Abstract
Background
Adenoviral infections cause morbidity and mortality in blood and marrow transplantation and pediatric oncology patients. Cidofovir is active against adenovirus, but must be used judiciously because of its nephrotoxicity and unclear indications. Therefore, before introducing cidofovir use during an adenoviral outbreak, we developed a clinical algorithm to distinguish low risk patients from those who merited cidofovir therapy because of significant adenoviral disease and high risk for death.
Objective
This study was conducted to determine whether the algorithm accurately predicted severe adenovirus disease and whether selective cidofovir treatment was beneficial.
Study Design
A retrospective analysis of a pediatric oncology/ blood and marrow transplantation cohort prealgorithm and postalgorithm implementation was performed.
Results
Twenty patients with adenovirus infection were identified (14 high risk and 6 low risk). All low-risk patients cleared their infections without treatment. Before algorithm implementation, all untreated high-risk patients died, 4 out of 5 (80%), from adenoviral infection. In contrast, cidofovir reduced adenovirus-related mortality in the high-risk group postalgorithm implementation (9 patients treated, 1 patient died; RR 0.14, P<0.05) and all treated high-risk patients cleared their virus.
Conclusions
The clinical algorithm accurately identified patients at high risk for severe fatal adenoviral disease who would benefit from selective use of cidofovir.
Keywords: adenovirus algorithm, hematopoietic stem cell transplant, cidofovir, hemolytic-uremic syndrome
Adenoviral infections present a threat to immunocompromised blood and marrow transplant (BMT) recipients and oncology patients.1–4 Pediatric and adult case series have suggested that the mortality of disseminated, untreated, adenoviral disease after BMT approaches 60%2,4,5 and fatalities have also been documented in other oncology patients.6,7 No single factor has identifed patients likely to die from adenovirus, although studies have associated allogeneic BMT, T cell depletion, acute or chronic graft-versus-host disease, intensive preparative regimens, young age, adenovirus seropositive donor, solid organ transplantation, positive blood polymerase chain reaction (PCR), and high viral load in the plasma with increased risk of morbidity and/or mortality from adenovirus. 1,4,8–10 Until recently, the lack of mortality predictors was of little significance due to an absence of treatment strategies,4,11–14 however, cidofovir has emerged as a therapy for adenoviral disease with reported clinical efficacy approaching 98%.15–20 Unfortunately, renal toxicity, with rates of up to 50%, has limited its use.1,16,21,22 Furthermore, adenoviral clearance without cidofovir treatment has been observed in some pediatric BMT patients, although adenovirus-related deaths did occur in the cohort.23 Taken together, the published experience suggests that cidofovir may be of value for the treatment of adenoviral disease in certain pediatric oncology and BMT patients. Given the potential toxicity of this therapy, it is important to identify the cohort at highest risk for adenoviral-related mortality, who would most benefit from cidofovir treatment. As significant is delineating patients less likely to suffer adenoviral-related mortality, in whom potentially toxic therapy can be deferred. We developed a clinical algorithm to identify those patients at high risk of fatal adenoviral infection. The objective of this study was to validate the clinical algorithm in a historical cohort. The risk stratification algorithm was developed to classify patients most likely to benefit from the treatment. We hypothesized that mortality would be high in untreated high-risk patients and low in untreated low-risk patients. To test this hypothesis, outcomes were assessed in a retrospective cohort with adenoviral infections during a 2-year period before and after algorithm implementation.
METHODS
Clinical Algorithm Adopted During the Adenoviral Outbreak
Between 2003 and 2004, an unusually high incidence of adenoviral disease in the pediatric oncology unit prompted the development and implementation in December 2003 of a clinical algorithm to identify patients at high risk for fatal therapy (Table 1). Published data on risk factors for adenoviral mortality and current definitions of adenoviral infection and disease were used in algorithm generation. 2,10,17 At our institution, cidofovir had not previously been used for the treatment of adenoviral disease. The presence of adenovirus was identified either by viral surveillance cultures (nasopharynx, urine, and stool) performed weekly on pediatric oncology unit patients or by site-directed workup prompted by symptoms including: fever, cough, rhinorrhea, sinusitis, pneumonia, hematuria, hematochezia, hematemesis, and elevated liver function tests.
TABLE 1.
Algorithm Classification of HRHI and LRHI Patients and Guide to Observation or Therapy With CDV
| High-risk Host? | High-risk Infection? | Algorithm Classification | Algorithm Recommendation |
|---|---|---|---|
| Yes* | Yes† | HRHI | Treat‡ |
| Yes | No | LRHI | Observe |
| No | Yes | LRHI | Observe |
| No | No | LRHI | Observe |
High-risk host describes patients with: (1) blood and marrow transplantation (allogeneic, <180 d posttransplant; autologous, or <60 d posttransplant), (2) acute or chronic graft-versus-host disease, or (3) patients receiving immunosuppressive regimens with functional immunodeficiencies (eg, acute myeloid leukemia therapy, acute lymphocytic leukemia induction, severe aplastic anemia, rituximab, or protracted leukopenia).
