Abstract
During ventral forebrain development, orthologs of the homeodomain transcription factor Nkx2.1 control patterning of hypothalamus, preoptic region, and ventral telencephalon. However, the relative contributions of Nkx2.1 and Nkx2.4 to prosencephalon development are poorly understood. Therefore, we analyzed functions of the previously uncharacterized nkx2.4-like zgc:171531 as well as of the presumed nkx2.1 orthologs nkx2.1a and nkx2.1b in zebrafish forebrain development. Our results show that zgc:171531 and nkx2.1a display overlapping expression patterns and a high sequence similarity. Together with a high degree of synteny conservation, these findings indicate that both these genes indeed are paralogs of nkx2.4. As a result, we name zgc:171531 now nkx2.4a, and changed the name of nkx2.1a to nkx2.4b, and of nkx2.1b to nkx2.1. In nkx2.1, nkx2.4a, and nkx2.4b triple morpholino knockdown (nkx2TKD) embryos we observed a loss of the rostral part of prosomere 3 and its derivative posterior tubercular and hypothalamic structures. Furthermore, there was a loss of rostral and intermediate hypothalamus, while a residual preoptic region still develops. The reduction of the ventral diencephalon was accompanied by a ventral expansion of the dorsally expressed pax6, revealing a dorsalization of the basal hypothalamus. Within the telencephalon we observed a loss of pallidal markers, while striatum and pallium are forming. At the neuronal level, nkx2TKD morphants lacked several neurosecretory neuron types, including avp, crh, and pomc expressing cells in the hypothalamus, but still form oxt neurons in the preoptic region. Our data reveals that, while nkx2.1, nkx2.4a, and nkx2.4b genes act partially redundant in hypothalamic development, nkx2.1 is specifically involved in the development of rostral ventral forebrain including the pallidum and preoptic regions, whereas nkx2.4a and nkx2.4b control the intermediate and caudal hypothalamus.
Keywords: hypothalamus, preoptic region, pallidum, diencephalon, telencephalon, neural patterning, neuroendocrine neurons, zebrafish
Introduction
Regional specification of the central nervous system (CNS) commences during gastrulation and at neural plate stage, when patterning along the anteroposterior as well as the dorsoventral axes is initiated. The anteroposterior patterning assigns rostrocaudal regional identities (Lumsden and Krumlauf, 1996), and together with the dorsoventral patterning influences cell fate specification and consequently the generation of different neuronal subtypes (Tanabe and Jessell, 1996). Several Nkx genes act during patterning of the ventral CNS, and contribute to a molecular code for neuronal differentiation (Shimamura et al., 1995; Ericson et al., 1997; Pabst et al., 1998). Nkx2.1 is a member of the vertebrate Nkx homeobox transcription factor family (Pera and Kessel, 1998; Small et al., 2000; van den Akker et al., 2008). It is also known as thyroid transcription factor 1 (TTF-1) or Thyroid-specific enhancer-binding protein (T/ebp) because of its involvement in thyroid development (Guazzi et al., 1990; Mizuno et al., 1991; Elsalini et al., 2003). Two nkx2.1 genes in the zebrafish genome have been previously described as paralogs, nkx2.1a and nkx2.1b (Rohr et al., 2001). Expression of nkx2.1a and b in the zebrafish CNS was reported to initiate toward the end of gastrulation in a rostrocaudal stripe in the medial anterior neural plate, giving rise to hypothalamus, preoptic region and ventral telencephalon (Rohr et al., 2001). In the medial neural plate and ventral neural tube, expression of mammalian nkx2.1 has distinct anteroposterior and dorsoventral boundaries (Shimamura et al., 1995; Briscoe et al., 2000; Puelles and Rubenstein, 2003). nkx2.1a and b expression domains display a common posterior border, which resides in the basal part of prosomere 3 located ventrally to the prethalamus. Both are expressed in the posterior tuberculum and basal hypothalamus. However, the preoptic region, the basal telencephalon around the anterior commissure, and the alar preoptic region exclusively express nkx2.1b (Rohr et al., 2001; Lauter et al., 2013). The medial hypothalamus is characterized by nkx2.1a expression and low or absent nkx2.1b expression.
In mice, development of ventral hypothalamus and telencephalic medial ganglionic eminence depend on NKX2.1. In homozygous Nkx2.1 knockout mice, ventral forebrain developmental abnormalities start anteriorly in the septal area and extend to the mammillary body of the hypothalamus (Kimura et al., 1996). Loss of NKX2.1 also causes a transformation of medial ganglionic eminence into lateral ganglionic eminence structures (Sussel et al., 1999). In addition, both in mouse and Xenopus embryogenesis removal of nkx2.1 causes a dorsalization of the basal plate forebrain (Sussel et al., 1999; van den Akker et al., 2008). Nkx genes are also involved in neuronal differentiation. In the spinal cord the combinatorial expression of Nkx transcription factors (Nkx6.1, Nkx2.2, and Nkx2.9) specifies ventral identity of neurons (Briscoe et al., 2000; Sander et al., 2000). Less is known about potential involvements of Nkx2 factors in forebrain neuronal differentiation, where Nkx2.1 has been shown to contribute to cortical interneuron subtype specification (Butt et al., 2008).
Here, we report the identification, expression and functional analysis of the zebrafish nkx2.4a gene. During embryogenesis nkx2.4a is expressed in the hypothalamus in a manner similar to the gene previously reported as nkx2.1a. Phylogenetic analysis suggests that nkx2.4a resulted from an nkx2.1 gene duplication event that is not restricted to teleosts (Price, 1993; Small et al., 2000; Wang et al., 2000). In zebrafish, like in mouse (Marín et al., 2002), nkx2.4a is expressed in a restricted area of the hypothalamus. In contrast to nkx2.1, nkx2.4a is not expressed in the telencephalon. Synteny and phylogenetic analysis reveals that the previously reported nkx2.1a is in fact a paralog of nkx2.4, and not of nkx2.1, and thus will be named nkx2.4b here in accordance with zebrafish nomenclature. Combined inactivation of nkx2.1, nkx2.4a, and nkx2.4b function by triple Morpholino knockdown (here called nkx2TKD) revealed their contribution to neural patterning and neuronal differentiation. In contrast to single knockdown, the ventral diencephalon of nkx2TKD morphants is dorsalized, revealing that nkx2.1, nkx2.4a, and nkx2.4b act redundantly in hypothalamic patterning. All three genes are required for development of specific subsets of neurosecretory neurons in the hypothalamus.
Materials and methods
Zebrafish husbandry
Zebrafish breeding and maintenance were carried out under standard conditions at 28.5°C (Westerfield, 2000). We used AB-TL wildtype zebrafish, the enhancer trap ETvmat2:GFP (Wen et al., 2008), and smob641 (Barresi et al., 2000), which were identified morphologically. To inhibit pigmentation, embryos were incubated in egg water containing 0.2 mM 1-phenyl-2-thiourea. Embryos were staged according to Kimmel et al. (1995).
Injection of morpholinos and RNA
The following morpholinos were used (Gene Tools LLC): nkx2.4a TBMO (ATG): 5′-GCTCAGCGACATGGTTCAGCCCGCA-3′, nkx2.4a SBMO (e1i1): 5′-TGCATTAGAAGAACTTACTTGTTGA-3′. The standard control SCMO, p53 ATG (Robu et al., 2007), nkx2.1 (previously published as nkx2.1b) and nkx2.4b (published as nk2.1a-1) ATG morpholinos have been described (Elsalini et al., 2003). Morpholinos were diluted in H2O containing 0.05% phenol red or 0.05% rhodamine dextran. nkx morpholinos were injected at the one cell stage in different combinations at a total amount of 7.5 ng per embryo, plus an additional 2.5 ng p53 MO. As controls, 7.5 ng standard control morpholino (SCMO) and 2.5 ng p53 MO per embryo were injected.
The efficiency of the nkx2.4a-e1i1-SBMO was verified by RT-PCR using cDNA synthesized from 1, 2, and 3 dpf embryos injected with SCMO or nkx2.4a-e1i1-SBMO+nkx2.4b-MO+nkx2.1-MO and p53-MO at 2.5 ng each (same conditions as used for all nkx2TKD; Figure 3B). The PCR products where sequenced using primers spanning the whole nkx2.4a coding region: nkx2.4a F: 5′-ATGTCGCTGAGCCCAAAG-3′, nkx2.4a R: 5′-CTACCACGTTCTGCCATAAAGC-3′.
