytokine signaling-1, a negative regulator of the JAK/STAT pathway, rendering it a positive regulator of the innate immune system potentiating the antiviral effects of interferon (IFN) (2,4). Due to its essential role in the immune system, a number of studies began to investigate the role of miR-155 in viral infections. Several viral infections were associated with miR-155 upregulation, such as vesicular stomatitis virus (VSV) infection within murine macrophages (2) and Epstein-Barr virus (EBV) infection within primary human B-cells (12).
Likewise, other studies have investigated miR-155 expression during hepatits C virus (HCV) infection, and a positive correlation was found between the posttreatment persistence of HCV RNA in the serum and peripheral blood mononuclear cells (PBMCs) of infected patients and the expression of the miR-155 precursor (BIC) in PBMCs (13). Another study has shown that HCV replication was positively correlated to the increased expression of mature miR-155 in PBMCs of HCV-infected patients (14). Furthermore, miR-155 was upregulated in liver tissues, serum and PBMCs of genotypes 1, 2 and 3 HCV-infected patients (3,10).
However, in a previous study, miR-155 showed no difference in expression in PBMCs of genotype 4 (GT4)-infected patients compared to the healthy controls (15). This suggested that miR-155 may show a unique pattern of expression during GT4-HCV infection. Given this discrepancy in miR-155 expression in GT4-HCV-infected PBMCs versus other genotypes and the fact that >90% of HCV-infected Egyptian patients were found to harbor this ‘difficult-to-treat’ GT4 genome (16), the present study aimed to evaluate miR-155 expression in PBMCs, serum and liver tissues of chronic GT4-HCV-infected patients and to correlate this to the patient treatment response.
Patients and methods
Study subjects
For all the patients, the HCV infection was diagnosed by the presence of anti-HCV antibodies and HCV RNA in the serum. All the patients were negative for the hepatitis B surface antigen. The patients were grouped into naïve chronic HCV (cHCV) patients who did not receive antiviral therapy (naïve, n=15), naïve patients who started IFN-based therapy following sample collection and were later found to be either sustained virological responders (SVRs) or non-responders (NRs) (naïve SVRs, n=9; naïve NRs n=10, respectively) and end of treatment-patients (EOT), whose samples were collected following completion of the treatment course and were in turn grouped as SVRs (EOT SVRs, n=10) or NRs (EOT NRs, n=10). All the naïve patients were candidates for pegylated-interferon/ribavirin (PEG-IFN/RBV) therapy. The samples from posttreatment NRs and SVRs were obtained after 12 and 72 weeks of PEG-IFN/RBV therapy, respectively. The patients exhibited an age range of 25–55 years. The healthy controls were volunteers with a similar age range as the patients. The patients and controls included in the present study provided their written informed consent. All the experiments were performed in compliance with the guidelines of the Institutional Review Board of Kasr El Aini Medical School in Cairo University (Cairo, Egypt) and in accordance with the World Medical Association Declaration of Helsinki.
Sample handling
From each patient and healthy control, 8 ml peripheral venous blood was withdrawn into tubes containing the anticoagulant, EDTA, for isolation of PBMCs. All the samples were processed within a few hours after collection.
In addition, 2 ml peripheral venous blood samples were collected from the patients and healthy controls for serum separation. Liver tissue samples were withdrawn from treatment-naïve cHCV patients by fine-needle aspiration and from healthy donors during the liver transplantation procedure, and were directly transferred to the laboratory for cryopreservation.
Serum separation and isolation of PBMCs from the total blood
Blood samples intended for serum separation were centrifuged at 1,500 x g for 10 min. The upper serum phase was collected and immediately frozen at −80°C until use. PBMCs were isolated from total blood using the Ficoll (Axis-Shield PoC AS, Oslo, Norway) density gradient centrifugation method as previously described (15).
Genotyping
Genotyping was performed at the National Cancer Institute. Briefly, the 5′ untranslated region (UTR) and the core region of the HCV RNA extracted from the serum were reverse transcribed into a biotinylated complimentary DNA (cDNA) product using the HCV Amplification 2.0 kit (LiPA; Bayer HealthCare, Tarrytown, NY, USA). This product was processed using the VERSANT HCV Genotype 2.0 assay (LiPA; Bayer HealthCare, Eragny, France), according to the manufacturer's instructions (17).
In vitro transcription of JFH1 and ED43/JFH1 full-length genome
The pJFH plasmid harboring the genotype 2a genome (kindly provided by Professor T. Wakita) (18) and the intergenotypic recombinant pED435′UTR-NS2/JFH1T827A, T977S harboring the genotype 4a genome (kindly provided by Professor J. Bukh) (19) were linearized using the XbaI restriction enzyme (Thermo Scientific, Waltham, MA, USA) and purified using the phenol-chloroform technique. The full length viral RNA was in vitro transcribed using the MEGAscript T7 polymerase kit (Ambion, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions.