High-risk infection describes patients with: invasive (symptoms of disease from a site with PCR/biopsy/culture evidence of adenovirus), disseminated adenovirus more than 2 sites positive for adenovirus (excluding urine and stool in children in diapers), or blood PCR positive.
CDV dose is 5mg/kg/dose IV weekly unless there is evidence of renal dysfunction (1mg/kg/dose IV 3 times weekly). Therapy should be continued until 3 negative results have been obtained from all obtainable sites of disease or until the patient is no long a high-risk host. Hydration should include 20 mL/kg normal saline pre-CDV and post-CDV infusion (or equivalent IV fluids). Probenecid should be administered as renal protection at a dose of 1 to 1.25 mg/m2/ dose 3 hours before and 1 and 8 hours at the end of CDV infusion to decrease nephrotoxicity (round to nearest 250 mg).
CDV indicates cidofovir; HRHI, high-risk host and infectious disease; IV, intravenously; LRHI, low-risk host and/or infectious disease; PCR, polymerase chain reaction.
Patients were deemed high risk for adenoviral mortality if (1) host features suggested severe immune compromise and (2) infectious disease features suggested disseminated or invasive adenoviral infection. These patients were termed high-risk host and infectious disease (HRHI). All other patients were designated low-risk host and/or infectious disease (LRHI) because either the host risk was low or the adenoviral disease risk was low, or both (Table 1). High-risk host features of severe immunocompromise included: <180 days after allogeneic BMT, patients with acute or chronic graft-versus-host disease, <60 days after autologous BMT, oncology patients receiving immunosuppressive regimens causing functional immunodeficiency (eg, alemtuzumab or rituximab), or protracted leukopenia. High-risk adenoviral disease was defined by evidence of dissemination or invasion. Invasive disease was defined as severe signs or symptoms from a site, with concordant culture, histology, or PCR. Qualifying signs and symptoms included sinusitis, lower respiratory tract disease, hematuria, hematochezia, hematemesis, and biopsy-proven hepatitis. Liver enzyme elevation alone did not constitute invasive adenoviral disease, as multiple confounders can elevate liver enzymes in this population. Fever was not sufficient to define invasive disease but may have prompted investigation for infection. Disseminated disease was defined as more than 2 sites positive for adenovirus (counting urine and stool as a single site in young children due to frequent cross-contamination) or positive PCR from blood. Patients were classified as LRHI or HRHI at the time of the first positive test for adenoviral infection. All patients identified as HRHI after the introduction of this algorithm received cidofovir. LRHI patients were monitored without cidofovir therapy.
Chart Review to Validate the Clinical Algorithm in Patients With Adenoviral Infections
To validate the ability of the algorithm to anticipate which patients would do poorly, an institutional review board approved retrospective medical chart review of clinical and microbiologic data on adenoviral infection cases occurring in pediatric oncology patients before algorithm implementation (July 2001 to November 2003) and postalgorithm implementation (December 2003 to July 2004) was conducted. Data were collected from electronic and paper hospital records. Patient data were assessed for at least 1 year from the date of the last identified infection or until death (1 to 3 y). Patient risk classification was assigned at the first positive adenoviral test and determined without knowledge of outcome in the retrospective cohort. No patients in the prealgorithm group received cidofovir for their adenoviral disease. All HRHI patients in the postalgorithm groups were treated with cidofovir.
Adenovirus Detection and Serotyping
Adenoviruses were detected from blood, urine, and relevant tissue by PCR using previously published methods, obtained weekly for postalgorithm patients.24 The assay detects at least 20 adenovirus serotypes and strains from all 5 adenovirus serogroups (A to E) with an analytical sensitivity of 500 copies/mL. Adenoviruses were detected from other sites (respiratory, conjunctiva, stool, and urine) by shell vial and tube culture with monoclonal antibody confirmation. Serotyping of randomly selected isolates was performed by amplification and sequencing of a 301 bp region of the hexon gene.25 After diagnosis, during the outbreak period, PCR of blood and urine samples were performed on a weekly basis.
Definition of Outcome
Adenovirus disease clearance was defined as symptom resolution, with lymphoid function reconstitution and negative virologic test results from all previously positive sites. Recrudescence of occult adenoviral infection was defined as clearance followed by renewed detection. Overall and adenovirus-specific mortality were determined in the cohort. Adenovirus-specific mortality was defined as (1) evidence of only adenoviral disease at autopsy, (2) listing of adenoviral disease in the clinical summary without other possible cause of death identified, or (3) microbiologic evidence of adenoviral disease at the time of death with no other identified cause. Adenovirus-related mortality was ascribed to any death with evidence of active adenoviral disease, regardless of the identification of other primary cause of death. When data on a site were unavailable, the site was presumed negative.
Cidofovir Regimen
Patients identified as HRHI after the introduction of the algorithm were treated once weekly with intravenous cidofovir (5 mg/kg/dose); patients with severe renal dysfunction (n=2) received a modified regimen (Table 1). All cidofovir doses were administered with probenecid and hydration to decrease nephrotoxicity (Table 1). Cidofovir was administered until patients either no longer met high-risk host criteria or had 3 negative tests separated by at least 1 week from all available prior sites of adenoviral disease. Change in renal function was calculated as the difference/change between baseline creatinine (1mo before identification of adenoviral infection) and average creatinine (assessed at weekly intervals) at completion of cidofovir therapy. Renal failure was defined as the need for dialysis. Supportive care for patients routinely included intravenous immunoglobulin; oral immunoglobulin was given to one infant with severe adenoviral gastrointestinal disease.