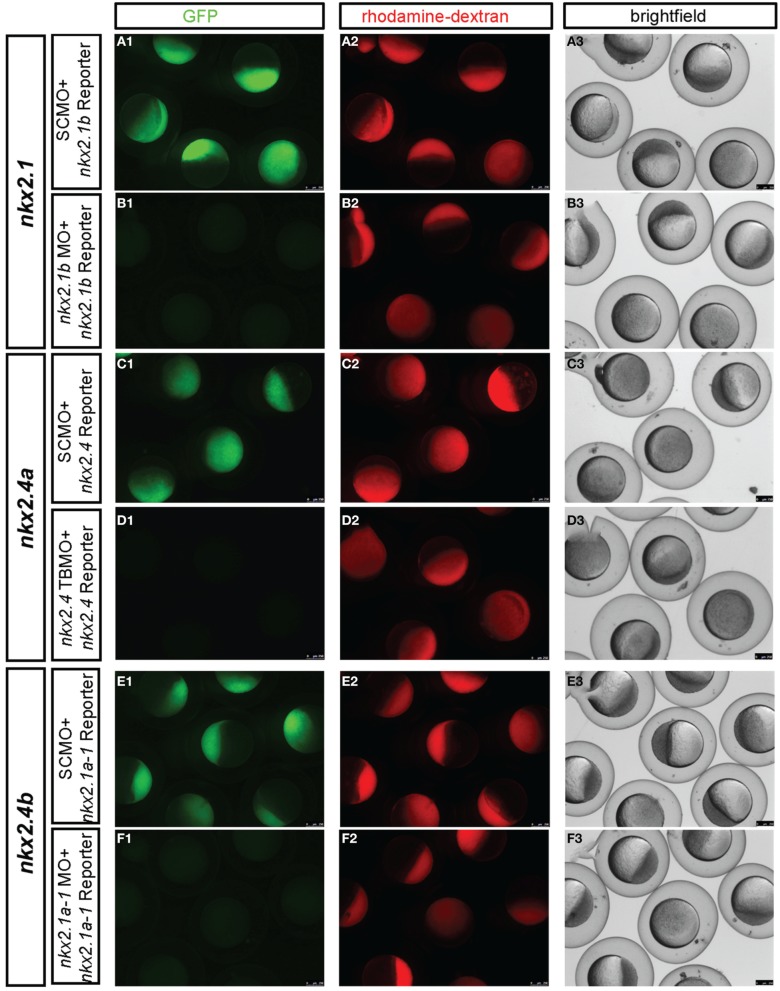
To analyse the specificity and efficacy of the translation blocking MOs (TBMOs) directed against the translation start sites of nkx2.1, nkx2.4a, or nkx2.4b, pCS2+-gfp-reporter plasmids were created which each harbor the respective morpholino target sequence, fused in frame to the GFP ORF. gfp-reporter mRNAs were generated from plasmids pCS2+-5′UTR-nkx2.4b-gfp, pCS2+-5′UTR-nkx2.1-gfp and pCS2+-5′UTR-nkx2.4a-gfp linearized with NotI, and transcribed using the SP6 mMessage mMachine kit (Ambion). gfp-reporter mRNAs were co-injected into one-cell stage embryos in combination with SCMO or the respective specific targeting morpholino. At epiboly stages, embryos were assayed for GFP fluorescence (Figure 4). We also attempted to validate the knockdown phenotypes by rescue experiments injecting mRNAs encoding the Nkx2.1 and Nkx2.4, however broad overexpression of Nkx2.1 or Nkx2.4 from injected mRNAs caused severely abnormal development likely because of gain-of-function effects due to ectopic expression (data not shown).
To verify the results obtained with the nkx2.4a splice blocking MO, we repeated the nkx2TKD substituting the nkx2.4a SBMO with the translation blocking nkx2.4a TBMO in the nkx2TKD mix, and analyzed tyrosine hydroxylase (th), lhx5, and lhx6 expression (Figure 6). Both nkx2.4a SBMO or TBMO at 2.5 ng per embryo in combination with nkx2.4b and nkx2.1 TBMO generated a similar phenotype.
Synthetic lefty1 mRNA (mMessage mMachine kit, Ambion) was injected as described (Barth and Wilson, 1995; Thisse and Thisse, 1999).
In situ hybridization and tunel staining
Whole-mount in situ hybridization (WISH) (Lauter et al., 2011) and fluorescent WISH with immunohistochemistry (Filippi et al., 2010) were performed as described. The following digoxigenin-labeled riboprobes were synthesized: dbx1a (Fjose et al., 1994), dlx5a (Akimenko et al., 1994), foxa2 (Strähle et al., 1993), lhx5 and lhx6 (Toyama et al., 1995), shha (Ekker et al., 1995), otpa (Del Giacco et al., 2006; Ryu et al., 2007), pax6 (Krauss et al., 1991b), pomca (Herzog et al., 2003), nkx2.2 (Barth and Wilson, 1995). For generation of nkx2.4b (RefSeq NM_131589.1), nkx2.1(b) (NM_131776.1), and nkx2.4a (NM_001111166.1) probes, the coding sequences were PCR amplified and cloned into the TOPO PCRII vector using the primers nkx2.4b F: 5′-ATGTCCTTGAGCCCCAAAC-3′; nkx2.4b R: 5′-TCACCATGTTCTGCCGTACA-3′; nkx2.1 F: 5′-ATGTCGATGAGCCCTAAGCA-3′; nkx2.1 R: 5′-TCACCACGTCCTGCCATA-3′ (for nkx2.4a see above).
TUNEL assay was performed using the Apoptag in situ apoptosis detection kit (Chemicon) (Ryu et al., 2005). Classification of signal: “absent”—no blue stained cell detected in specific brain region; “few cells”—small number of blue stained cell (1–10) dispersed in specific brain region (example telencephalic region in Figures 3I,J); “many cells” accumulation of >10 blue stained cells, often clustered, in specific brain region.
Microscopy and image analysis
Transmitted light images were acquired using a Zeiss Axioskop compound microscope. Fluorescently labeled embryos were documented by confocal image stacks using a Zeiss LSM510. Images shown are z-projections of defined sets of consecutive focal planes assembled with the Zeiss Zen 2012 software.
Sequence alignments and synteny
NKX protein sequences were aligned and analyzed with CLC Genomics Workbench 5 (www.clcbio.com) using the distance based method to generate a phylogenetic tree. The NCBI accession numbers are: NK2.4b [Danio rerio] NP_571664.1, NK2.1(b) [Danio rerio] NP_571851.1, Nkx-2.1 [Mus musculus] NP_001139670.1, Nkx-2.1 isoform 2 [Homo sapiens] NP_003308.1, Nkx-2.1 isoform 1 [Xenopus tropicalis] XP_002935383.1, Nkx-2.4a [Danio rerio] NP_001104636.1, Nkx-2.4 [Mus musculus] NP_075993.1, Nkx-2.4 [Homo sapiens] NP_149416.1, Nkx-2.4 [Xenopus tropicalis] XP_002939478.1, Nkx-2.2 [Homo sapiens] NP_002500.1, Nkx2-2 protein [Mus musculus] AAI38160.1, Nkx-2.2 isoform X1 [Xenopus tropicalis] XP_002939477.1, Nkx-2.2a [Danio rerio] NP_571497.1, Nkx-2.3 [Homo sapiens] NP_660328.2, Nkx2-3 [Mus musculus] CAA72002.1, Nkx-2.3 [Xenopus tropicalis] XP_002937234.1, Nkx2.3 [Danio rerio] AAC05228.1, Nkx-2.5 [Mus musculus] NP_032726.1, Nkx-2.5 isoform 1 [Homo sapiens] NP_004378.1, Nkx2.5 [Danio rerio] AAC05229.1, Nkx2-5 [Xenopus tropicalis] AAI60531.1
Synteny between the mouse and zebrafish nkx2 genes was visualized using Cinteny (http://cinteny.cchmc.org).
Results
Nkx2.1 and Nkx2.4 genes in zebrafish
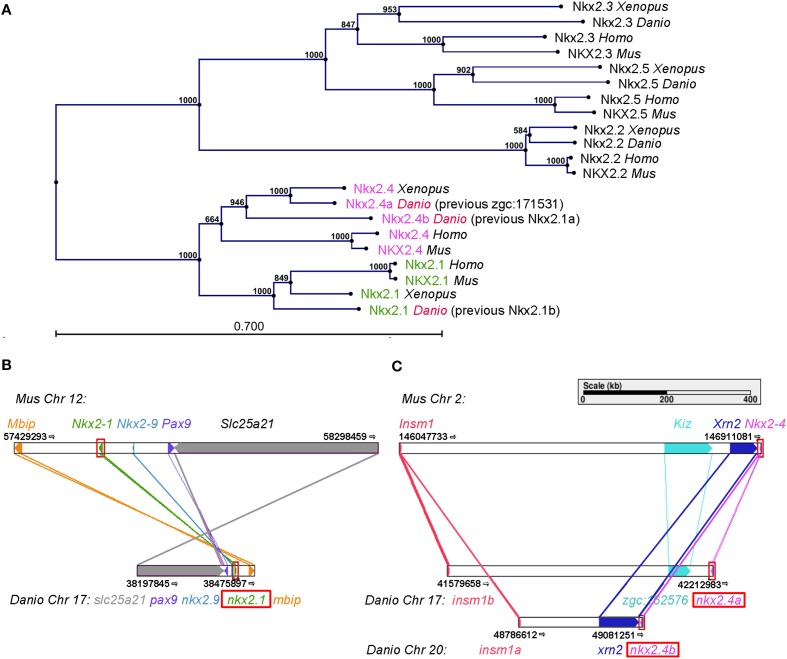
We systematically searched for zebrafish genes that may interact and be co-expressed with nkx2.1 transcription factors during ventral diencephalon development, and noticed that the zgc:171531 gene (ENSDARG00000075107) encodes an NKX2 family member. zgc:171531 has recently been reported to be expressed in the hypothalamus during late somitogenesis stages (Armant et al., 2013), but its function has not been studied. We analyzed the phylogenetic relationship of Zgc:171531 to other members of the Nkx protein family, and could confirm the suggestion by Armant et al. (2013) that Zgc:171531 is an ortholog of mouse NKX2.4 (Figure 1). Phylogenetic tree analysis showed that zebrafish Zgc:171531 is the closest relative of mammalian NKX2.4, and that Zgc:171531and Nkx2.1 are more closely related to each other than to any other Nkx2 transcription factor family member (Figure 1A). Therefore, we refer to Zgc:171531 as Nkx2.4a.
Figure 1.
Positioning nkx2.1 and nkx2.4 within the nkx2 homeobox transcription factor family. (A) Phylogenetic tree of five members of the Nkx2 homeobox transcription factor family in zebrafish, Xenopus, mouse, and human. Distance based tree calculations used full-length protein sequences (Materials and Methods). The numbers at the nodes are bootstrap confidence levels from 1000 replicates. The scale bar represents 0.7 substitutions per position. (B,C) Genetic synteny at the nkx2.1 (B) and nkx2.4 (C) loci in mouse and zebrafish analyzed using Cinteny (cinteny.cchmc.org). The arrows indicate transcriptional direction.