Transfection of the transcribed viral genome into Huh7 cell lines
The transcribed viral genome was delivered into hepatoma Huh7 cell lines using lipofection (SuperFect; Qiagen, Hilden, Germany) according to the manufacturer's instructions. The supernatant containing the released HCV particles was collected at day 3 post-transfection, filtered through 0.45-µm pore size syringe filters and stored at −80°C for further use.
Cell culture and infection of Huh7 cells
Naïve Huh7 cells were seeded in Dulbecco's modified Eagle's medium (Invitrogen, Karlsruhe, Germany) supplemented with L-glutamine, non-essential amino acids, 1% penicillin/streptomycin and 10% fetal bovine serum at 37°C with 5.0% CO2 in a 24-well plate at a density of 200,000 cells/well. After 24 h, filtered HCV-containing culture supernatants were added to the Huh7 cells and incubated for 24 h.
RNA processing
Total RNA was extracted from serum, PBMCs and liver tissues using the Biozol extraction reagent (Bioer Technology Co., Ltd., Hangzhou, China) according to the manufacturer's instructions as first described by Chomczynski and Sacchi (20). miR-155 and the reference miRNA, RNU6B, were reverse transcribed into single-stranded cDNA using specific stem loop primers and the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA) following manufacturer's instructions.
Quantitative polymerase chain reaction (qPCR)
The expression of miR-155 and RNU6B was quantified using qPCR (StepOne Real-Time PCR; Applied Biosystems) using the TaqMan MicroRNA assay (Applied Biosystems). For the Huh7 cell lines, PBMCs and tissue samples, the amount of miR-155 was calculated relative to the amount of RNU6B miRNA present in the same sample.
Statistical analysis
miRNA expression in PBMCs, liver tissues and Huh7 cell lines was calculated using the ∆∆CT method [relative quantitation (RQ)=2−∆∆CT]. In serum, miRNA expression was calculated using the ∆CT method (RQ=2−∆CT) as described previously (21). The results are expressed as the mean of the RQ ± standard error of the mean. Data were analyzed using Prism 5 software, version 5.00 (GraphPad Software, San Diego, CA, USA). Unpaired t-test was used to draw comparisons between groups. Pearson test was used for correlation studies. P<0.05 were considered to indicate statistically significant differences.
Results
Expression pattern of miR-155 in serum, PBMCs and liver tissues of naïve GT4-cHCV-infected patients
All the patients were shown to be infected with the HCV GT4 genome. The expression of miR-155 was quantified in PBMCs, serum and liver tissue samples of treatment-naïve cHCV-infected patients and healthy controls using RT-qPCR. In PBMCs, no difference in miR-155 expression was observed between naïve GT4-cHCV patients (n=15) (6.122±3.039) and healthy controls (n=12) (3.983±1.694) (P=0.5715) (Fig. 1A). Similarly in the serum, no difference in miR-155 expression was observed between naïve GT4-cHCV patients (n=15) (1.431±0.2512) and healthy controls (n=12) (1.489±0.2592) (P=0.8747) (Fig. 1B). However, naïve cHCV-infected liver tissues (n=10) (0.3926±0.1183) showed a significant downregulation in miR-155 expression compared to the healthy controls (n=10) (2.286±0.7954) (P=0.0301) (Fig. 1C).
Figure 1.

Expression pattern of microRNA-155 (miR-155) in serum and peripheral blood mononuclear cells (PBMCs) of genotype 4 (GT4)-hepatits C virus (HCV)-infected patients and healthy controls. miR-155 expression was quantified in PBMCs and serum of GT4-HCV-infected patients (n=15) and healthy controls (n=12), and in liver tissue samples of GT4-HCV-infected patients (n=10) and healthy controls (n=10). Expression of miR-155, as represented in relative quantitation (RQ), did not differ between patients and healthy controls (A) at the level of PBMCs (P=0.5715) or (B) at the level of the serum (P=0.8747). (C) In chronic HCV-infected liver tissues, miR-155 expression was significantly downregulated compared to healthy controls (*P=0.0301). Results are expressed as mean of relative quantitation ± standard error of the mean. ns, not significant.