Statistical Analyses
We evaluated 3 groups: HRHI treated, HRHI untreated (prealgorithm), and LRHI (prealgorithm and postalgorithm). The primary outcome was adenoviral-related mortality. An odds ratio (OR) was obtained by univariate logistic regression to assess the relationship of risk categorization with mortality in the absence of cidofovir treatment. Univariate logistic regression was performed to assess the relationship of our clinical risk categorization with the following objective predictor variables: age in years, prior BMT, absolute lymphocyte count (ALC), absolute neutrophil count (ANC), and days post-BMT. Because ALC and ANC were significantly associated with risk categorization, the ability of these objective variables to replace our clinical risk categorization was tested using stepwise multivariate logistic regression for the primary outcome of adenoviral-related mortality, using a significance level of 0.2 for removal of a factor from the model. Overall survival curves were generated by the method of Kaplan-Meier. Stata Version 9 (College Station, TX) for Windows was used for statistical analysis.
RESULTS
Assessment of the Adenoviral Algorithm
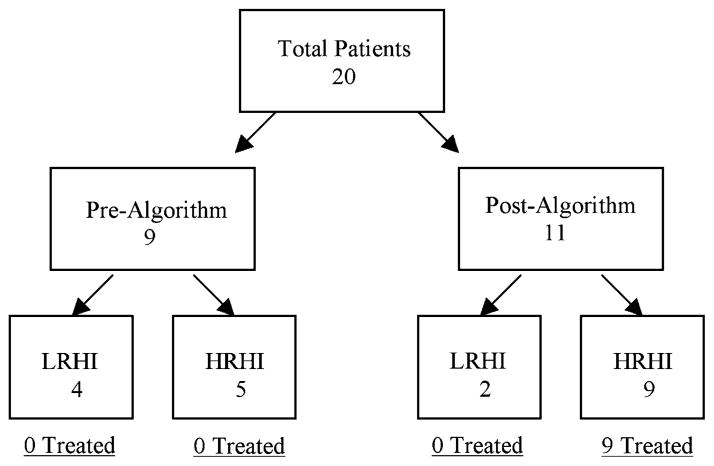
Twenty pediatric oncology and BMT patients with adenoviral infection were identified (Fig. 1). Using the algorithm, 5 HRHI patients were identified preimplementation (Fig. 1; Table 2, patients A to E). These untreated patients died primarily of adenoviral-related disease (100% overall mortality; 80% [4 out of 5] due to adenovirus). In contrast, all 6 LRHI patients (Table 2 patients O to T) initially cleared their infection without treatment. Logistic regression indicated that the clinical algorithm correctly identified patients at high risk of adenoviral-specific mortality (OR=20 in the HRHI-untreated vs. LRHI groups, P=0.056).
FIGURE 1.
Branching diagram of patients identified prealgorithm and postalgorithm.
TABLE 2.
Clinical Data, Risk Classification, and Treatment of Study Subjects
| ID | Number | Clinical Setting at Diagnosis of Adenovirus | HR Host? | Diagnostic Test Results | Evidence of Invasive Disease at Diagnosis | HR Infection? | HI Risk | Cidofovir |
|---|---|---|---|---|---|---|---|---|
| A | 14 | Allo BMT d121 +GVHD | Yes | Blood PCR+* Stool cx+ † Urine PCR+ | Respiratory failure, GI bleed, Sepsis | Yes | High | No |
| B | 16 | Allo BMT d88, +GVHD | Yes | Urine PCR+ | Hematuria, oxygen requirement | Yes | High | No |
| C | 7 | Allo BMT d73 +GVHD | Yes | Urine cx+ | DIC, renal failure, septic shock | Yes | High | No |
| D | 19 | Wilm tumor, renal transplant, +immunosuppression‡ | Yes | NP cx+ BAL cx+ | Hypoxia, ventilated | Yes | High | No |
| E | 20 | Allo BMT d210, +GVHD | Yes | Liver biopsy PCR+ | Elevated LFTs, respiratory failure | Yes | High | No |
| F | 1 | Allo BMT d6 | Yes | Stool cx+ | Bloody diarrhea | Yes | High | Yes |
| G | 3 | Auto BMT d5 | Yes | Stool cx+PCR blood+ | Bloody diarrhea | Yes | High | Yes |
| H | 4 | AML d4 chemo | Yes | Blood PCR+ | Viremia§ | Yes | High | Yes |
| I | 6 | Allo BMT d49 +GVHD | Yes | Pleural fluid cx+ | Respiratory failure, pneumonia | Yes | High | Yes |
| J | 8 | Auto BMT d4 | Yes | Blood PCR+ stool cx+ urine PCR+, Np cx − | Viremia§ | Yes | High | Yes |
| K | 9 | Allo BMT d47 − GVHDJ | Yes | NP cx +, rising antibody | Sinusitis | Yes | High | Yes |
| L | 10 | Allo BMT | Yes | Urine cx+ | Hematuria/ hematochezia, respiratory distress | Yes | High | Yes |
| M | 11 | Auto BMT d1 | Yes | Throat cx+ urine PCR/cx+ stool cx+ blood PCR+ | GI bleed/ulcer, shock | Yes | High | Yes |
| N | 13 | AML chemo d8 | Yes | Urine cx+ stool cx+ | Bloody diarrhea | Yes | High | Yes |
| O | 2§ | Allo BMT d178, +GVHD | Yes | Urine PCR+ | None (U/A −, blood −, BAL −) | No | Low | No |
| P | 12 | Nbl, pretreatment | No | NP cx+ | Pneumonia | Yes | Low | No |
| Q | 5 | Allo BMT d2 | Yes | NP cx+ | None | No | Low | No |
| R | 15 | Allo BMT d100, − GVHD | No | Stool cx+ | Non-bloody diarrhea | No | Low | No |
| S | 17 | Allo BMT d255, +GVHD | Yes | Stool cx+ | None | No | Low | No |
| T | 18 | AML chemo d2 | Yes | Throat cx+ | None | No | Low | No |
A-E: HRHI patients were identified prealgorithm era.