To explore the evolutionary history of the nkx2.1 and nkx2.4 genes, we searched for synteny conservation. We found that the gene previously reported as zebrafish nkx2.1b is located in a microsyntenic chromosomal region related to mouse Nkx2.1 (Figure 1B), thus nkx2.1b is indeed the ortholog of mouse Nkx2.1. Zebrafish zgc:171531 resides in a microsyntenic chromosomal region related to mouse Nkx2.4. Importantly, also zebrafish nkx2.4b (previously named nkx2.1a) shares synteny with mouse Nkx2.4 (Figure 1C). Thus, we conclude that nkx2.4a and nkx2.4b are indeed paralogous NKX2.4 genes derived from the teleost specific genome duplication. In contrast, the previous nkx2.1a and nkx2.1b are not paralogs, and zgc:171531 is now named nkx2.4a, while nkx2.1a is named nkx2.4b, and nkx2.1b is named nkx2.1. This change in nomenclature was approved by the zebrafish gene nomenclature committee (see www.zfin.org).
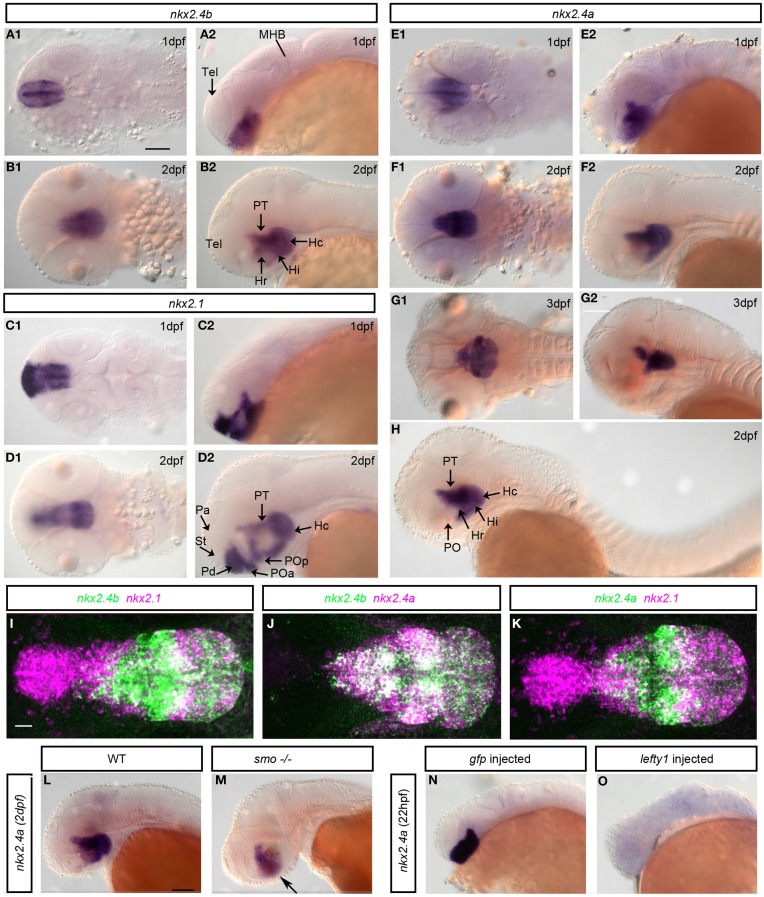
We analyzed expression of nkx2.4a in comparison to nkx2.1 (Figure 2) (Rohr et al., 2001; Tessmar-Raible et al., 2007). At 1 dpf nkx2.4a is expressed within the forebrain in basal prosomere 3 including the hypothalamus and the posterior tuberculum, and with decreasing expression levels into the preoptic hypothalamic areas of the basal secondary prosencephalon (Figure 2E). In contrast to nkx2.1 (Figures 2C,D), expression of nkx2.4a was not detected in the alar hypothalamic region (anterior preoptic region) or the pallidum of the secondary prosencephalon. At 2 dpf nkx2.4a expression is downregulated in the basal hypothalamic preoptic area, but continues in the posterior tuberculum and caudal hypothalamus (Figure 2F). At 3 dpf nkx2.4a expression persists in the posterior tuberculum and hypothalamus (Figure 2G), similar to nkx2.4b (Figures 2A,B). This expression pattern is maintained until 4 dpf (data not shown). Thus, all three nkx2 genes are expressed in the posterior tuberculum as well as in the basal hypothalamus. However, nkx2.1 is also expressed in the basal telencephalon around the anterior commissure as well as in the alar hypothalamus preoptic region. nkx2.4a, in contrast to nkx2.4b (Elsalini et al., 2003), is not expressed in the thyroid gland (Figure 2H). Double fluorescent WISH of nkx2.4a expression in combination with nkx2.4b or nkx2.1 revealed broad coexpression of nkx2.4a and nkx2.4b (Figure 2J), whereas nkx2.4a and nkx2.1 coexpression is restricted mainly to the intermediate hypothalamus (Figures 2I,K).
Figure 2.
Expression and regulation of nkx2.1, nkx2.4a, and nkx2.4b. (A,B) nkx2.4b, (C,D) nkx2.1 and (E–H) nkx2.4a mRNA expression analyzed by whole mount in situ hybridization at indicated stages. (I–K) Expression of nkx2.4a and nkx2.4b and nkx2.1 were detected by double fluorescent whole mount in situ hybridization of 2 dpf embryos, magenta and green channels as indicated in panel headings. Shown are 140–190 μm Z-projections of confocal image stacks. (L,M) Expression of nkx2.4a in WT and smo homozygous mutant embryo. Arrow: residual nkx2.4a expressions (M, n = 10). nkx2.4a expression in controls (N) and embryos with Nodal signaling inactivated by lefty1 mRNA injection (O: 65% of embryos showed a complete loss, 25% a reduction, 10% normal nkx2.4a expression; n = 25). (A1–G1,L-O) lateral views; (A2–G2,I–K) dorsal views. Anterior to left. Scale bar in A1 = 100 μm (for A–H), in I = 20 μm (for I–K), in L = 100 μm (for L–O).
Nodal and Shh signaling control Nkx2.4a expression
Nodal signaling has been revealed essential for initiation, and Shh for maintenance of nkx2.1 and nkx2.4b expression (Rohr et al., 2001). Smoothened is a key transmembrane effector essential for transmitting Hedgehog signals into the cell (Rohatgi and Scott, 2007). smoothened homolog (smo−/−) zygotic mutant embryos have severely reduced Shh signaling, although no complete loss occurs due to residual maternal Smo activity (Barresi et al., 2000; Rohr et al., 2001). We detected nkx2.4a expression at reduced levels in smo−/− embryos (Figures 2L,M), indicating that Hedgehog signaling activity is required to maintain nkx2.4a expression. However, Shh is not required for nkx2.4a initial induction, similar to what has been shown for nkx2.4b. We then overexpressed lefty1 mRNA, encoding an inhibitor of Nodal activity, which resulted in complete loss of nk2.4 expression in >50% of experimental embryos (Figures 2N,O). Thus, Nodal is strictly required to induce nkx2.4a expression, while Shh contributes to its maintenance.
Nkx2.1 and Nkx2.4a/4b combined knockdown severely affects the hypothalamus
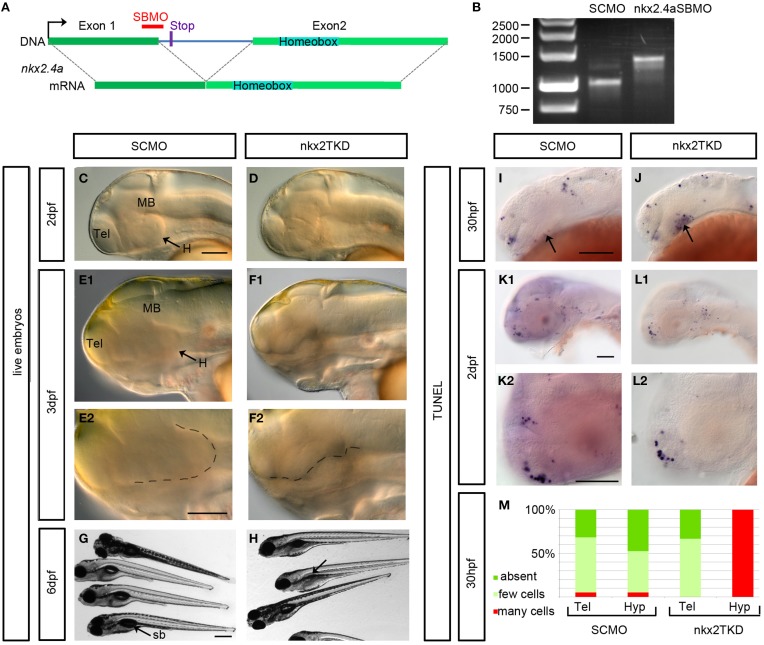
To examine the role of Nkx2.4a we performed gene knockdown using antisense MOs. The nkx2.4a SBMO is complementary to the exon1-intron1 boundary (Figure 3A). We have amplified nkx2.4a mRNA by RT-PCR from nkx2.4a SBMO injected embryos, and found intron1 to be completely retained in the mature mRNA (Figure 3B). Sequencing of the splice-blocked mRNA revealed a premature stop codon truncating the Nkx2.4a protein before the homeodomain. To determine the efficiency of the nkx2.4a TBMO a nkx2.4a-gfp reporter mRNA with the MO binding site at the GFP ATG position was co-injected with the nkx2.4a TBMO and shown to completely suppress GFP expression (Figure 4). Thus, nkx2.4a SBMO and TBMO efficiently inhibit Nkx2.4a expression. Knocking down nkx2.4a alone did not result in any detectable morphological phenotype (data not shown).
Figure 3.
