Abstract
Aeromonas strains isolated from sediments upstream and downstream of a water resource recovery facility (WRRF) over a two-year time period were tested for susceptibility to thirteen antibiotics. Incidence of resistance to antibiotics, antibiotic resistance phenotypes, and diversity (based on resistance phenotypes) were compared in the two populations. At the beginning of the study, the upstream and downstream Aeromonas populations were different for incidence of antibiotic resistance (p < 0.01), resistance phenotypes (p < 0.005), and diversity. However, these differences declined over time and were not significant at the end of the study. These results (1) indicate that antibiotic resistance in Aeromonas in stream sediments fluctuates considerably over time and (2) suggest that WRRF effluent does not, when examined over the long term, affect antibiotic resistance in Aeromonas in downstream sediment.
Keywords: water resource recovery facility effluent, antibiotic resistance, Aeromonas, stream sediments
Introduction
Resistance to antibiotics poses a serious threat to human health. Resistance in pathogens increases morbidity and mortality. It also increases healthcare costs due to the need for alternatives to standard antibiotic treatment (McGowan 2001). Antibiotic resistance in bacteria from clinical and community settings has been the subject of intensive study. However, less is known about antibiotic resistance outside these settings.
It has been shown that resistance to antibiotics is not unusual in environmental bacteria, occurring in bacteria from wastewater, surface water, groundwater, and in soils and sediments (Baquero et al. 2008; Faria et al. 2009; Goni-Urriza et al. 2000; Kummerer 2004; Mispagel and Gray 2005; Schwartz et al. 2003; Watkinson et al. 2007). It has been suggested that injudicious use of antibiotics is linked to the spread of resistance in the environment and to human pathogens. It has also been suggested that antibiotics in the environment promote the development and dissemination of resistance in environmental bacteria, which can then be transferred to human pathogens or commensals (Cabello 2006; Witte 1998; Rhodes et al. 2000). Antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes can enter the environment through anthropogenic inputs such as agriculture, septic systems, and water resource recovery facilities (Rosenblatt-Farrell 2009).
Water resource recovery facilities are generally very effective at reducing excess nutrients and toxic compounds associated with raw wastewater. However, treated wastewater has been shown to contain trace amounts of antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes (Kim and Aga 2007; Zhang et al. 2009a). Since the discharge volume can be substantial, exposure to water resource recovery facility effluent may affect resistance in downstream bacteria.
Bacteria from the genus Aeromonas are ubiquitous in both natural and man-made aquatic ecosystems (Holmes et al. 1996; Martone-Rocha et al. 2010; Poffe and Op de Beeck 1991). They are planktonic in water, but also form biofilms in sediment in freshwater streams, drinking water systems, and water resource recovery facilities (Andersson et al. 2008; Chauret et al. 2001; Keevil 2003; Zalmum et al. 1998; Peduzzi et al. 1992; Szabo et al. 2011). Aeromonas represent 9-20% of cultivable bacteria in biofilms from freshwater sediment (Peduzzi et al. 1992; Szabo et al. 2011). Clonal lineages of Aeromonas can persist in the environment for 3 years (Rahman et al. 2007). In addition, Aeromonas strains have been linked to a variety of illnesses in humans, particularly in immunocompromised individuals (Janda and Abbott 2010; Parker and Shaw 2011). Because of their persistence in the environment and their medical relevance, Aeromonas is ideally suited for studies concerning the effect of water resource recovery facility effluent on the development and persistence of antibiotic resistance in the environment and on the dissemination of resistance from the environment to human pathogens and commensals. In this study, conducted over a two-year period, the incidence and patterns of antibiotic resistance in Aeromonas strains from sediments upstream and downstream of a water resource recovery facility were compared. Aeromonas strains were isolated from creek sediments rather than water because Aeromonas in biofilms in sediment are more likely to be resident in the ecosystem than bacteria transiting through the sampling site in the water and, therefore, more appropriate for a long-term study.
Materials and Methods
Study sites and sample collection
The Tahlequah water resource recovery facility (WRRF) began operating at its present location in 1972. It is a tertiary treatment facility that processes primarily domestic wastewater including a small amount of hospital waste that is not pre-treated. Wastewater treatment consisted of i) screening and grit removal, ii) biological nutrient removal in aeration tanks, iii) precipitation of phosphate by addition of aluminum sulfate (Al2(SO4)3) followed by filtration through sand and anthracite, and iv) disinfection by ultraviolet (UV) light. The facility began disinfecting wastewater with UV in 1992 and during the study used a UV3000plus disinfection system (Trojan Technologies, Ontario, Canada). Monthly averages of total coliforms/100ml (October-April) or fecal coliforms/100ml (May-September) present in Tahlequah WRRF effluent in 2007, 2008, and 2009 are shown in Table 1. Treated wastewater (570-760m3) was discharged into Tahlequah Creek every 90 minutes. Each discharge took approximately 45 minutes and during discharge the stream volume and flow rate increased 2- to 3-fold over base flow.