F-N: HRHI patients were identified postalgorithm era.
O-P: LRHI patients were identified postalgorithm era.
Q-T: LRHI patients were identified prealgorithm era.
Positive.
Culture.
Immunosuppression included mycophenolate mofeitil, sirolimus, and prednisone.
Viremia was detected during a workup for fever during the time of the outbreak.
No evidence of GVHD.
Allo indicates allogeneic; AML, acute myelogenous leukemia; Auto, autologous; BAL, bronchoalveolar lavage; BMT, blood and marrow transplant; chemo, chemotherapy; d, day after therapy (eg, post-BMT); DIC, disseminated intravascular coagulopathy; GI, gastrointestinal; GVHD, graft-versus-host disease; HRHI, high-risk host and infectious disease; LFTs, liver function tests; LRHI, low-risk host and/or infectious disease; Nbl, neuroblastoma; NP, nasopharynx; PCR, polymerase chain reaction; U/A, urinalysis.
Evaluation of Treatment Efficacy and Toxicity in HRHI Patients
To evaluate the potential benefit of cidofovir, we compared the outcomes of HRHI patients before (Fig. 1, n=5; Table 2, patients A to E) and after (Fig. 1, n=9; Table 2, patients F to N) algorithm implementation. Notably, the treated HRHI patients did not differ significantly from the historical untreated HRHI control patients (Table 3). Compared with 80% adenoviral-specific mortality in untreated HRHI patients, only 11% (1 out of 9) of cidofovir-treated HRHI patients died of adenoviral-related disease (Table 4). Overall survival was superior with cidofovir in the HRHI patients (Fig. 2). Cidofovir treatment was associated with significantly lower adenovirus-related mortality in HRHI patients [relative risk (RR) 0.14, P<0.05] (Table 4). Of the 9 treated HRHI patients, all initially cleared their adenovirus. One died of disseminated fungal disease shortly after adenovirus was detected in bronchoalveolar lavage fluid (patient 1); however, no evidence of adenovirus infection was found postmortem.
TABLE 3.
Patient Baseline Characteristics by Risk Designation*
| Variable | No. Patients
|
||
|---|---|---|---|
| Low Risk
|
High Risk
|
||
| Untreated | Untreated | Treated | |
| Underlying disease | |||
| Acute leukemia | 3 | 2 | 5 |
| Chronic myelogenous leukemia | 1 | ||
| Solid tumor | 2 | 3 | |
| Other | 1 | 2 | 1 |
| Clinical Setting | |||
| Autologous | 3 | ||
| Allogeneic | 4 | 4 | 4 |
| Chemotherapy | 1 | 2 | |
| Solid organ transplant | 1 | ||
| Other | 1 | ||
| Period post-BMT | |||
| <30 d | 1 | 5 | |
| 30-90 d | 2 | 2 | |
| >90 d | 3 | 2 | |
| Period postchemotherapy | |||
| <30 d | 1 | 2 | |
| Period postkidney transplant | |||
| >90 d | 1 | ||
| Other | 1 | ||
| Median age, y (range) | 5 (2-17) | 14 (5-17) | 3 (0.2-18) |
| Absolute lymphocyte count median (x103)w | 1.2 (0.3-1.9) | 0.5 (0.1-1.9) | 0.2 (0-1.6) |
| Absolute neutrophil count median (x103)z | 6.8 (1.2-21.1) | 2.1 (0.7-24.1) | 0 (0-9.7) |
| GVHD (percentage of total number) | 2 (33) | 4 (80) | 2 (22) |
| CMV status (patient) | |||
| Positive | 2 | 1 | |
| Negative | 3 | 5 | 8 |
| Unknown | 1 | ||
P=Not significant for all values except for those below.
OR=0.04, P<0.05.
OR=0.16, P=0.06.
BMT indicates blood and marrow transplant; CMV, cytomegalovirus; GVHD, graft-versus-host disease
TABLE 4.