Phenotype of nkx2.1, nkx2.4a, nkx2.4b triple (nkx2TKD) morphants. (A) Schematic representation of the nkx2.4a gene with the morpholino binding site and the stop codon (12 bases into intron) terminating the protein when intron excision is blocked by the SBMO. (B) RT-PCR for nkx2.4a mRNA prepared from 3 dpf embryos injected with 7.5 ng SCMO or with 2.5 ng each of nkx2.4a-e1i1-MO, nkx2.4b and nkx2.1 ATG MOs. The nkx2.4a-e1i1 MO effectively blocks the splice donor site at the exon1-intron1 boundary, giving rise to a longer cDNA. The mature nkx2.4a mRNA is nearly completely eliminated under these conditions. (C–F) Morphological in vivo phenotype of nkxTKD knockdown combined with p53MO knockdown documented using DIC transmitted light microscopy at indicated stages. (E2–F2) Close-ups with the ventral border of the diencephalon outlined by dotted lines. (G,H) nkxTKD larvae survived at least until 6 dpf (swim bladder is not inflated in fish in H indicated by an arrow). (I–M) TUNEL staining for apoptotic cells in 30 hpf and 2 dpf SCMO plus p53MO-injected control and nkxTKD plus p53MO morphant embryos (arrows in I and J: hypothalamus) (K–L,K2,L2 show close-ups). (M) Quantification of number of TUNEL stained apoptotic cells in nkxTKD+p53 (n = 12) morphant larvae compared to SCMO+p53MO injected larvae (n = 19). Lateral views, anterior at left. Abbreviations see Table 1. Scale bars = 100 μm except G: scale bar = 400 μm.
Figure 4.
Experimental validation of in vivo knockdown efficiency of translation blocking morpholinos used in this study. MO knockdown efficiency was demonstrated by co-injection of GFP reporter mRNAs with the morpholino binding sites engineered at the start ATG of GFP. The following GFP reporter were injected at 100 pg mRNA per embryo: (A,B) 5′UTR-nkx2.1-gfp mRNA, (C,D) 5′UTR-nkx2.4a-gfp mRNA, and (E,F) 5′UTR-nkx2.4b-gfp mRNA. Morpholinos used are indicated in boxes at left of each panel and were injected at 5 ng each, controls containing the same amount of SCMO. nkx2.1 ATG MO and nkx2.4b ATG MO morpholinos have been described previously (Elsalini et al., 2003). All embryos were also co-injected with rhodamine-dextran, which was used to sort embryos after 4–5 h. of development for homogenous amounts and distribution of injected material. Knockdown efficiency was then documented between 4 and 5 hpf by pictures taking with the epifluorescence dissecting microscope in the red channel to verify that embryo were injected, in the green channel to control efficiency of GFP knockdown, and in the transmitted light channel to control morphology and viability of embryos. nkx2.4b and nkx2.1 and nkx2.4aTBMO morphants were rhodamine-dextran fluorescence levels similar to control-injected embryos but showed no expression of the respective GFP reporters, indicating that these TBMOs effectively bind and block translation of target mRNAs in vivo.
Table 1.
Anatomical abbreviations.
| 1 | Basal prosomer 1 |
| 2 | Basal prosomer 2 |
| 3c | Caudal portion of basal prosomer 3 (dbx1+) |
| 3r | Rostral portion of basal prosomer 3 (Pt) |
| Arc | Arcuate nucleus |
| H | Hypothalamus |
| HB | Hindbrain |
| Hc | Caudal hypothalamus |
| Hi | Intermediate hypothalamus |
| Hr | Rostral hypothalamus |
| MB | Midbrain |
| OB | Olfactory bulb |
| P3c | Caudal Prosomere 3 |
| Pa | Pallium |
| Pd | Pallidum |
| pit | Pituitary |
| PO | Preoptic region |
| POa | Anterior preoptic region |
| POp | Posterior preoptic region |
| Pr | Pretectum |
| PT | Posterior tuberculum |
| pTh | Prethalamus |
| sb | Swim bladder |
| SP | Subpallium |
| SPV | Supraoptoparaventricular region |
| St | Striatum |
| Tel | Telencephalon |
| Th | Thalamus |
| zli | Zona limitans intrathalamica |
The similarity in amino acid sequences and the overlapping expression patterns suggest that nkx2.1, nkx2.4a, and nkx2.4b may act redundantly. Therefore, we aimed at combined knockdown of all three nkx2 genes. We decided to use Morpholino antisense knockdown to study the combined activity of these three genes, because mutations in none of these genes are available, and even if mutations may become available at some point, the genetic analysis of triple mutations providing the desired phenotype only in one out of 64 embryos is cumbersome. ATG-morpholinos specific to both nkx2.1 genes have been published (Elsalini et al., 2003). We confirmed their efficacy by knockdown of GFP expression from injected reporter mRNAs (Figure 4). nkx2.4b TBMO, nkx2.1 TBMO, and nkx2.4a SBMO were co-injected at 2.5 ng each per embryo in combination with p53 morpholino (for exact amounts used in each figure panel see Supplemental Table 1). Given that some morpholinos cause cell death irrespective of knockdown of specific gene function (Robu et al., 2007), we included p53 MO in our nkx2TKD knockdown mix in all experiments. Live nkx2TKD embryos and larvae were analyzed for morphological abnormalities using transmitted light microscopy (Figures 3C–H). At 2 dpf nkx2TKD embryos developed a severe loss of hypothalamic structures, as well as abnormalities in the preoptic region (Figures 3C,D). At 3 dpf the massive loss of ventral forebrain tissue became more prominent (Figures 3E,F). With the exception of the ventral forebrain, nkx2TKD embryos and larvae developed anatomical structures morphologically similar to control WT embryos, and overall progress of development was not affected. nkx2TKD larvae established blood circulation and survived at least until 6 dpf (Figures 3G,H), but frequently did not fully inflate the swim bladder.
To test whether nkx2TKD embryos may develop apoptosis, we performed TUNEL assays. Apoptosis was clearly increased in the hypothalamus of morphants compared to SCMO injected embryos at 30 hpf (Figures 3I,J,M). However, at 2 dpf apoptosis levels in nkx2TKD morphants were similar to SCMO injected control larvae (Figures 3K,L). Thus, nkx2TKD causes increased apoptosis in those brain regions of nkx2.1/2.4 expression specifically when the hypothalamus forms, but not at later stages.
Knockdown of Nkx2.1/4a/4b severely affects ventral forebrain pattern formation
Shh is a key determinant of ventral pattern formation in the neural plate. Studies have shown that its expression in the basal telencephalon depends on nkx2.1 function in mice (Sussel et al., 1999). In wild-type zebrafish embryos shha (Figure 5A) is expressed in floor plate and underlying axial mesoderm, as well as in the zona limitans intrathalamica (ZLI; Scholpp et al., 2006), which is a narrow transverse region between the prethalamus and thalamus (Shimamura et al., 1995; Kiecker and Lumsden, 2004). In nkx2.1 morphants shha expression was mostly normal, with only a minor reduction in the hypothalamus (Figure 5B). In nkx2.4a/4b double morphants (Figure 5C) we detected a more severe reduction of the hypothalamic shha expression. In nkx2TKD embryos shha expression in the hypothalamus was almost completely eliminated (Figure 5D), while in the ZLI and caudal to it shha expression appeared normal. Thus, all three nkx2 genes redundantly regulate shha expression in the hypothalamus. Caudal to prosomer 3 shha expression was not affected in nkx2TKD morphants. This is consistent with the finding that the expression pattern of foxa2, a marker for medial and lateral floor plate caudal to prosomere 2 (Odenthal and Nusslein-Volhard, 1998), appeared unaffected in nkx2TKD morphants with some enlargement of basal prosomeres 1 and 2 (Figures 5O,P). With respect to basal prosomere 3, the nkx2.1 expression extends only into the rostral half of prosomere 3, while dbx1a is expressed in the caudal part of basal prosomere 3 (Lauter et al., 2013). dbx1a expression was not affected in nkx2TKD morphants, indicating normal development of the caudal portion of prosomer 3 (Figures 5M,N). This finding demonstrates that brain regions caudal to the limit of nkx2.1/4a/4b expression are not affected upon nkx2TKD.
Figure 5.
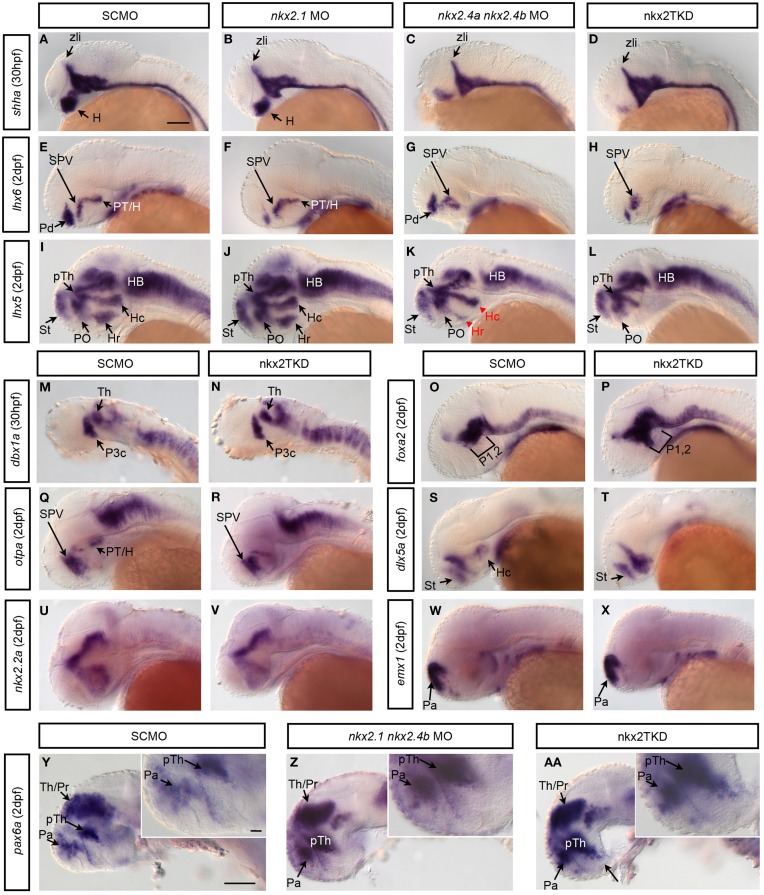
nkx2.1, nkx2.4a, and nkx2.4b knockdown affects forebrain patterning. Zygotes were injected with single or combinations of Nkx2 gene morpholinos as indicated in boxes above the panels, and incubated until 30 hpf (A–D,M,N) or 2 dpf (E–L,O–AA). Expression of patterning genes were analyzed by WISH. (A–D) Hypothalamic shha expression in (B) nkx2.1 morphants is largely normal, with a minor reduction (n = 10.10); (C) nkx2.4a/4b double morphants (n = 10.10) is more severely reduced; (D) nkx2TKD embryos is severely reduced in the rostral forebrain (n = 29.29). (E–H) lhx6, and (I–L) lhx5 expression in morphants at 2 dpf (E, n = 42.42; F, n = 11.11; G, n = 4.4; H, n = 67.67; I, n = 30.30; J, n = 7.7; K, n = 9.9; L, n = 23.23) (red arrowhead in K indicates affected area/reduced expression). (M,N) dbx1a expression was not perturbed by nkx2TKD at 30 hpf. (O,P) nkxTKD leads to a slight anterior expansion of foxa2 expression in basal prosomeres 1 and 2 (O, n = 22.22; P, n = 11.11). (Q,R) otpa and (S,T) dlx5a expression were affected in the posterior tuberculum and hypothalamus of nkx2TKD morphants at 2 dpf (Q, n = 34.34; R, n = 60.60; S, n = 16.16; T, n = 12.12). (U,V) nkx2TKD affects the expression pattern of nkx2.2 only in the caudal hypothalamus (n = 14.14). (W,X) The pallial telencephalic emx1 expression is not affected in nkx2TKD. (Y,AA) In nkx2TKD morphants the prethalamic pax6 expression domain expands into the ventral hypothalamus (inset in AA, n = 13.13), while double nkx2.1 and nkx2.4b morphant have a less severe ventral pax6 expansion (inset in Z, n = 25.25). Lateral views, anterior is to the left. Abbreviations see Table 1. lateral views. Black arrows point at positions of anatomical structures labeled by abbreviation at start of arrow. Scale bar = 100 μm in (A) for (A–X), in (Y) for (Y–AA).