Impact of GT2 and GT4 infection on miR-155 expression in Huh7 cells
miR-155 expression was assessed in the Huh7 cell lines infected with JFH1 (GT2-) (n=4) and ED43/JFH1 (GT4-) HCV (n=4) and was compared to its expression in uninfected Huh7 cells (n=4) to confirm the impact of the HCV genotype on miR-155 expression. GT2-infection induced a significant upregulation in miR-155 expression (44.20±14.20) (P=0.0229), whereas GT4-infection failed to induce miR-155 expression (4.544±3.840) (P=0.3998) when compared to uninfected Huh7 cells (1.059±0.2072). miR-155 expression was found to be significantly upregulated in GT2-infected compared to GT4-infected Huh7 cells (P=0.0358) (Fig. 2).
Figure 2.
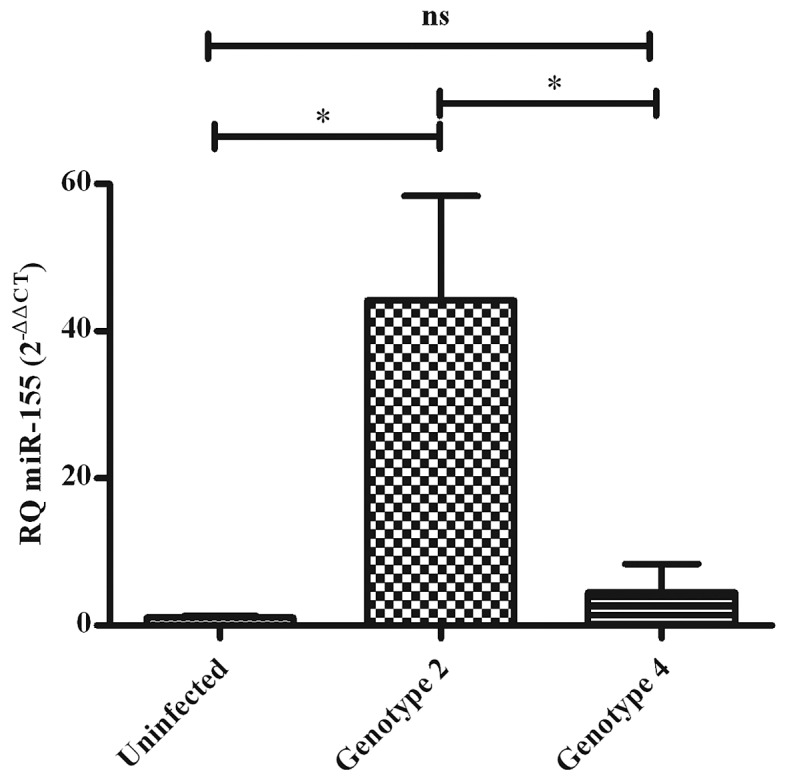
Expression pattern of microRNA-155 (miR-155) in hepatits C virus-infected and uninfected Huh7 cells. miR-155 expression was quantified in uninfected (n=4) and genotype 2 (GT2)- (n=4) or genotype 4 (GT4)- (n=4) infected Huh7 cells. Expression of miR-155, as represented in relative quantitation (RQ), did not differ between GT4-infected and uninfected Huh7 cells (P=0.3998). GT2-infected Huh7 cells showed a significant upregulation of miR-155 expression compared to the uninfected (*P=0.0229) and GT4-infected Huh7 cells (*P=0.0358). Results are expressed as mean of relative quantitation ± standard error of the mean. ns, not significant.
Correlation analysis between miR-155 expression and liver transaminases
miR-155 expression in PBMCs, serum and liver tissues was correlated with the corresponding levels of liver transaminases, alanine transaminase (ALT) and aspartate transaminase (AST), in naïve GT4-HCV-infected patients. No correlation was observed between miR-155 expression in PBMCs (n=15), and the ALT (P=0.6393) and AST (P=0.8337) levels (Fig. 3A). Similarly, in the liver tissues (n=10) there was no correlation to the ALT (P=0.3186) and AST (P=0.2710) levels observed (Fig. 3C). However, a positive correlation was found between the serum miR-155 expression (n=15) and ALT (Pearson r=0.6298) (P=0.0199) and AST (Pearson r=0.6223) (P=0.0132) (Fig. 3B).
Figure 3.