Outcomes of Adenoviral Infection in Pediatric Oncology and Blood and Marrow Transplantation Patients Based on Clinical Algorithm Risk Stratification and Treatment
| Risk N (%) | Cidofovir Therapy | n | Adenovirus Clearance* N (%) | Adenoviral-related Mortality† N (%)/RR | Overall Mortality N (%) |
|---|---|---|---|---|---|
| High 14 (70) | Treated | 9 | 8 (89) | 1 (11)/0.14‡ | 3 (33) |
| Untreated | 5 | 1 (20) | 4 (80)/1§ | 5 (100) | |
| Low 6 (30) | Treated | 0 | NA | NA | NA |
| Untreated | 6 | 6 (100) 1 recrudescence | 1 (17) | 2 (33) |
Adenovirus clearance was defined as symptom resolution with immune reconstitution and negative microbiologic test results from all previously positive sites.
Adenovirus-related mortality was defined as death with evidence of active adenoviral disease, regardless of the identification of other causes of death.
P<0.05.
Patient O (Table 2) cleared his adenoviral disease. He recrudesced 2mo after initial presentation with aggressive disease, met criteria for high-risk host and infectious disease at recrudescence but died of complications before therapy could be instituted. At the time of recrudescence, the patient had had a flare in chronic graft-versus-host disease, a decline in absolute lymphocyte count, and hemolytic-uremic syndrome, all factors that are identified with severe disease
FIGURE 2.
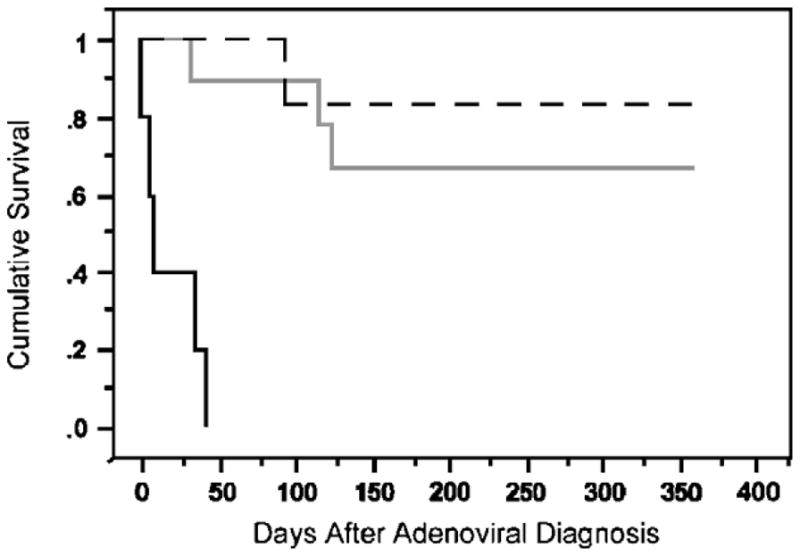
Kaplan-Meier survival curve of high-risk and low-risk patients, stratified according to treatment. Black dashed line indicates untreated LRHI patients; black solid line, untreated HRHI patients; grey solid line, treated HRHI patients.
Surprisingly, treated HRHI patients had less renal dysfunction than untreated HRHI patients. Treated HRHI patients had a median change in creatinine of 0.06 mg/dL (0 to 0.2 mg/dL, SD 0.08), excluding the 2 patients on hemodialysis at baseline. Fewer patients developed renal failure with cidofovir than without: 22% of treated (2 out of 9) compared with 80% of untreated (4 out of 5, P=0.05) and untreated HRHI patients had significantly higher baseline creatinine values than treated HRHI patients (P<0.01). Only 2 of the treated HRHI patients required dose reductions of cidofovir for renal insufficiency and both had multiple coincident renal complications including hemolytic-uremic syndrome (HUS). Patients received a median of 8 doses of cidofovir (3 to 32). Notably, renal dysfunction did not tend to progress with increasing numbers of (or subsequent) cidofovir doses.
Analyses of Risk Factors in HRHI Patients
Although 4 out of 6 LRHI patients were deemed high-risk hosts, because they exhibited low-risk patterns of adenoviral infection, the algorithm identified them as LRHI. Demographic data for HRHI and LRHI were similar with respect to patient age, transplant type, and days post-therapy (Table 3). We explored the relationship of other clinical variables with adenovirus-related mortality to identify potential ways to improve risk stratification. No potentially confounding viral coinfections [cytomegalovirus and other respiratory viruses (influenza, respiratory syncytial virus, parainfluenza, rhinovirus)] were identified in this cohort. Using molecular serotyping, a single, circulating causative adenoviral strain was not detected (data not shown). The median ANC for LRHI patients was 6.8 (1.2 to 26.1) ×109 versus 0.44 (0.0 to 24.1) ×109 neutrophils/L for the HRHI patients (P=0.06, OR=0.16). The median ALC for HRHI was significantly lower than that of the LRHI group 0.24 (0.35 to 1.87) ×109 versus 1.150 (0 to 1.90) ×109 lymphocytes/L (OR=0.04, P<0.05, Table 3). To address the concern that superior outcomes in the treated HRHI group may have resulted from differences in host immunity, we compared the leukocyte counts of the treated and untreated HRHI groups. The median ALC and ANC were higher in the untreated HRHI group versus the treated group (0.54×109 vs. 0.20×109 lymphocytes/L and 0×109 vs. 2.06×109 neutrophils/L).