A target of mouse NKX2.1 in the telencephalon is Lhx6 (Sussel et al., 1999; Du et al., 2008). In our study, in nkx2.1 morphants pallidal lhx6 expression was strongly reduced, but not the diencephalic lhx6 expression (Figures 5E,F). In nkx2.4a and nkx2.4b double morphants however pallidal lhx6 expression was maintained, whereas in the diencephalon the caudal posterior tubercular expression was lost (Figure 5G). In nkx2TKD morphants, both the pallidal and posterior tubercular lhx6 expression were severely reduced, while lhx6 expression in the preoptic supraoptoparaventricular (SPV) region was retained (Figure 5H). These results were confirmed by analysing the more broadly expressed lhx5 (Figure 5I). The lhx5 domains in posterior tuberculum and caudal and rostral hypothalamus were depleted in nkx2TKD morphants, while expression in the alar preoptic region was still detectable (Figure 5L). In the telencephalon, the dorsal lhx5 expression domain expanded ventrally into the pallidal area, from which lhx5 was absent in wildtype. This ventral expansion of the telencephalic lhx5 expression was also visible in nkx2.1 morphants (Figure 5J), which otherwise had an unaltered lhx5 expression pattern. Double nkx2.4a and nkx2.4b morphants did not develop the telencephalic lhx5 expansion, but exhibited a reduction of the hypothalamic expression pattern (Figure 5K). The nkx2TKD expression changes of lhx5 and lhx6 were also validated in triple knockdown embryos using the nkx2.4a TBMO (Figure 6). The expression of otpa in the posterior tuberculum was strongly reduced in nkx2TKD morphants, while expression was still detected in the anterior preoptic area (Figures 5Q,R). Analysis of dlx5a reveals that the dorsal subpallium (striatum) still forms in nkx2TKD morphants (Figures 5S,T). In contrast, the caudal hypothalamic dlx5a domain was absent in triple morphants. Similar to what was observed for lhx6 expression in nkx2TKD morphants, the dlx5a expression domain in the alar preoptic region appears expanded ventro-caudally (Figures 5S,T). nkx2.2a has a rostro-caudal domain extending approximately along the alar-basal boundary of the fore- and midbrain (Puelles and Rubenstein, 1993; Hauptmann and Gerster, 2000). This boundary remained unaffected in the triple morphants. Also, the prethalamic and thalamic as well as preoptic nkx2.2a expression domains appeared largely normal, while the caudal hypothalamic expression was reduced (Figures 5U,V). Analysing emx1 expression, we did not observe any effect of nkx2TKD on the pallium (Figures 5W,X). In summary, these results show that loss of nkx2.1/4a/4b activity mainly affects three regions of the forebrain. In the caudal diencephalon, there is a loss of the rostral part of basal prosomer 3 and derivative posterior tubercular and hypothalamic structures, while the alar basal boundary appears non-shifted. Further rostrally, there is a loss of rostral and intermediate hypothalamic markers, while a residual preoptic region including the supraoptoparaventricular region develops in nkx2TKD morphants. Within the telencephalon, there is a loss of pallidal markers, while striatum and pallium are forming.
Figure 6.
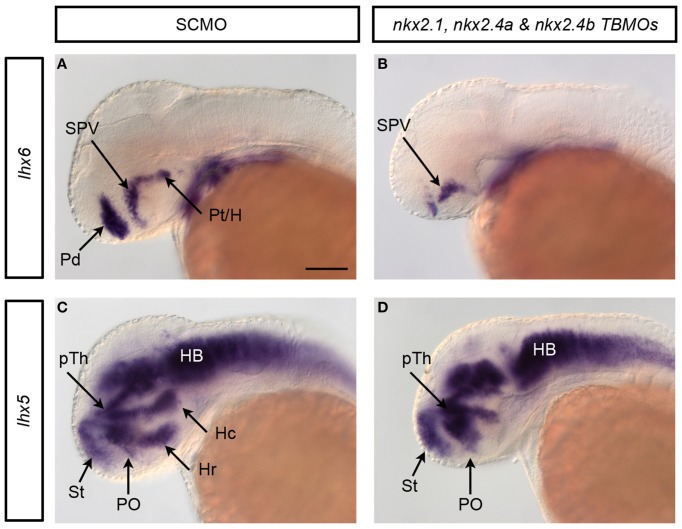
Validation of nkx2TKD phenotype by independent nkx2.4a TBMO. By designing a second, non-overlapping MO against nkx2.4a (nkx2.4a TBMO) concerns with off-target phenotypes can be addressed. The probability of both nkx2.4a MOs yielding a similar off-target phenotype is significantly lower compared to a single MO only (Eisen and Smith, 2008). The triple knockdown experiments with the nkx2.4a TBMO were analyzed for changes in lhx6 (A,B) and lhx5 (C,D) expression. The observed phenotypes were very similar to the nkx2TKD with the SBMO: the respective expression domains in the pallidum, the posterior tuberculum, the caudal and rostral hypothalamus were depleted in nkx2TKD with the nkx2.4a TBMO, while expression in other anatomical domains, including the alar preoptic region, was not significantly affected. Therefore, the analysis of nkx2TKD with the nkx2.4a TBMO confirmed the results obtained in our study with the nkx2TKD with the nkx2.4a SBMO. Black arrows point at positions of anatomical structures labeled by abbreviation at start of arrow.
Knockdown of nkx2.1/4a/4b leads to a dorsalization of the ventral diencephalon
Studies in mice suggested that Nkx2.1 knockout causes a ventral expansion of dorsal diencephalic marker genes (Kimura et al., 1996). Therefore, we tested whether similar fate shifts occur in nkx2TKD embryos by analysing pax6a expression, which is predominantly expressed in dorsal parts of the diencephalon (Krauss et al., 1991a), as well as in cells at the pallial-subpallial boundary (Wullimann and Rink, 2001). In nkx2TKD embryos, pax6 expression was expanded ventrally in its prethalamic domain, suggesting a loss of basal prosomere 3 (Figures 5Y–AA). Together with the data obtained for dbx1a expression (Figures 5M,N; caudal part of prethalamus and caudal part of basal prosomer 3; Lauter et al., 2013), it appeared that the rostral part of basal prosomere 3 was completely dorsalized, while the caudal dbx1a expressing part of prosomere 3 was not affected. Also, it appears that alar pax6a expression did not invade into the basal prosomere 1 and 2 regions, which in morphants appeared more pronounced caudal to prosomere 3 (see also 2 dpf foxa2 expression Figures 5O,P). In contrast, when only nkx2.1 and nkx2.4b were knocked down, no obvious ventral expansion of the pax6a domain into the hypothalamus was detectable. Thus, nkx2.4a can compensate loss of nkx2.1 and nkx2.4b in hypothalamic patterning. In summary these results suggest that the loss of nkx2.1/4a/4b activity leads to a dorsalization of the basal prosomer 3 as well as of the pallidum.