Table 1. Monthly averages for total coliforms or fecal coliforms in Tahlequah water resource recovery facility effluent 2007-2009.
| Year | Month | Total coliforms/100 mla | Fecal coliforms/100mlb |
|---|---|---|---|
| 2007 | Jan | 16 | |
| Feb | 25 | ||
| Mar | 11 | ||
| Apr | 80 | ||
| May | 10 | ||
| Jun | 14 | ||
| Jul | 4 | ||
| Aug | 37 | ||
| Sep | 33 | ||
| Oct | 215 | ||
| Nov | 185 | ||
| Dec | 217 | ||
| 2008 | Jan | 189 | |
| Feb | 318 | ||
| Mar | 134 | ||
| Apr | 127 | ||
| May | 41 | ||
| Jun | 34 | ||
| Jul | 3 | ||
| Aug | 26 | ||
| Sep | 15 | ||
| Oct | 101 | ||
| Nov | 1118 | ||
| Dec | 464 | ||
| 2009 | Jan | 177 | |
| Feb | 622 | ||
| Mar | 1700 | ||
| Apr | 3313 | ||
| May | 51 | ||
| Jun | 27 | ||
| Jul | 12 | ||
| Aug | 14 | ||
| Sep | 19 | ||
| Oct | 99 | ||
| Nov | 352 | ||
| Dec | 241 |
Tahlequah WRRF Oklahoma/National Pollutant Discharge Elimination System (OPDES/NPDES) permit limitations: Daily Maximum Discharge of 20,000 coliforms/100 ml and an Average (geometric mean) Monthly Discharge of 5,000 coliforms/100ml.
Tahlequah WRRF Oklahoma/National Pollutant Discharge Elimination System (OPDES/NPDES) permit limitations: Daily Maximum Discharge of 400 fecal coliforms/100 ml and an Average (geometric mean) Monthly Discharge of 200 fecal coliforms/100ml.
Tahlequah Creek is a 2.1 km second-order stream that flows into the Illinois River. Its two branches (Ross Branch and Town Branch, 7.3 km and 10.1 km in length, respectively) have multiple springs feeding into them. The confluence of the two branches and the beginning of Tahlequah Creek is roughly 1.3 km upstream of the discharge point of the Tahlequah WRRF (EPA My WATERS Mapper, http://watersgeo.epa.gov). Land use in the Town Branch watershed is primarily urban with some agricultural uses in the headwaters north of Tahlequah. Land use in the Ross Branch watershed is a mix of agricultural, forest, and urban (in descending order). Land use in the Tahlequah Creek watershed is a mix of forest, agricultural, and the WRRF.
Water and sediment samples were collected approximately 200 m upstream and 100 m downstream of the Tahlequah WRRF discharge site. The upstream and downstream sampling sites were very similar with the exception that the downstream location was exposed to treated wastewater from the plant. For the purposes of this study, the upstream site served as a control.
In November 2007 upstream and downstream water samples were collected and assayed for the presence of antibiotics with the assistance of the United States Geological Survey (USGS). Downstream water samples were taken during discharge of treated wastewater. Water was collected from a single vertical point in Tahlequah Creek, filtered with a 0.7-micron pore size baked glass-fiber filter, and shipped in amber baked-glass bottles chilled to 4°C. Samples were analyzed for antibiotics and antibiotic residues using solid-phase extraction and liquid chromatography/mass spectrometry (LC/MS) at the USGS Kansas Water Science Center.
Sediment samples were collected from Tahlequah Creek in November 2007 to coincide with the antibiotic analysis. Subsequent sediment sampling was brought forward to May in 2008 and repeated in May 2009. Analysis of the May data sets revealed that these results differed from that of November 2007. Accordingly, a fourth sampling date was added, November 2009, in order to investigate possible seasonal effects.
Sterile 60cc Teflon scoops were used to collect upstream and downstream sediment samples, and the sediment samples were transferred into sterile containers. Samples were placed on ice and processed within 6 hours as recommended for environmental samples containing microorganisms (Hurst et al. 2002). Sediment samples were taken from approximately the same locations on each date. Sediments at both sampling sites were similar. They were composed of chert (microcrystalline quartz, > 95%) and limestone (< 5%). They were primarily gravel-sized (> 95%) with some clay-sized particles (< 3%) and very little sand or silt (< 1% each). Grains were primarily very angular to subangular (approximately 40% each) with some subround (approximately 20%). Sediments were poorly sorted (i.e., the sediment particles varied in size).
Precipitation in the Tahlequah Creek watershed varied on the different sampling dates: in November 2007 and May 2008 0.5-1.0 inches of rain fell the week prior to sampling, in May 2009 > 1.5 inches of rain fell the week prior to sampling, and in November 2009 no precipitation was measured the week prior to sampling (Oklahoma Climatological Survey Mesonet website, www.mesonet.org, and Tahlequah WRRF records).