Four HRHI patients developed HUS and died (A, F, I, and O at recrudescence). The diagnosis of HUS preceded the diagnosis of adenoviral disease (thus before the introduction of cidofovir) in 3 of 4 patients. All but one HRHI patients with HUS received cidofovir.
DISCUSSION
Because of the risk of renal toxicity associated with cidofovir, an algorithm would be helpful to identify those patients at highest risk for severe adenoviral disease who will benefit from cidofovir therapy. Currently, there is no consensus cidofovir treatment strategy. Our data suggest that this algorithm is a multifaceted clinical strategy taking into consideration patient factors, symptoms, and adenovirus diagnostics that may help guide clinical decision making regarding cidofovir treatment.
The algorithm was able to correctly identify patients at highest risk for adenoviral-related mortality as evidenced by the high adenoviral-related mortality (80%) in untreated HRHI as compared with LRHI and the benefit of cidofovir therapy is reflected in lower mortality (11%) in treated HRHI patients. Treatment offered a survival advantage for HRHI patients without increased nephrotoxicity, presumably due to the high rate of adenovirus-associated renal dysfunction in this population. The observation of low mortality in LRHI patients was as important as low mortality in cidofovir-treated HRHI patients. Furthermore, 83% of LRHI patients achieved sustained viral clearance without treatment. Finally, although one LRHI patient recrudesced and died, HRHI criteria were met at recrudescence and were associated with severe disease; however, death occurred before treatment could be initiated. Our findings are similar to those for a prospective trial that tested a high-risk adenoviral disease classification. 26 Our study further explores the addition of patient risk factors, to assist in clinical decision-making, and is one of the first to suggest a benefit in survival with preemptive treatment over historical controls.
A low ANC, ALC, and HUS were poor prognostic factors in our cohort. Although a correlation between lymphopenia and adenoviral disease severity has been reported,23,27 our data demonstrate a statistically significant association with HRHI patients, likely because adenoviral clearance is dependent on effective T-cell immunity.28 Further support of these data is that adoptive immunotherapy and removal of immunosuppression has been associated with clearance of adenoviral disease in 4 BMT patients.29,30 Although ALC was not part of our initial HRHI criteria, we propose that cidofovir should be considered in high-risk hosts with lymphopenia and adenoviral infection even without dissemination or invasion.
All 4 patients with HUS were identified as HRHI and died. Although there are many etiologies of HUS, there may be a link between severe adenoviral disease and HUS,31 and our findings suggest there should be a lower threshold for treatment initiation in the setting of HUS and adenoviral infection. Atypical HUS has been previously associated with diminished surface CD46 receptor expression, 32 which certain adenoviral strains use for attachment and internalization.33 An attractive hypothesis is that CD46 may be down modulated by adenovirus entry into the cell, leading to complement hyperactivation and atypical HUS.
This study is limited by its small size, inherent heterogeneity among the patient cohort, and retrospective design. Because patients with more severe disease are more likely to have received a comprehensive workup, it is conceivable that this led to the capture of sicker HRHI patients prealgorithm than postalgorithm during the outbreak. However, untreated (prealgorithm) HRHI patients were similar to treated HRHI patients in extent of disease and other clinical risk factors, including ALC at diagnosis. Although aggressive surveillance during the outbreak may have identified more LRHI patients, the greater LRHI number in the prealgorithm group does not indicate a strong ascertainment bias. Finally, variability of lymphoid reconstitution among the cohort could have contributed to differences in clearance of adenoviral disease. However, it is unlikely that differences in lymphoid reconstitution account for the observed effect of cidofovir on survival among HRHI patients, who continued to display profound T-cell deficiency and would typically require months to years for immunologic recovery.34
The absence of a consistent surveillance strategy in the prealgorithm patients may have limited the number of patients captured and limits our ability to comment on clearance without treatment. Because our retrospective analysis was limited by the diagnostic work-up completed at the time of infection, we assigned prealgorithm patients the minimal risk possible. Finally, although improvements in supportive care could increase the apparent benefit of cidofovir, there were no other major institutional changes in BMT or chemotherapy protocols that would account for the magnitude of the observed difference in outcome in the cidofovir-treated cohort. In addition, as adenovirus-related mortality was ascribed to any death in the presence of active adenoviral disease, regardless of the identification of other primary cause of death, it is possible that adenovirus might not be directly responsible for all of the cases of adenovirus-related mortality.
These data suggests that the algorithm identifies pediatric oncology/BMT patients at high risk for adenoviral mortality and thereby most likely to benefit from cidofovir treatment. In a larger patient cohort, a prospective study to verify the utility of this algorithm, evaluate the appropriate length of time to continue adenoviral screening, duration of therapy, and benefit of cidofovir in lower risk patients may be of value.