Nkx2.1/4a/4b control expression of nkx2.1 and nkx2.4 genes
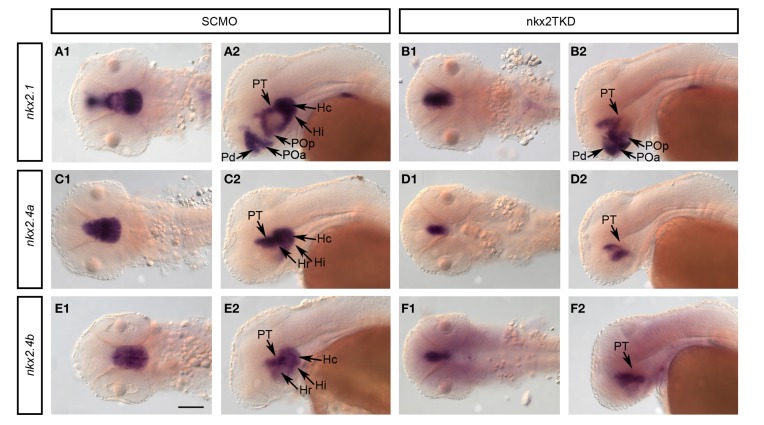
We next analyzed whether Nkx2.1/4a/4b activity is required for continued expression of these genes during embryonic development. Compared to controls, in nkx2TKD morphants the expression domains of all three genes were restricted to more medial forebrain regions, most pronounced for nkx2.4a and nkx2.4b (Figure 7). At 2 dpf, expression of all three genes was maintained in the posterior tubercular portion of the prosomer 3 derived basal hypothalamus in nkx2TKD embryos. In contrast, expression of all three nkx2 genes was depleted in the intermediate and caudal hypothalamus. nkx2.1/4a/4b expression was reduced in the preoptic region. nkx2.1 expression domains in the alar plate preoptic telencephalon and pallidum were maintained, albeit the size of the domain and the expression level appeared slightly reduced (Figures 7A,B). These results suggest that nkx2.1 and nkx2.4a/b activity may be required to maintain their expression in specific domains of the developing forebrain. An alternate explanation may be that some hypothalamic tissue may get lost due to apoptosis (see Figures 3I,J).
Figure 7.
Analysis of nkx2 gene expression in nkx2.1, nkx2.4a, and nkx2.4b morphants. Expression of nkx2.1, nkx2.4a, and nkx2.4b was analyzed in SCMO (A,C,E) and nkx2TKD morphants (B,D,F) at 2 dpf. (A1–F2) dorsal views, (A2–F2) lateral views; anterior is to the left. Abbreviations see Table 1. Black arrows point at positions of anatomical structures labeled by abbreviation at start of arrow. Scale bar = 100 μm in A1 for all panels.
nkx2TKD affects hypothalamic neurosecretory populations
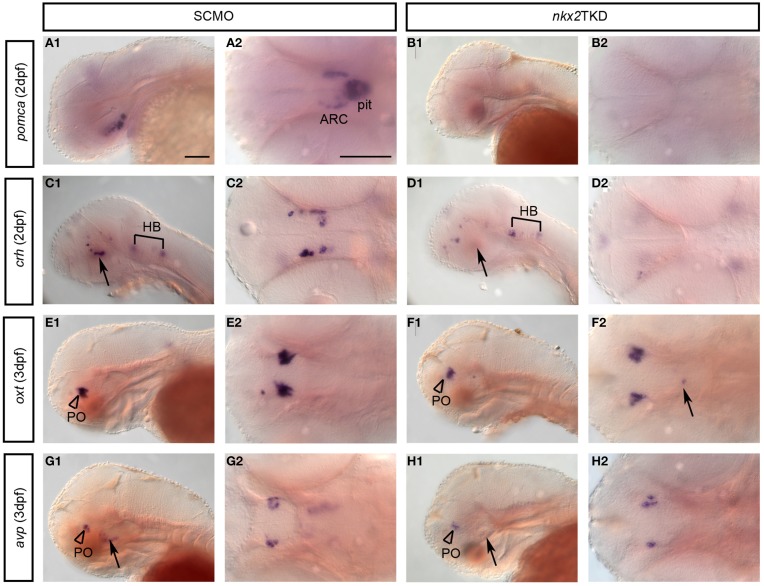
We next investigated which neuroendocrine systems were affected by loss of nkx2.1 or nkx2.4 activity. Expression of pomca (Hansen et al., 2003; Lohr and Hammerschmidt, 2011) is lost in the arcuate nucleus as well as the pituitary of nkx2TKD morphants (Figures 8A,B). To further characterize differential effects of nkx2TKD on ventral neuroendocrine vs. preoptic hypothalamus, we analyzed expression of the neuroendocrine hormone genes corticotropin releasing hormone (crh), oxytocin (oxt), and arginine vasopressin (avp) at 2 and 3 dpf. crh expression (Chandrasekar et al., 2007) in the posterior tuberculum and hypothalamus of nkx2TKD morphants was completely absent (Figures 8C,D), while crh expressing neurons in all other regions developed normally. oxt is exclusively expressed in the preoptic region (Figures 8E,F), and similar to the preoptic domain of avp (Figures 8E,F) was not affected in triple morphants. In contrast, avp expression in the neuroendocrine ventral hypothalamus was strongly reduced or eliminated in nkx2TKD morphants (Figures 8G,H). We also occasionally observed oxt-expressing cells at ectopic locations posterior to the domain in the anterior hypothalamus in several nkx2TKD morphants (Figure 8F, arrow). In summary, nkx2TKD affects neuroendocrine development selectively in the hypothalamus, but not in the preoptic region.
Figure 8.
Neuronal differentiation in the hypothalamus of nkx2TKD embryos. Expression of differentiation markers for neuroendocrine neurons analyzed by WISH at 2, 3, and 4 dpf, stages as indicated. (A,B) Loss of pomca expression in the arcuate nucleus of the hypothalamus (arrowheads) and in the pituitary in nkx2TKD morphants. (C,D) crh neurons in the posterior tuberculum and hypothalamus (arrows) are reduced in nkx2TKD morphants (n = 7.7). (E,F) Formation of oxt-expressing cells in the PO (arrowheads) is not significantly affected in nkx2TKD morphants. However, in nkx2TKD morphants, oxt-expressing cells are detected at ectopic locations within the diencephalon (arrow) (n = 10.10). (G,H) In nkx2TKD morphants, avp-expressing cells in the PO (arrowheads) are still present, but lost in the hypothalamus (n = 9.9). (A1–H1) lateral views; (A2–H2) dorsal views, anterior to the left. The arrows point to the hypothalamus. Abbreviations see Table 1. Scale bars = 100 μm in (A1) for (A1–J1) and in (A) for (A2–J2).
Discussion
The hypothalamus harbors many highly conserved neuroendocrine and neuromodulatory systems vital for the control of fundamental behavioral patterns and physiology. Hypothalamus organization and major developmental control centers have been well described (Puelles and Rubenstein, 2003; Moreno and Gonzalez, 2011). However, the complex organization and dynamic morphogenesis of the hypothalamus have hindered a more detailed understanding of molecular mechanisms controlling patterning and neuronal differentiation. Nkx2.1 genes have a crucial role in development of the ventral forebrain, but analysis of mutant mice (Kimura et al., 1996; Sussel et al., 1999; Marín et al., 2002) and knockdown in Xenopus (van den Akker et al., 2008) have resulted in phenotypes that differentially affect the alar preoptic region and the basal hypothalamus. Here, we characterize a second nkx2.1-related gene in zebrafish, nkx2.4a. Partial redundancies between nkx2.1 and nkx2.4a, which has not been previously knocked out in mice, may explain difficulties in understanding the role of NKX2.1 factors in ventral forebrain development.
zgc:171531 / nkx2.4a encodes a novel zebrafish Nkx2 homeobox transcription factor of high sequence similarity to Nkx2.1 and Nkx2.4. In light of synteny and phylogenetic tree analysis, the previously reported zebrafish nkx2.1a is more closely related to nkx2.4a than to nkx2.1. Recent phylogenetic analysis of rainbow trout Nkx2-4 also suggested zebrafish the previously nkx2.1a named gene to be a nkx2.4 paralog (Uemae et al., 2014). Analysis of nkx2.4a expression in relation to the nkx2.1 genes also reveals a strong similarity to the expression pattern in the brain of the gene previously reported as nkx2.1a. nkx2.4a is expressed in the posterior tuberculum and hypothalamus. nkx2.4a is not expressed in the preoptic region or pallidum, as nkx2.1 is. For zebrafish, our data indicate that the gene previously named nkx2.1a is indeed a paralog of nkx2.4, and the two paralogous genes will be named nkx2.4a and nkx2.4b as orthologs of mammalian Nkx2.4, while nkx2.1 is the only Nkx2.1 ortholog in zebrafish.
Expression patterns similar to zebrafish have been reported for nkx2.4 in Xenopus (Small et al., 2000; Ermakova et al., 2007). Mouse Nkx2.4 expression (Price, 1993) is also restricted to the hypothalamus and absent from the preoptic region (Marín et al., 2002). Thus, it appears that the nkx2.4 expression pattern is conserved throughout evolution, and based on the high sequence similarity in the homeodomain, may contribute to hypothalamic development in a redundant manner with nkx2.1. The two “parallel” nkx2.1 and nkx2.4 genes may also explain other riddles, for example LhjTTF-1/LjNkx2.1 reported for lampreta to lack telencephalic expression (Ogasawara et al., 2001) may indeed be a nkx2.4 ortholog.
Analysis of nkx2TKD embryos revealed that most nkx2.1/4a/4b expression in the basal hypothalamus depends on nkx2.1/4a/4b activity, except for the posterior tubercular domain. In contrast, it appears that the preoptic and pallidal domain do not strictly depend on nkx2.1/4a/4b activity. All three genes are reduced in the medio-lateral extent of their expression in nkx2TKD embryos, which would be in line with a reduced midline-derived activity like Shh required to establish and maintain these domains. Our findings are consistent with reports that nkx2.1 expression is maintained in a smaller more medial-ventral domain in mice (Sussel et al., 1999), and in Xenopus (van den Akker et al., 2008) nkx2.1 loss-of-function embryos. However, in mouse, expression of nkx2.4 appears to strictly depend on nkx2.1 activity (Marín et al., 2002), which would suggest changes in regulatory interactions in evolution.
While morpholino antisense oligonucleotide based knockdown of individual nkx2.1/4 genes had only subtle effects on hypothalamus development, the combined knockdown of nkx2.1, nkx2.4a and nkx2.4b expression revealed that these genes together control specification and patterning of the basal hypothalamus. Figure 9 gives an overview of anatomical changes as revealed by marker gene analysis in nkx2TKD morphants. Triple morphant embryos were devoid of the basal hypothalamus including the rostral half of prosomere 3 and extending to the optic commissure. The defects in the nkx2TKD basal hypothalamus are thus limited to the region of nkx2.1/4a/4b expression, which extends caudally into the rostral half of basal prosomer 3 (Lauter et al., 2013). In nkx2TKD morphants, the caudal half of prosomer 3 expresses dbx1a normally, revealing that the nkx2.1/4 activity has no effects caudal to its expression limits.