Isolation and identification of Aeromonas from sediment
Sterile distilled water (100ml) was added to the sediment samples described above, samples were shaken for 3 minutes, and large particulates were allowed to settle. One ml of water from the prepared sediment samples (both undiluted and diluted 10-fold in sterile water) was added directly to the differential media Coliscan® or ECA Check® EasyGel (Micrology Laboratories, Goshen, IN) per the manufacturer's instructions. In addition, as most Aeromonas spp. are intrinsically resistant to ampicillin (Clinical and Laboratory Standards Institute 2006; Rossolini et al. 1996), ampicillin was added to the differential media at a concentration of 32μg/ml. Five plates each were prepared using undiluted and diluted sediment samples per sampling site. Plates were incubated at 35°C for 36 hours, and 50 putative Aeromonas colonies were selected from both upstream sediment and downstream sediment samples for additional analysis. Cultures were purified by sub-culturing on BBL™ Mueller Hinton II Agar (BD, Franklin Lakes, NJ) containing 32 μg/ml ampicillin and stored at -80°C (Microbank®, Pro-Lab Diagnostics, Austin, TX).
Total DNA was extracted from overnight bacterial cultures using a PurElute™ Bacterial Genomic Kit (Edge BioSystems, Gaithersburg, MD) or an UltraClean® Microbial DNA Isolation Kit (MoBio Laboratories, Inc., Carlsbad, CA). DNA was quantitated using a Qubit® fluorometer and Quant-iT™ dsDNA Broad Range Assay Kit (Invitrogen Corporation, Carlsbad, CA). 16S rRNA gene sequences were amplified using universal primers, 8F and 805R (Lee et al. 2007). Amplification reactions were performed in a volume of 50μl containing 100 ng DNA, 1 mM MgSO4, 0.3 mM of each dNTP, 0.3 μM of each primer, 1× Pfx amplification buffer, and 1 unit Platinum® Pfx DNA polymerase (Invitrogen Corporation, Carlsbad, CA). The amplification program consisted of an initial denaturation step of 95°C for 5 min, followed by 35 cycles of 15 sec at 95°C, 30 sec at 55°C, 68°C for 1 min, and a final extension step at 68°C for 10 min. PCR products were purified for sequencing using E.Z.N.A.® Cycle Pure or Gel Extraction Kits (Omega Bio-Tek Inc., Norcross, GA). Isolates were identified from these sequences using the 16S rRNA gene database, GreenGenes (DeSantis et al. 2006).
Antibiotic susceptibility testing
Antibiotic susceptibility tests were performed using Etest strips (bioMérieux, Durham, NC) and BBL™ Sensi-Discs (BD, Franklin Lakes, NJ) according to the manufacturer's instructions and CLSI recommendations (Clinical and Laboratory Standards Institute 2006). E. coli strains ATCC 25922 and 35218 were included as controls. Aeromonas isolates were tested for susceptibility to thirteen antibiotics (type of test, antibiotic abbreviation, and antibiotic concentration where applicable are indicated in parentheses): tetracycline (Etest, TET), ciprofloxacin (Sensi-Disc, CIP, 5 μg), gentamicin (Etest, GM), chloramphenicol (Etest, CHL), ampicillin (Sensi-Disc, AM, 10 μg), amoxicillin plus clavulanic acid (Sensi-Disc, AMC, 30 μg), cefazolin (Sensi-Disc, CZ, 30 μg), ceftazidime (Sensi-Disc, CAZ, 30 μg), cefotaxime (Sensi-Disc, CTX, 30 μg), cefepime (Sensi-Disc, FEP, 30 μg), cefoxitin (Sensi-Disc, FOX, 30 μg), aztreonam (Sensi-Disc, ATM, 30 μg), and imipenem (Sensi-Disc, IPM, 10 μg). After incubation at 35°C for 18 hours isolates were defined as resistant when the inhibition zone diameters surrounding the Sensi-Discs were less than or equal to 15mm for CIP, 13 mm for AM, 13 mm for AMC, 14 mm for CZ, 14 mm for CAZ, 14 mm for CTX, 14 mm for FEP, 14 mm for FOX, 15 mm for ATM, and 13 mm for IPM or the minimum inhibitory concentrations (MICs) were greater than or equal to 16 μg/ml for TET, 16 μg/ml for GM, and 32 μg/ml for CHL when using Etest strips (Clinical and Laboratory Standards Institute 2006).
Antibiotics from five groups were tested including aminoglycosides (gentamicin), quinolones (ciprofloxacin), phenicols (chloramphenicol), tetracyclines (tetracycline), and beta-lactams. The beta-lactam antibiotics used in these tests belong to the following classes: penicillins (ampicillin), cephems (cephalosporin I-cefazolin, cephalosporin II-cefoxitin, cephalosporin III-cefotaxime and ceftazidime, and cephalosporin IV-cefepime), monobactams (aztreonam), penems (imipenem), and beta-lactams with beta-lactamase inhibitors (amoxicillin with clavulanic acid).
Analysis of Aeromonas populations
Dendrograms representing strain relatedness were determined using the unweighted pair group method using arithmetic means (UPGMA) based on 16S rRNA gene sequences (Bionumerics 6.5 software, Applied Maths, Austin, TX). Aeromonas isolates from upstream and downstream of the water resource recovery facility were grouped and analyzed by sampling date or combined for analysis. In addition, antibiotic resistance phenotypes in the upstream and downstream Aeromonas isolates were compared.