CONCLUSIONS
We have designed a clinical algorithm that identified patients at high risk for severe adenoviral disease likely to benefit from cidofovir therapy and correctly predicted which patients could be managed conservatively with close observation and no treatment. Using this algorithm to select patients for therapy, our data suggests that cidofovir seems to be safe and effective for pediatric oncology and BMT patients. Furthermore, we identified ALC as a significant risk factors for mortality in this population. This clinical algorithm may provide practitioners with a practical tool to distinguish those pediatric oncology and BMT patients with adenoviral disease likely to receive a potential survival benefit from therapy and deserves further evaluation in a larger prospective cohort.
Acknowledgments
The authors thank Michael Forman for his expert performance of molecular serotyping.
Supported by the funds from NIH-NIAID/SSS contract No. 204VC004 and The HIV Prevention Trials Network (HPTN) sponsored by NIAID, NIDA, NIMH, and the Office of AIDS Research of the NIH, DHHS (U01-AI-068613) (AV).
Footnotes
None of the authors have a conflict of interest with this manuscript.
References
- 1.Ljungman P. Treatment of adenovirus infections in the immunocompromised host. Eur J Clin Microbiol Infect Dis. 2004;23:583–588. doi: 10.1007/s10096-004-1165-x. [DOI] [PubMed] [Google Scholar]
- 2.Hale GA, Heslop HE, Krance RA, et al. Adenovirus infection after pediatric bone marrow transplantation. Bone Marrow Transplant. 1999;23:277–282. doi: 10.1038/sj.bmt.1701563. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti S, Collingham KE, Stevens RH, et al. Isolation of viruses from stools in stem cell transplant recipients: a prospective surveillance study. Bone Marrow Transplant. 2000;25:277–282. doi: 10.1038/sj.bmt.1702164. [DOI] [PubMed] [Google Scholar]
- 4.La Rosa AM, Champlin RE, Mirza N, et al. Adenovirus infections in adult recipients of blood and marrow transplants. Clin Infect Dis. 2001;32:871–876. doi: 10.1086/319352. [DOI] [PubMed] [Google Scholar]
- 5.Wang WH, Wang HL. Fulminant adenovirus hepatitis following bone marrow transplantation. A case report and brief review of the literature. Arch Pathol Lab Med. 2003;127:e246–e248. doi: 10.5858/2003-127-e246-FAHFBM. [DOI] [PubMed] [Google Scholar]
- 6.Hough R, Chetwood A, Sinfield R, et al. Fatal adenovirus hepatitis during standard chemotherapy for childhood acute lymphoblastic leukemia. J Pediatr Hematol Oncol. 2005;27:67–72. doi: 10.1097/01.mph.0000153958.95486.6f. [DOI] [PubMed] [Google Scholar]
- 7.Kaur B, Gottardo NG, Keil AD, et al. A rare case of adenoviral fulminant hepatic necrosis after chemotherapy. Pediatr Hematol Oncol. 2002;19:361–371. doi: 10.1080/08880010290057390. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin A, Kingman H, Darville M, et al. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 2000;26:1333–1338. doi: 10.1038/sj.bmt.1702716. [DOI] [PubMed] [Google Scholar]
- 9.Claas EC, Schilham MW, de Brouwer CS, et al. Internally controlled real-time PCR monitoring of adenovirus DNA load in serum or plasma of transplant recipients. J Clin Microbiol. 2005;43:1738–1744. doi: 10.1128/JCM.43.4.1738-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lion T, Baumgartinger R, Watzinger F, et al. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood. 2003;102:1114–1120. doi: 10.1182/blood-2002-07-2152. [DOI] [PubMed] [Google Scholar]
- 11.Abe S, Miyamura K, Oba T, et al. Oral ribavirin for severe adenovirus infection after allogeneic marrow transplantation. Bone Marrow Transplant. 2003;32:1107–1108. doi: 10.1038/sj.bmt.1704276. [DOI] [PubMed] [Google Scholar]
- 12.Arav-Boger R, Echavarria M, Forman M, et al. Clearance of adenoviral hepatitis with ribavirin therapy in a pediatric liver transplant recipient. Pediatr Infect Dis J. 2000;19:1097–1100. doi: 10.1097/00006454-200011000-00016. [DOI] [PubMed] [Google Scholar]
- 13.Gavin PJ, Katz BZ. Intravenous ribavirin treatment for severe adenovirus disease in immunocompromised children. Pediatrics. 2002;110 (1 Pt 1):e9. doi: 10.1542/peds.110.1.e9. [DOI] [PubMed] [Google Scholar]
- 14.Lankester AC, Heemskerk B, Claas EC, et al. Effect of ribavirin on the plasma viral DNA load in patients with disseminating adenovirus infection. Clin Infect Dis. 2004;38:1521–1525. doi: 10.1086/420817. [DOI] [PubMed] [Google Scholar]
- 15.Yusuf U, Hale GA, Carr J, et al. Cidofovir for the treatment of adenoviral infection in pediatric hematopoietic stem cell transplant patients. Transplantation. 2006;81:1398–1404. doi: 10.1097/01.tp.0000209195.95115.8e. [DOI] [PubMed] [Google Scholar]