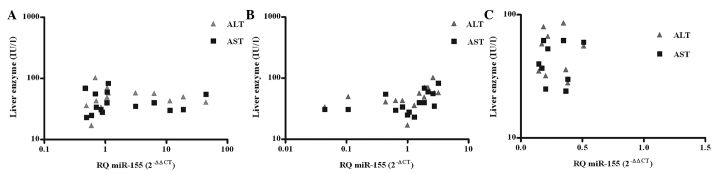
Correlation analysis between microRNA-155 (miR-155) and liver transaminases. miR-155 expression, as represented in relative quantitation (RQ), in the serum and peripheral blood mononuclear cells (PBMCs) (n=15) and liver tissues (n=10) of chronic genotype 4-hepatits C virus treatment-naïve patients was correlated to levels of alanine transaminase (ALT) and aspartate transaminase (AST). (A) In PBMCs, no correlation was observed to ALT (P=0.6393) or AST (P=0.8337), whereas (B) serum miR-155 expression positively correlated with ALT (Pearson r=0.6298) (*P=0.0199) and AST (Pearson r=0.6223) (*P=0.0132) levels. (C) miR-155 expression in liver tissues did not correlate to ALT (P=0.3186) and AST (P=0.2710) levels.
Assessment of the variability of miR-155 expression in the serum of NR and SVR chronic GT4-HCV patients prior and subsequent to IFN therapy
The expression level of serum miR-155 was quantified retrospectively in the NRs and SVRs of GT4-HCV patients prior (naïve NRs and SVRs) and subsequent (EOT NRs and SVRs) to IFN-based therapy and healthy controls using RT-qPCR. Comparing naïve patients and healthy controls, miR-155 showed a significant downregulation in naïve NRs (0.4707±0.08375), compared to the healthy controls (1.489±0.2592) (P=0.0025) and naïve SVRs (1.778±0.5679) (P=0.0280) without exhibiting a significant difference in expression between the naïve SVRs and healthy controls (P=0.6196). Similarly, comparing miR-155 of the EOT patients and healthy controls showed a significant downregulation in EOT NRs (0.3217±0.05878), compared to the healthy controls (1.489±0.2592) (P=0.0007) and EOT SVRs (9.067±3.386) (P=0.0188), however this was accompanied by an upregulation of miR-155 in EOT SVRs compared to healthy controls (P=0.0234) (Fig. 4).
Figure 4.

Expression pattern of microRNA-155 (miR-155) in the serum of chronic genotype 4 (GT4)-hepatits C virus (HCV)-infected patients with different responses prior (naïve) and subsequent to treatment (end of treatment, EOT). The expression of miR-155, as represented in relative quantitation (RQ), was quantified in the serum of naïve and EOT GT4-HCV-infected patients classified into non-responders (NRs) and sustained virological responders (SVRs). Naïve NRs (n=10) showed a significant downregulation in miR-155 expression compared to the healthy controls (n=12) (**P=0.0025) and naïve SVRs (n=9) (*P=0.0280). EOT NRs (n=10) also showed a significant downregulation in miR-155 expression compared to the healthy controls (***P=0.0007) and EOT SVRs (n=10) (*P=0.0188). Naïve SVRs did not show any significant difference in miR-155 expression compared to the healthy controls (P=0.6196), whereas EOT SVRs showed a significant upregulation (*P=0.0234). Results are expressed as mean of relative quantitation ± standard error of the mean. ns, not significant.
Discussion
In recent studies, miR-155 has shown an increase in the serum, PBMCs and liver tissues of genotypes 1, 2 and 3 cHCV-infected patients (3,10). However, it was also reported in a previous study that miR-155 expression did not vary in the PBMCs of GT4-cHCV patients compared to healthy controls (15), which triggered an inquiry to investigate the genotype-dependent expression of the immunoregulatory miR-155. Therefore, the present study aimed to further study miR-155 expression in a wide variety of human samples, including PBMCs, serum and liver tissues of GT4-HCV-infected patients.
The study was initiated by screening for miR-155 in PBMCs of GT4-HCV patients to increase the pool of investigated samples, in which miR-155 expression did not vary between infected patients and healthy controls (Fig. 1A), which confirms the previous data (15). This pattern of equal miR-155 expression in the patients and controls was also obtained by screening for miR-155 expression in the serum of GT4-infected patients (Fig. 1B). These findings contradict the reported upregulation of miR-155 expression in viral infections, such as VSV infection within murine macrophages (2), EBV infection within primary human B-cells (12) and more notably, genotypes 1, 2 and 3 HCV infection within the serum and PBMCs (10). However, by screening for miR-155 in naïve GT4-HCV-infected liver tissues, it was found to be significantly downregulated compared to healthy tissues (Fig. 1C), which opposes the previous finding, in which miR-155 was upregulated in genotypes 1, 2 and 3 HCV-infected liver tissues (3). This repressed induction of miR-155 that is attributed to GT4-HCV infection was suggested to be due to interference of this genotype in the JAK/STAT pathway activation (15).