Statistical analysis
Fisher's exact test of independence was used to compare the incidence of antibiotic resistance in the two Aeromonas populations (isolates from sediments upstream and from sediments downstream of the water resource recovery facility) on each sampling date, for sampling dates combined, and for comparison of incidence of antibiotic resistances in the two seasons (November vs. May). χ2 and paired t-tests were used to compare antibiotic resistance phenotypes of upstream and downstream Aeromonas isolates on each sampling date, for sampling dates combined, and for comparison of resistance phenotypes present in November vs. May. Statistical analysis was performed using Microsoft® Excel® 2008 for Mac (Microsoft Corporation, Redmond, WA). p-values < 0.05 were considered to be significant. Diversity of the upstream and downstream Aeromonas populations was calculated using Simpson's index of diversity according to the formula Di = 1-Σ[Ni × (Ni-1)/[N × (N-1)] where Ni equals the number of isolates assigned to the ith antibiotic resistance phenotype and N is the total number of isolates examined (Kuhn et al. 1997; Hurst et al. 2002).
Results
Antibiotics in Tahlequah Creek water samples
In November 2007 water samples taken upstream and downstream of the Tahlequah, OK water resource recovery facility were analyzed for 25 antibiotic residues by the Kansas Water Science Center (Kolpin et al. 2002) (Table 2). No antibiotics were detected in the upstream water sample. The downstream water sample contained 0.042 μg/L azithromycin, 0.006 μg/L ciprofloxacin, 0.039 μg/L ofloxacin, and 0.024 μg/L trimethoprim.
Table 2. Antibiotics detected in water samples collected upstream and downstream of the Tahlequah water resource recovery facility in November 2007.
| Antibiotic | Detection limit | Upstream | Downstream |
|---|---|---|---|
| azithromycin | <0.005 μg/L | NDa | 0.042 μg/L |
| chloramphenicol | <0.1 μg/L | ND | ND |
| chlorotetracycline | <0.01 μg/L | ND | ND |
| ciprofloxacin | <0.005 μg/L | ND | 0.006 μg/L |
| doxycycline | <0.01 μg/L | ND | ND |
| enrofloxacin | <0.005 μg/L | ND | ND |
| erythromycin | <0.008 μg/L | ND | ND |
| lincomycin | <0.005 μg/L | ND | ND |
| lomefloxacin | <0.005 μg/L | ND | ND |
| norfloxacin | <0.005 μg/L | ND | ND |
| ofloxacin | <0.005 μg/L | ND | 0.039 μg/L |
| ormetoprim | <0.005 μg/L | ND | ND |
| oxytetracycline | <0.01 μg/L | ND | ND |
| roxithromycin | <0.005 μg/L | ND | ND |
| sarafloxacin | <0.005 μg/L | ND | ND |
| sulfachloropyridazine | <0.005 μg/L | ND | ND |
| sulfadiazine | <0.1 μg/L | ND | ND |
| sulfadimethoxine | <0.005 μg/L | ND | ND |
| sulfamethazine | <0.005 μg/L | ND | ND |
| sulfamethoxazole | <0.005 μg/L | ND | ND |
| sulfathiazole | <0.02 μg/L | ND | ND |
| tetracycline | <0.01 μg/L | ND | ND |
| trimethoprim | <0.005 μg/L | ND | 0.024 μg/L |
| tylosin | <0.005 μg/L | ND | ND |
| virginiamycin | <0.005 μg/L | ND | ND |
ND = none detected
Incidence of antibiotic resistance in Aeromonas isolates from sediments upstream and downstream of the Tahlequah WRRF
Aeromonas strains isolated from sediments upstream and downstream of the Tahlequah water resource recovery facility in November 2007, May 2008, May 2009, and November 2009 were tested for susceptibility to thirteen antibiotics. Resistance to six antibiotics (not including ampicillin) from five classes was detected: ciprofloxacin, tetracycline, chloramphenicol, amoxicillin plus clavulanic acid, cefazolin, and cefoxitin (Table 3). The most commonly encountered resistance was to cefazolin, followed by resistance to amoxicillin plus clavulanic acid, cefoxitin, tetracycline, and ciprofloxacin and chloramphenicol (one strain each). None of the Aeromonas strains were resistant to gentamicin, ceftazidime, cefotaxime, cefepime, aztreonam, or imipenem. Significant differences in the incidence of antibiotic resistance in the upstream and downstream Aeromonas populations were found for three antibiotics: amoxicillin plus clavulanic acid (AMC), cefazolin (CZ), and cefoxitin (FOX) (Table 3). The incidence of resistance to AMC and to CZ was higher in the downstream population than in the upstream population (AMC, sampling dates combined and November 2007, p< 0.001; CZ, sampling dates combined, p< 0.01 and CZ, November 2007, p< 0.0001). In contrast, the incidence of resistance to FOX was higher in the upstream population than in the downstream population (sampling dates combined and May 2008, p<0.05). In May 2009 and in November 2009 the incidence of antibiotic resistance in the two populations was not significantly different for any of the antibiotics tested.