- 16.Nagafuji K, Aoki K, Henzan H, et al. Cidofovir for treating adenoviral hemorrhagic cystitis in hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2004;34:909–914. doi: 10.1038/sj.bmt.1704682. [DOI] [PubMed] [Google Scholar]
- 17.Legrand F, Berrebi D, Houhou N, et al. Early diagnosis of adenovirus infection and treatment with cidofovir after bone marrow transplantation in children. Bone Marrow Transplant. 2001;27:621–626. doi: 10.1038/sj.bmt.1702820. [DOI] [PubMed] [Google Scholar]
- 18.Muller WJ, Levin MJ, Shin YK, et al. Clinical and in vitro evaluation of cidofovir for treatment of adenovirus infection in pediatric hematopoietic stem cell transplant recipients. Clin Infect Dis. 2005;41:1812–1816. doi: 10.1086/498151. [DOI] [PubMed] [Google Scholar]
- 19.Sivaprakasam P, Carr TF, Coussons M, et al. Improved outcome from invasive adenovirus infection in pediatric patients after hemopoietic stem cell transplantation using intensive clinical surveillance and early intervention. J Pediatr Hematol Oncol. 2007;29:81–85. doi: 10.1097/MPH.0b013e318030875e. [DOI] [PubMed] [Google Scholar]
- 20.Neofytos D, Ojha A, Mookerjee B, et al. Treatment of adenovirus disease in stem cell transplant recipients with cidofovir. Biol Blood Marrow Transplant. 2007;13:74–81. doi: 10.1016/j.bbmt.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 21.Seidemann K, Heim A, Pfister ED, et al. Monitoring of adenovirus infection in pediatric transplant recipients by quantitative PCR: report of six cases and review of the literature. Am J Transplant. 2004;4:2102–2108. doi: 10.1111/j.1600-6143.2004.00631.x. [DOI] [PubMed] [Google Scholar]
- 22.Machado CM, Boas LS, Mendes AV, et al. Low mortality rates related to respiratory virus infections after bone marrow transplantation. Bone Marrow Transplant. 2003;31:695–700. doi: 10.1038/sj.bmt.1703900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls T, Hawrami K, Ushiro-Lumb I, et al. Adenovirus infection after pediatric bone marrow transplantation: is treatment always necessary? Clin Infect Dis. 2005;40:1244–1249. doi: 10.1086/429235. [DOI] [PubMed] [Google Scholar]
- 24.Echavarria MS, Ray SC, Ambinder R, et al. PCR detection of adenovirus in a bone marrow transplant recipient: hemorrhagic cystitis as a presenting manifestation of disseminated disease. J Clin Microbiol. 1999;37:686–689. doi: 10.1128/jcm.37.3.686-689.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allard A, Albinsson B, Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J Clin Microbiol. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson EJ, Guzman-Cottrill JA, Kletzel M, et al. High-risk adenovirus-infected pediatric allogeneic hematopoietic progenitor cell transplant recipients and preemptive cidofovir therapy. Pediatr Transplant. 2008;12:219–227. doi: 10.1111/j.1399-3046.2007.00851.x. [DOI] [PubMed] [Google Scholar]
- 27.van Tol MJ, Claas EC, Heemskerk B, et al. Adenovirus infection in children after allogeneic stem cell transplantation: diagnosis, treatment and immunity. Bone Marrow Transplant. 2005;35 (suppl 1):S73–S76. doi: 10.1038/sj.bmt.1704852. [DOI] [PubMed] [Google Scholar]
- 28.Heemskerk B, Lankester AC, van Vreeswijk T, et al. Immune reconstitution and clearance of human adenovirus viremia in pediatric stem-cell recipients. J Infect Dis. 2005;191:520–530. doi: 10.1086/427513. [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti S, Collingham KE, Fegan CD, et al. Adenovirus infections following haematopoietic cell transplantation: is there a role for adoptive immunotherapy? Bone Marrow Transplant. 2000;26:305–307. doi: 10.1038/sj.bmt.1702508. [DOI] [PubMed] [Google Scholar]
- 30.Kampmann B, Cubitt D, Walls T, et al. Improved outcome for children with disseminated adenoviral infection following allogeneic stem cell transplantation. Br J Haematol. 2005;130:595–603. doi: 10.1111/j.1365-2141.2005.05649.x. [DOI] [PubMed] [Google Scholar]
- 31.Hale GA, Bowman LC, Rochester RJ, et al. Hemolytic uremic syndrome after bone marrow transplantation: clinical characteristics and outcome in children. Biol Blood Marrow Transplant. 2005;11:912–920. doi: 10.1016/j.bbmt.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Goodship TH, Liszewski MK, Kemp EJ, et al. Mutations in CD46, a complement regulatory protein, predispose to atypical HUS. Trends Mol Med. 2004;10:226–231. doi: 10.1016/j.molmed.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Marttila M, Persson D, Gustafsson D, et al. CD46 is a cellular receptor for all species B adenoviruses except types 3 and 7. J Virol. 2005;79:14429–14436. doi: 10.1128/JVI.79.22.14429-14436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams KM, Hakim FT, Gress RE. T cell immune reconstitution following lymphodepletion. Semin Immunol. 2007;19:318–330. doi: 10.1016/j.smim.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]