In order to confirm that the repressed induction of miR-155 in GT4-infected patients is attributed to genotype specificity; miR-155 expression was assessed following infection of the Huh7 cell lines with the JFH1 (GT2) and ED43/JFH1 (GT4) viral genomes and compared to naïve Huh7 cells. To the best of our knowledge, for the first time, GT4 infection was not shown to induce miR-155 in Huh7 cells, whereas GT2 infection induced miR-155 expression in the cell lines (Fig. 2). The latter is in concordance with previous data reporting an induction of miR-155 upon infection of Huh7 cells with the JFH1 genome (3).
The observed miR-155 expression in PBMCs, serum and liver tissues was then correlated to the liver enzymes as an indication for hepatic inflammation (22). No correlation was found in PBMCs (Fig. 3A) and liver tissues (Fig. 3C), whereas serum miR-155 expression showed a positive correlation with the ALT and AST levels (Fig. 3B). This correlation may highlight the role of miR-155 in hepatic inflammation during GT4-cHCV infection.
All the previously mentioned results were for naïve patients or cell lines. However, it was also important to examine the role of miR-155 as a non-invasive prognostic marker in GT4-HCV patients, particularly as this has never been investigated previously. Therefore, serum miR-155 expression was investigated in association with different treatment responses of GT4-HCV patients. Notably, naïve SVRs showed a significant upregulation of miR-155 compared to naïve NRs (Fig. 4). This data may suggest miR-155 as a good predictor of response to treatment, with the ability to distinguish between SVR and NR GT4-HCV patients as early as week zero, which is prior to the start of the therapy. Posttreatment SVRs and NRs showed a similar pattern to naïve SVRs and NRs, with a downregulation in miR-155 expression in EOT NRs compared to EOT SVRs and healthy controls, suggesting that miR-155 may be a good prognostic marker in GT4-cHCV infection. This finding contradicts miR-155 expression pattern in genotypes 1, 2 and 3 SVRs, where miR-155 was found to decrease in the serum (3) and PBMCs (10) posttreatment following viral clearance.
In conclusion, miR-155 showed a unique expression pattern in GT4-cHCV-infected PBMCs, serum and liver tissues confirming that the repressed induction of miR-155 is genotype-specific (10). It was also positively correlated to the ALT and AST levels. Finally, miR-155 was downregulated in the serum of NRs compared to SVRs pre- and posttreatment, rendering it a promising predictor of response and a prognostic marker specific for GT4-HCV infection.
Acknowledgements
The authors would like to acknowledge Professor Jens Bukh (Copenhagen University Hospital, Hvidovre, Denmark) and Professor Takaji Wakita (Department of Virology II, National Institute of Infectious Diseases, Tokyo, Japan) for providing the HCV vectors.
References
- 1.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang P, Hou J, Lin L, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, Sun S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 4.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Huang X, Cui H, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 7.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haasch D, Chen YW, Reilly RM, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 9.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 10.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Hooks JJ, Redmond TM. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem Biophys Res Commun. 2010;402:390–395. doi: 10.1016/j.bbrc.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidorkiewicz M, Grek M, Jozwiak B, Majda-Stanislawska E, Piekarska A, Bartkowiak J. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from Hepatitis C patients after antiviral treatment. Acta Virol. 2010;54:75–78. doi: 10.4149/av_2010_01_75. [DOI] [PubMed] [Google Scholar]
- 14.Grek M, Piekarska A, Bartkowiak J, et al. Coordinated increase of miRNA-155 and miRNA-196b expression correlates with the detection of the antigenomic strand of hepatitis C virus in peripheral blood mononuclear cells. Int J Mol Med. 2011;28:875–880. doi: 10.3892/ijmm.2011.748. [DOI] [PubMed] [Google Scholar]
- 15.El-Ekiaby N, Hamdi N, Negm M, Ahmed R, Zekri AR, Esmat G, Abdelaziz AI. Repressed induction of interferon-related microRNAs miR-146a and miR-155 in peripheral blood mononuclear cells infected with HCV genotype 4. FEBS Open Bio. 2012;2:179–186. doi: 10.1016/j.fob.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerra J, Garenne M, Mohamed MK, Fontanet A. HCV burden of infection in Egypt: results from a nationwide survey. J Viral Hepat. 2012;19:560–567. doi: 10.1111/j.1365-2893.2011.01576.x. [DOI] [PubMed] [Google Scholar]
- 17.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 19.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1–6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 22.de Bruijne J, Buster EH, Gelderblom HC, et al. Treatment of chronic hepatitis C virus infection - Dutch national guidelines. Neth J Med. 2008;66:311–322. [PubMed] [Google Scholar]