Table 3. Incidence of antibiotic resistance in Aeromonas strains isolated from sediments upstream and downstream of the Tahlequah water resource recovery facility.
| Class of antibiotics | Antibiotic | November 2007 | May 2008 | May 2009 | November 2009 | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Upstream (n = 44) % Resistant | Downstream (n = 24) % Resistant | Upstream (n = 42) % Resistant | Downstream (n = 43) % Resistant | Upstream (n = 49) % Resistant | Downstream (n = 46) % Resistant | Upstream (n = 51) % Resistant | Downstream (n = 53) % Resistant | Upstream (n = 186) % Resistant | Downstream (n = 166) % Resistant | ||
| quinolones | ciprofloxacin | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.6 |
| tetracyclines | tetracycline | 2 | 8 | 0 | 0 | 0 | 2 | 0 | 2 | 0.5 | 2.4 |
| chloramphenicol | chloramphenicol | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.6 |
| β-lactam/β-lactamase inhibitor | amoxicillin/clavulanic acid | 41a | 88 a | 40 | 56 | 74 | 83 | 76 | 83 | 59d | 77d |
| cephems | cefazolin | 50 b | 100 b | 98 | 86 | 96 | 98 | 98 | 96 | 86e | 95e |
| cefoxitin | 7 | 4 | 26c | 7 c | 31 | 17 | 0 | 2 | 16f | 7.8f | |
All of the Aeromonas strains were resistant to ampicillin. None of the Aeromonas strains tested were resistant to gentamicin, ceftazidime, cefotaxime, cefepime, aztreonam, or imipenem. Differences between upstream and downstream populations that were statistically significant according to Fisher's exact test of independence are indicated by superscripts.
p = 0.0002,
p < 0.0001,
p = 0.021,
p = 0.001,
p = 0.008,
p = 0.032
Seasonal differences in the incidence of antibiotic resistance in the upstream and downstream Aeromonas populations were also observed. The incidence of resistance to cefoxitin (FOX) in both the upstream and downstream populations was higher in May than in November (p<0.05). The incidence of resistance to cefazolin (CZ) in the upstream population only was higher in May than November (p<0.0005). The incidence of resistance to amoxicillin plus clavulanic acid (AMC) in the downstream population only was higher in November than in May (p<0.05).
Comparison of Aeromonas upstream and downstream sediment populations
Dendrograms based on 16S rRNA gene sequences showed no evidence of clustering of the upstream and downstream Aeromonas isolates (data not shown). The same or very similar (≥ 97% sequence identity) 16S rRNA alleles were present in both populations. Therefore, there was no evidence of differences between the upstream and downstream Aeromonas populations by this criterion.
Antibiotic resistance phenotypes of Aeromonas isolates from sediments upstream and downstream of the water resource recovery facility were also compared (Table 4). Ten different resistance phenotypes were observed among the 352 Aeromonas isolates. All ten phenotypes were present at some time point in the downstream Aeromonas population. Seven of the ten phenotypes were detected in the upstream population. Three of the phenotypes were detected only once over the two-year period. In all cases this was in the downstream Aeromonas population only. Based on their resistance phenotypes the upstream and downstream Aeromonas populations were significantly different on November 2007, May 2008, and when the sampling dates were combined (p<0.005, p<0.05, and p<0.005, respectively). However, the upstream and downstream populations were not statistically different in May 2009 and November 2009.
Table 4.
Antibiotic resistance phenotypes of Aeromonas isolates from sediments upstream and downstream of the Tahlequah water resource recovery facilitya
| AMC | AM | CHL | CIP | CZ | FOX | TET | November 2007 | May 2008 | May 2009 | November 2009 | % Total (n = 352) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Upstream (n = 44) | % Downstream (n = 24) | % Upstream (n = 42) | % Downstream (n = 43) | % Upstream (n = 49) | % Downstream n (n = 46) | % Upstream (n = 51) | % Downstream (n = 53) | ||||||||
| R | R | R | 30 | 75 | 33 | 44 | 45 | 61 | 76 | 81 | 55.7 | ||||
| R | R | 11 | 8 | 40 | 35 | 20 | 15 | 22 | 13 | 21.0 | |||||
| R | R | R | R | 5 | 0 | 7 | 7 | 24 | 17 | 0 | 2 | 8.2 | |||
| R | 45 | 0 | 0 | 9 | 0 | 0 | 2 | 2 | 7.4 | ||||||
| R | R | R | 2 | 4 | 19 | 0 | 6 | 2 | 0 | 0 | 4.0 | ||||
| R | R | 5 | 0 | 0 | 5 | 4 | 2 | 0 | 0 | 2.0 | |||||
| R | R | R | R | 2 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 0.9 | |||
| R | R | R | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.3 | ||||
| R | R | R | R | R | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | ||
| R | R | R | R | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | |||
Gray boxes indicate that the isolate(s) is resistant to that antibiotic based on the Clinical Laboratory Standards Institute guidelines. Numbers are percentages of isolates with that resistance phenotype. Percentages in columns may not equal 100 due to rounding. AMC = amoxicillin plus clavulanic acid, AM = ampicillin, CHL=chloramphenicol, CIP=ciprofloxacin, CZ=cefazolin, FOX=cefoxitin, TET=tetracycline
Seasonal differences were observed for four of the ten antibiotic resistance phenotypes. The AMCR AMR CZR FOXR phenotype was more abundant in May than in November in both the upstream and downstream populations (p<0.005). The AMR CZR FOXR phenotype was more abundant in the upstream population in May than in November (p<0.005). The AMR CZR phenotype was more abundant in the downstream population in May than in November (p<0.05). The AMR phenotype was more abundant in the upstream population in November than in May (p<0.001).