Figure 9.
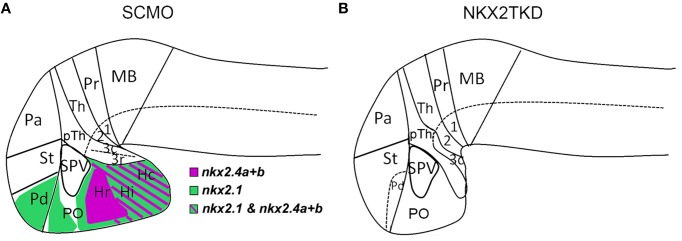
Forebrain patterning abnormalities in nkx2TKD morphants at 2 dpf. (A,B) Schematic sagittal representation of the anatomical domains in the brains of wildtype (A) and nkx2TKD morphant (B) embryos at 2 dpf. The expression domains of nkx2.1, nkx2.4a, and nkx2.4b are indicated in the wildtype scheme (A), for color code see legend in figure. Abbreviations see Table 1.
The loss of basal hypothalamic structures upon nkx2TKD is clearly revealed by loss or reduction of the expression domains of lhx6, lhx5, and dlx5a in this region. This basal plate tissue is dorsalized, as the prethalamic expression domain of pax6a expands to the ventral edge of the forebrain. However, the ventral expansion of dorsal fates does not reflect a global expansion of alar territories, as the dorsoventral positioning of the nkx2.2 expression domain, which correlates with the alar-basal boundary, is not shifted. Thus, our data are consistent with the previously published dorsalization of ventral prethalamus in nkx2.1 mutant mice (Sussel et al., 1999; Marín et al., 2002). They potentially explain discrepancies reported between mouse and Xenopus patterning mechanisms. The xnkx2.1 knockdown has been reported to develop a much weaker basal hypothalamus phenotype (van den Akker et al., 2008) as compared to mice. This could be due to the fact that the Xenopus nkx2.4 gene (Ermakova et al., 2007) could compensate loss of nkx2.1, while in mice nkx2.4 expression depends on nkx2.1 and thus the nkx2.1 phenotype is much stronger.
In nkx2TKD morphants the preoptic region is less severely affected, and the SPV appears to develop normally. Both the basal hypothalamus and the preoptic region express nkx2.1, except for a small alar plate region of the SPV area characterized by Orthopedia (otp) expression (Puelles and Rubenstein, 2003). otp is also expressed in the posterior tuberculum and in the arcuate nucleus region. Interestingly, in nkx2TKD morphants the otpa expressing domain in the preoptic SPV area was not affected, while the posterior tubercular otpa domain was absent. This is consistent with otpa in the preoptic SPV not being co-expressed with nkx2.1/4a/4b, while in the posterior tuberculum nkx2.1 is coexpressed (Ryu et al., 2007). The differentiation of neuroendocrine cells in the preoptic region has been studied in detail (Kurrasch et al., 2009; Machluf et al., 2011; Herget et al., 2014) and aids in dissecting the neuroanatomical structures effected in nkx2.1 mutants. The less severe preoptic phenotype is in line with our finding that neuroendocrine cells develop in the preoptic region of nkx2TKD morphants. In contrast, development of essentially all tested basal hypothalamic neuroendocrine cells depends on nkx2.1/4a/4b activity.
The major subdivisions of the larval zebrafish telencephalon, pallium, striatum (dorsal part of area ventralis telencephali, Sdd) and pallidum (ventral part of area ventralis telencephali, Sdv), have been defined by marker gene expression including lhx6 and dlx2a (Mueller et al., 2008; Ganz et al., 2011). Based on loss of lhx6 expression, triple morphants have a severely reduced pallidum. Mammalian lhx6 contains a highly conserved Nkx2.1 binding site in its promoter (Du et al., 2008), suggesting a direct and evolutionary conserved regulation. The ventral expansion of the striatal lhx5 expression domain suggests that in the absence of nkx2.1 activity, striatal fate expands into the pallidum, resulting in a dorsalization of the ventral telencephalon similar to that observed in nkx2.1 mutant mice (Sussel et al., 1999; Marín et al., 2002). When we analyzed emx expression, we did not observed an expansion of the pallium, again similar to nkx2.1 mutant mice (Sussel et al., 1999; Marín et al., 2002), but distinct from reports for nkx2.1 knockdown in Xenopus (van den Akker et al., 2008).
In summary, our data reveals that nkx2.1, nkx2.4a, and nkx2.4b genes act partially redundant in zebrafish hypothalamic development. nkx2.1 is specifically involved in the development of rostral ventral forebrain including the pallidum and preoptic regions, but its function in the basal hypothalamus appears redundant with both nkx2.4 genes. In contrast, nkx2.4a and nkx2.4b control aspects of basal hypothalamus development including the intermediate and caudal hypothalamus, where loss of nkx2.4 activity is not fully compensated by nkx2.1.
Author contributions
Wolfgang Driever and Martha Manoli designed the study. Martha Manoli performed all experiments and documentation. Martha Manoli and Wolfgang Driever carried out data analysis and wrote and edited the manuscript. Wolfgang Driever obtained funding and supervised the project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the zebrafish community for plasmids and probes. We thank Thomas Müller, Jörn Schweitzer, Meta Rath, and Alida Filippi for discussion and comments on the manuscript. S. Götter and R. Schlenvogt provided expert care of the fish. This work was supported by the DFG (EXC294-BIOSS), GRK1104 (Martha Manoli), and by the European Commission (FP7, mesDANEURODEV 222999, DOPAMINET 223744, ZFHEALTH 242048) (Wolfgang Driever).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnana.2014.00145/abstract
References
- Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armant O., März M., Schmidt R., Ferg M., Diotel N., Ertzer R., et al. (2013). Genome-wide, whole mount in situ analysis of transcriptional regulators in zebrafish embryos. Dev. Biol. 380, 351–362. 10.1016/j.ydbio.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi M. J., Stickney H. L., Devoto S. H. (2000). The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127, 2189–2199. [DOI] [PubMed] [Google Scholar]
- Barth K. A., Wilson S. W. (1995). Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Pierani A., Jessell T. M., Ericson J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–445. 10.1016/S0092-8674(00)80853-3 [DOI] [PubMed] [Google Scholar]
- Butt S. J. B., Sousa V. H., Fuccillo M. V., Hjerling-Leffler J., Miyoshi G., Kimura S., et al. (2008). The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron 59, 722–732. 10.1016/j.neuron.2008.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar G., Lauter G., Hauptmann G. (2007). Distribution of corticotropin-releasing hormone in the developing zebrafish brain. J. Comp. Neurol. 505, 337–351. 10.1002/cne.21496 [DOI] [PubMed] [Google Scholar]
- Del Giacco L., Sordino P., Pistocchi A., Andreakis N., Tarallo R., Di Benedetto B., et al. (2006). Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev. Biol. 6:50. 10.1186/1471-213X-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T., Xu Q., Ocbina P. J., Anderson S. A. (2008). NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development 135, 1559–1567. 10.1242/dev.015123 [DOI] [PubMed] [Google Scholar]
- Eisen J. S., Smith J. C. (2008). Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743. 10.1242/dev.001115 [DOI] [PubMed] [Google Scholar]
- Ekker S. C., Ungar A. R., Greenstein P., Kessler D. P. V., Porter J. A., Moon R. T., et al. (1995). Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr. Biol. 5, 944–955. 10.1016/S0960-9822(95)00185-0 [DOI] [PubMed] [Google Scholar]
- Elsalini O. A., Gartzen J., V., Cramer M., Rohr K. B. (2003). Zebrafish hhex, nk2.1a, and pax2.1 regulate thyroid growth and differentiation downstream of Nodal-dependent transcription factors. Dev. Biol. 263, 67–80. 10.1016/S0012-1606(03)00436-6 [DOI] [PubMed] [Google Scholar]
- Ericson J., Rashbass P., Schedl A., Brenner-Morton S., Kawakami A., van Heyningen V., et al. (1997). Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180. 10.1016/S0092-8674(00)80323-2 [DOI] [PubMed] [Google Scholar]
- Ermakova G. V., Solovieva E. A., Martynova N. Y., Zaraisky A. G. (2007). The homeodomain factor Xanf represses expression of genes in the presumptive rostral forebrain that specify more caudal brain regions. Dev. Biol. 307, 483–497. 10.1016/j.ydbio.2007.03.524 [DOI] [PubMed] [Google Scholar]
- Filippi A., Mahler J., Schweitzer J., Driever W. (2010). Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J. Comp. Neurol. 518, 423–438. 10.1002/cne.22213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A., Izpisúa-Belmonte J. C., Fromental-Ramain C., Duboule D. (1994). Expression of the zebrafish gene hlx-1 in the prechordal plate and during CNS development. Development 120, 71–81. [DOI] [PubMed] [Google Scholar]
- Ganz J., Kaslin J., Freudenreich D., Machate A., Geffarth M., Brand M. (2011). Subdivisions of the adult zebrafish subpallium by molecular marker analysis. J. Comp. Neurol. 520, 633–655. 10.1002/cne.22757 [DOI] [PubMed] [Google Scholar]
- Guazzi S., Price M., Felice M., D., Damante G., Mattei M. G., Di Lauro R. (1990). Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 9, 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen I. A., To T. T., Wortmann S., Burmester T., Winkler C., Meyer S. R., et al. (2003). The pro-opiomelanocortin gene of the zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 303, 1121–1128. 10.1016/S0006-291X(03)00475-3 [DOI] [PubMed] [Google Scholar]