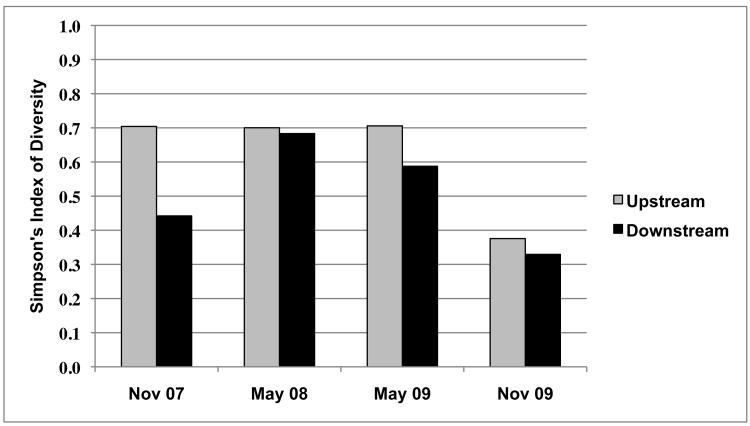
Diversity in the upstream and downstream Aeromonas populations was compared over time based on antibiotic resistance phenotypes (Figure 1). The upstream population had relatively high levels of diversity in November 2007, May 2008 and May 2009 (0.7040, 0.7003, and 0.7058, respectively). However, in November 2009 the diversity level of the upstream population decreased almost two-fold (0.3757). The downstream population exhibited greater variation. Diversity levels were low in November 2007 and November 2009 (0.4420 and 0.3295, respectively), but were relatively high in May 2008 and May 2009 (0.6833 and 0.5874, respectively). Diversity levels in the upstream and downstream Aeromonas populations were similar except in November 2007 when diversity in the downstream population was much lower compared to the upstream population.
Figure 1. Comparison of diversity in upstream and downstream Aeromonas populations based on antibiotic resistance phenotypes (Simpson's index of diversity).
Discussion
It has been shown that antibiotics and other pharmaceuticals are present in water resource recovery facility (WRRF) effluent and in surface waters that receive effluent (this study and Fick et al. 2009; Galloway et al. 2005; Kolpin et al. 2002; Lindberg et al. 2005; Miao et al. 2004). This has led to debate about the impact of these, usually low level, pollutants on the environment and on human health (Cooper et al. 2008; Kim and Aga 2007; Rodriguez-Mozaz and Weinberg 2010). Several studies have been published that examine the association between WRRF effluent and antibiotic resistance (Akiyama and Savin 2010; Goni-Urriza et al. 2000; Zhang et al. 2009b). They show that bacteria downstream of water resource recovery facilities are more likely to be resistant to antibiotics and to greater numbers of antibiotics than upstream bacteria. For example, Goni-Urriza et al. have reported that Aeromonas isolates from water downstream of an urban water resource recovery facility were more likely to be resistant to antibiotics than Aeromonas isolated from water upstream of the plant (Goni-Urriza et al. 2000). The expectation in this study was that any differences in antibiotic resistance observed between upstream and downstream Aeromonas sediment populations would be at least as great if not greater than those observed from water for two reasons. Firstly, it has been shown that in urban playa lakes greater numbers of antibiotic resistant Aeromonas were isolated from sediments than water (Huddleston et al. 2006). Secondly, it has been shown that antibiotics adsorb to soils and stream sediments (Cordova-Kreylos and Scow 2007; Figueroa et al. 2004; ter Laak et al. 2006; Luo et al. 2011; Zhou et al. 2011; Kim and Carlson 2007; Massey et al. 2010) and that some antibiotics can persist in sediments for hundreds of days (Lai et al. 2011; Lin et al. 2010). This adsorption may generate microenvironments in the sediment that promote resistance in bacteria at that location. Indeed, it has been shown that sediments downstream of water resource recovery facilities have higher concentrations of antibiotics than water samples taken at the same location (Kim and Carlson 2007; Massey et al. 2010). Our findings contradicted our expectations and previous reports on the impact of WRRF effluent on downstream bacteria (Akiyama and Savin 2010; Goni-Urriza et al. 2000; Zhang et al. 2009b). However, previous studies, although similar, were not identical. Akiyama et al. (2010) studied Escherichia coli and Zhang et al. (2009b) studied Acinetobacter rather than Aeromonas. Goni-Urriza et al. and Zhang et al. examined antibiotic resistance in Aeromonas and Acinetobacter, respectively, over time periods of less than one year. Furthermore, Goni-Urriza et al. isolated their Aeromonas spp. from water rather than from sediments as in this study. One possible explanation for the lack of effect of WRRF effluent on downstream Aeromonas is that this population is continuously being replenished in sediments. However, Aeromonas forms biofilms in sediment (Keevil 2003; Zalmum et al. 1998) and it has been shown that clonal lineages of Aeromonas can persist in the environment for 3 years (Rahman et al. 2007) indicating that the turnover rate of this genus is low.