- Hauptmann G., Gerster T. (2000). Regulatory gene expression patterns reveal transverse and longitudinal subdivisions of the embryonic zebrafish forebrain. Mech. Dev. 91, 105–118. 10.1016/S0925-4773(99)00277-4 [DOI] [PubMed] [Google Scholar]
- Herget U., Wolf A., Wullimann M. F., Ryu S. (2014). Molecular neuroanatomy and chemoarchitecture of the neurosecretory preoptic-hypothalamic area in zebrafish larvae. J. Comp. Neurol. 522, 1542–1564. 10.1002/cne.23480 [DOI] [PubMed] [Google Scholar]
- Herzog W., Zeng X., Lele Z., Sonntag C., Ting J.-W., Chang C.-Y., et al. (2003). Adenohypophysis formation in the zebrafish and its dependence on sonic hedgehog. Dev. Biol. 254, 36–49. 10.1016/S0012-1606(02)00124-0 [DOI] [PubMed] [Google Scholar]
- Kiecker C., Lumsden A. (2004). Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat. Neurosci. 7, 1242–1249. 10.1038/nn1338 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kimura S., Hara Y., Pineau T., Fernandez-Salguero P., Fox C. H., Ward J. M., et al. (1996). The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69. 10.1101/gad.10.1.60 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. (1991a). Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature 353, 267–270. 10.1038/353267a0 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Moens U., Ericson J. U., Fjose A. (1991b). Zebrafish pax[zf-a]: a paired box-containing gene expressed in the neural tube. EMBO J. 10, 3609–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch D. M., Nevin L. M., Wong J. S., Baier H., Ingraham H. A. (2009). Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural Dev. 4:22. 10.1186/1749-8104-4-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter G., Söll I., Hauptmann G. (2011). Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev. 6:10. 10.1186/1749-8104-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter G., Söll I., Hauptmann G. (2013). Molecular characterization of prosomeric and intraprosomeric subdivisions of the embryonic zebrafish diencephalon. J. Comp. Neurol. 521, 1093–1118. 10.1002/cne.23221 [DOI] [PubMed] [Google Scholar]
- Lohr H., Hammerschmidt M. (2011). Zebrafish in endocrine systems: recent advances and implications for human disease. Annu. Rev. Physiol. 73, 183–211. 10.1146/annurev-physiol-012110-142320 [DOI] [PubMed] [Google Scholar]
- Lumsden A., Krumlauf R. (1996). Patterning the vertebrate neuraxis. Science 274, 1109–1115. 10.1126/science.274.5290.1109 [DOI] [PubMed] [Google Scholar]
- Machluf Y., Gutnick A., Levkowitz G. (2011). Development of the zebrafish hypothalamus. Ann. N.Y. Acad. Sci. 1220, 93–105. 10.1111/j.1749-6632.2010.05945.x [DOI] [PubMed] [Google Scholar]
- Marín O., Baker J., Puelles L., Rubenstein J. L. R. (2002). Patterning of the basal telencephalon and hypothalamus is essential for guidance of cortical projections. Development 129, 761–773. [DOI] [PubMed] [Google Scholar]
- Mizuno K., Gonzalez F. J., Kimura S. (1991). Thyroid-specific enhancer-binding protein (T/EBP): cDNA cloning, functional characterization, and structural identity with thyroid transcription factor TTF-1. Mol. Cell. Biol. 11, 4927–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno N., Gonzalez A. (2011). The non-evaginated secondary prosencephalon of vertebrates. Front. Neuroanat. 5:12. 10.3389/fnana.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller T., Wullimann M. F., Guo S. (2008). Early teleostean basal ganglia development visualized by Zebrafish Dlx2a, Lhx6, Lhx7, Tbr2 (eomesa), and GAD67 gene expression. J. Comp. Neurol. 507, 1245–1257. 10.1002/cne.21604 [DOI] [PubMed] [Google Scholar]
- Odenthal J., Nusslein-Volhard C. (1998). fork head domain genes in zebrafish. Dev. Genes Evol. 208, 245–258. 10.1007/s004270050179 [DOI] [PubMed] [Google Scholar]
- Ogasawara M., Shigetani Y., Suzuki S., Kuratani S., Satoh N. (2001). Expression of thyroid transcription factor-1 (TTF-1) gene in the ventral forebrain and endostyle of the agnathan vertebrate, Lampetra japonica. Genesis 30, 51–58. 10.1002/gene.1032 [DOI] [PubMed] [Google Scholar]
- Pabst O., Herbrand H., Arnold H. H. (1998). Nkx2-9 is a novel homeobox transcription factor which demarcates ventral domains in the developing mouse CNS. Mech. Dev. 73, 85–93. 10.1016/S0925-4773(98)00035-5 [DOI] [PubMed] [Google Scholar]
- Pera E. M., Kessel M. (1998). Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev. Genes Evol. 208, 168–171. 10.1007/s004270050170 [DOI] [PubMed] [Google Scholar]
- Price M. (1993). Members of the Dlx- and Nkx2-gene families are regionally expressed in the developing forebrain. J. Neurobiol. 24, 1385–1399. 10.1002/neu.480241010 [DOI] [PubMed] [Google Scholar]
- Puelles L., Rubenstein J. L. (1993). Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 16, 472–479. 10.1016/0166-2236(93)90080-6 [DOI] [PubMed] [Google Scholar]
- Puelles L., Rubenstein J. L. R. (2003). Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 26, 469–476. 10.1016/S0166-2236(03)00234-0 [DOI] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., et al. (2007). p53 activation by knockdown technologies. PLoS Genet. 3:e78. 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Scott M. P. (2007). Patching the gaps in Hedgehog signalling. Nat. Cell Biol. 9, 1005–1009. 10.1038/ncb435 [DOI] [PubMed] [Google Scholar]
- Rohr K. B., Barth K. A., Varga Z. M., Wilson S. W. (2001). The nodal pathway acts upstream of hedgehog signaling to specify ventral telencephalic identity. Neuron 29, 341–351. 10.1016/S0896-6273(01)00210-0 [DOI] [PubMed] [Google Scholar]
- Ryu S., Holzschuh J., Erhardt S., Ettl A. K., Driever W. (2005). Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 102, 18467–18472. 10.1073/pnas.0506187102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S., Mahler J., Acampora D., Holzschuh J., Erhardt S., Omodei D., et al. (2007). Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr. Biol. 17, 873–880. 10.1016/j.cub.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Sander M., Paydar S., Ericson J., Briscoe J., Berber E., German M., et al. (2000). Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 14, 2134–2139. 10.1101/gad.820400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S., Wolf O., Brand M., Lumsden A. (2006). Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development 133, 855–864. 10.1242/dev.02248 [DOI] [PubMed] [Google Scholar]
- Shimamura K., Hartigan D. J., Martinez S., Puelles L., Rubenstein J. L. (1995). Longitudinal organization of the anterior neural plate and neural tube. Development 121, 3923–3933. [DOI] [PubMed] [Google Scholar]
- Small E. M., Vokes S. A., Garriock R. J., Li D., Krieg P. A. (2000). Developmental expression of the Xenopus Nkx2-1 and Nkx2-4 genes. Mech. Dev. 96, 259–262. 10.1016/S0925-4773(00)00400-7 [DOI] [PubMed] [Google Scholar]
- Strähle U., Blader P., Henrique D., Ingham P. W. (1993). Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 7, 1436–1446. 10.1101/gad.7.7b.1436 [DOI] [PubMed] [Google Scholar]
- Sussel L., Marin O., Kimura S., Rubenstein J. L. (1999). Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370. [DOI] [PubMed] [Google Scholar]
- Tanabe Y., Jessell T. M. (1996). Diversity and pattern in the developing spinal cord. Science 274, 1115–1123. 10.1126/science.274.5290.1115 [DOI] [PubMed] [Google Scholar]
- Tessmar-Raible K., Raible F., Christodoulou F., Guy K., Rembold M., Hausen H., et al. (2007). Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129, 1389–1400. 10.1016/j.cell.2007.04.041 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (1999). Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development 126, 229–240. [DOI] [PubMed] [Google Scholar]
- Toyama R., Curtiss P. E., Otani H., Kimura M., Dawid I. B., Taira M. (1995). The LIM class homeobox gene lim5: implied role in CNS patterning in Xenopus and zebrafish. Dev. Biol. 170, 583–593. 10.1006/dbio.1995.1238 [DOI] [PubMed] [Google Scholar]
- Uemae Y., Sakamoto J., Hidaka Y., Hiratsuka A., Susa T., Kato Y., et al. (2014). Gene expression, function, and diversity of Nkx2-4 in the rainbow trout, Oncorhynchus mykiss. Gen. Comp. Endocrinol. 206, 193–202. 10.1016/j.ygcen.2014.07.007 [DOI] [PubMed] [Google Scholar]
- van den Akker W. M. R., Brox A., Puelles L., Durston A. J., Medina L. (2008). Comparative functional analysis provides evidence for a crucial role for the homeobox gene Nkx2.1/Titf-1 in forebrain evolution. J. Comp. Neurol. 506, 211–223. 10.1002/cne.21542 [DOI] [PubMed] [Google Scholar]
- Wang C.-C., Brodnicki T., Copeland N. G., Jenkins N. A., Harvey R. P. (2000). Conserved linkage of NK-2 homeobox gene pairs Nkx2-2/2-4 and Nkx2-1/2-9 in mammals. Mamm. Genome 11, 466–468. 10.1007/s003350010089 [DOI] [PubMed] [Google Scholar]
- Wen L., Wei W., Gu W., Huang P., Ren X., Zhang Z., et al. (2008). Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev. Biol. 314, 84–92. 10.1016/j.ydbio.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book: A Guide to the Laboratory Use of Zebrafish (Danio rerio): Eugene, OR: Univ. of Oregon Press. [Google Scholar]
- Wullimann M. F., Rink E. (2001). Detailed immunohistology of Pax6 protein and tyrosine hydroxylase in the early zebrafish brain suggests role of Pax6 gene in development of dopaminergic diencephalic neurons. Brain Res. Dev. Brain Res. 131, 173–191. 10.1016/S0165-3806(01)00270-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.