There are several mechanisms by which WRRF effluent could contribute to antibiotic resistance in downstream sediments. The effluent could contain resistant bacteria that colonize the sediment or transfer genes to autochthonous strains, or it could contain antibiotics that are adsorbed to sediment and result in direct selection of antibiotic resistance in downstream bacteria. The antibiotics ofloxacin, ciprofloxacin (both quinolones), azithromycin (a macrolide), and trimethoprim (a dihydrofolate reductase inhibitor) were detected in water downstream of the Tahlequah WRRF in November 2007 (this study) and have also been reported in nearby streams (Haggard et al. 2006). A small percentage of Aeromonas strains in this study were found to be resistant to ciprofloxacin. Fluoroquinolones such as ciprofloxacin are completely synthetic compounds, not based on natural compounds with antimicrobial activity. Therefore, resistance in environmental bacteria is unexpected, as they would not naturally be exposed to this type of compound. Quinolones have been shown to be adsorbed by soils and stream sediments (Cordova-Kreylos and Scow 2007; Luo et al. 2011; Zhou et al. 2011; Massey et al. 2010). Hence, a possible source of resistance is selection due to absorption of ciprofloxacin from the WRRF effluent by sediment. However, Tahlequah Creek sediments contain very little clay, sand, and silt and consequently are likely to be relatively poor at adsorbing antibiotics. This may be a reason why the numbers of ciprofloxacin resistant Aeromonas isolated were low and, more generally, why the incidence of antibiotic resistance in the upstream and downstream Aeromonas populations was similar at the end of the study although it does not explain the differences observed in November 2007.
Upstream and downstream Aeromonas populations were first compared based on the incidence of antibiotic resistance. The results from the November 2007 sampling were consistent with the hypothesis that exposure to WRRF effluent affects antibiotic resistance in downstream bacteria. The downstream Aeromonas population had a higher incidence of resistance to two antibiotics (amoxicillin plus clavulanic acid, AMC, and cefazolin, CZ) compared to the upstream population. However, on the second sampling date the results were quite different. The downstream population did not have a higher incidence of resistance for any of the antibiotics tested. The upstream population, however, had a higher incidence of resistance to cefoxitin (FOX) compared to the downstream population. On subsequent sampling dates no significant differences in the incidence of antibiotic resistance were observed between the upstream and downstream populations. The data show that differences in the incidence of antibiotic resistance could be detected between the two populations, but the differences were short-lived. At the end of two years the two populations were not significantly different.
Comparison of the antibiotic resistance phenotypes present in the upstream and downstream Aeromonas populations revealed a trend similar to the incidences of antibiotic resistance in the populations. The two populations were statistically different on November 2007 and on May 2008, but the differences were again short-lived. In May 2009 and November 2009 the two populations were statistically the same. By the end of the two-year period the resistance phenotypes of Aeromonas strains in the two populations were nearly identical. When the data from all sampling dates was combined there were statistical differences between the populations, but they appeared to be driven by differences between the populations in November 2007 and May 2008.
Diversity levels based on the antibiotic resistance phenotypes present in the upstream and downstream Aeromonas populations followed a similar pattern over time. The two populations were different on November 2007, but on subsequent sampling dates diversity levels in the populations were similar. Interestingly, the general trend in antibiotic resistance phenotypes in both Aeromonas populations over the two-year time period was towards reduced diversity. By November 2009 two resistance phenotypes were dominant in both populations: >90% of Aeromonas isolates from both populations had one of two resistance phenotypes.
For all comparisons, incidence of antibiotic resistance, resistance phenotypes, and diversity, differences were observed between the upstream and downstream Aeromonas populations in November 2007. Interestingly, on this date the ampicillin resistant bacterial population in downstream sediment differed from subsequent samples. Fewer than 60% of ampicillin resistant bacteria isolated from the downstream sediment in November 2007 were Aeromonas. Greater than 40% of the bacteria were coliforms. On all other dates for both upstream and downstream sediment samples, greater than 85% of ampicillin resistant bacteria isolated were Aeromonas. Tahlequah WRRF staff were consulted and they stated that the plant had not been modified nor, had there been major changes in operations at the plant during the months prior to initiation of the study or over the course of the study. The data suggest, therefore, that prior to the November 2007 sampling date the downstream Aeromonas population was perturbed by an unknown event but, over time, the population stabilized becoming more similar to the upstream Aeromonas population. This indicates the importance of sampling over a prolonged period in studies of this type.
The Aeromonas upstream and downstream sediment populations were also compared using 16S rRNA gene sequences for each sampling date and for all isolates combined. During the two-year period gene sequences in the two populations were ≥ 97% similar, and the same or very similar alleles were present in both the upstream and downstream populations. The high degree of 16S rRNA gene sequence similarity observed between the two populations could be due to three factors. Firstly, 16S rRNA gene sequences in Aeromonas may lack discretionary power. While use of 16S rRNA gene sequences to identify and distinguish bacterial species is an accepted methodology, it is not without disadvantages (Janda and Abbott 2007). It has been reported that Aeromonas species are difficult to distinguish from each other and that misidentification is not unusual (Janda and Abbott 2010; Parker and Shaw 2011; Janda and Abbott 2007). Secondly, WRRF effluent may not have had a selective effect on the downstream sediment population. Thirdly, unidentified selective pressure(s) on both populations may have caused the populations to be more similar than different. The fact that the 16S rRNA alleles were similar in the upstream and downstream populations suggests that the observed differences in antibiotic resistance phenotypes might be due at least in part to horizontal transfer of resistance genes.
In this study, Aeromonas strains were isolated from Tahlequah Creek sediments during different seasons, in the months of November and May, over the two-year period. Previous studies have indicated that Aeromonas species are present at higher densities in water in both natural and man-made ecosystems, such as water resource recovery facilities, during the warmer months of the year (Gavriel et al. 1998; Warren et al. 2004; Monfort and Baleux 1991; Villarruel-Lopez et al. 2005). Furthermore, seasonal effects on the isolation of antibiotic resistant Aeromonas have also been reported (Warren et al. 2004). In this study, statistical differences were observed when comparing antibiotic resistance in isolates collected in November to isolates collected in May. However, the data lacked a consistent pattern. For example, the incidence of resistance to cefoxitin in both the upstream and downstream populations was greater in May than in November. In contrast, the incidence of resistance to amoxicillin plus clavulanic acid was significantly different only in the downstream population: greater numbers of resistant strains were present in November than in May. Similar inconsistencies were observed when comparing resistance phenotypes in the two populations in the different seasons. It is likely that these results are due to complex interactions between the bacterial populations, treated wastewater, and environmental factors such as rainfall, streambed disturbances, and/or ambient temperature that are not addressed by our study. For example, one limitation of this study is that it is restricted to a single genus. An alternative approach is to assay antibiotic resistance genes in ecosystems directly. This approach avoids the need to isolate bacteria and is not restricted to a single genus. Studies using this approach suggest that human and agricultural inputs into streams do affect the downstream incidence of antibiotic resistance genes (Pei et al. 2006; Pruden et al. 2006; Storteboom et al. 2010; Chen et al. 2013; Luo et al. 2010). In these studies, however, samples were collected and assayed over a period of one year or less, raising the possibility that, as observed in this report, longer-term sampling might show different results.
Conclusions
Results from this study indicate that WRRF effluent did not have a long-term effect on antibiotic resistance in Aeromonas spp. downstream of the treatment facility although differences between the upstream and downstream populations were observed on early sampling dates. Factors such as bacterial species and microbial habitat (e.g. transiting in water versus resident in sediment) may play important roles in the development and spread of antibiotic resistance in the environment. In addition, the time period over which observations are made appears to be important. Therefore, studies that examine multiple bacterial species and/or multiple antibiotic resistance genes over long periods of time may be required to accurately determine the effects, if any, of wastewater treatment plant effluent on antibiotic resistance in downstream bacteria.
Acknowledgments
The authors thank Ms. Wanda Jones and the staff of the Tahlequah water resource recovery facility for their cooperation and assistance during the study, Mark F. Becker, USGS Oklahoma Water Science Center and Michael T. Meyer, USGS Kansas Water Science Center for assistance with antibiotic residue analysis of water samples, William Murry for technical support, Mark Paulissen and Jonathan Fisher for assistance with statistical analysis, John de Banzie for review of the manuscript, Deborah Hyde for assistance with the description of Tahlequah Creek sediment, and Jerry Fuller of Tahlequah, OK for allowing access to the upstream sampling site. The project described was supported by Grant Number P20RR016478 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and by the Oklahoma Center for the Advancement of Science and Technology, OHRS award HR07-124.
References
- Akiyama T, Savin MC. Populations of Antibiotic-Resistant Coliform Bacteria Change Rapidly in a Wastewater Effluent Dominated Stream. Sci Total Environ. 2010;408:6192–6201. doi: 10.1016/j.scitotenv.2010.08.055. [DOI] [PubMed] [Google Scholar]
- Andersson S, Kuttuva Rajarao G, Land CJ, Dalhammar G. Biofilm Formation and Interactions of Bacterial Strains Found in Wastewater Treatment Systems. FEMS Microbiol Lett. 2008;283:83–90. doi: 10.1111/j.1574-6968.2008.01149.x. [DOI] [PubMed] [Google Scholar]
- Baquero F, Martinez JL, Canton R. Antibiotics and Antibiotic Resistance in Water Environments. Curr Opin Biotechnol. 2008;19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Cabello FC. Heavy Use of Prophylactic Antibiotics in Aquaculture: A Growing Problem for Human and Animal Health and for the Environment. Environ Microbiol. 2006;8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- Chauret C, Volk C, Creason R, Jarosh J, Robinson J, Warnes C. Detection of Aeromonas Hydrophila in a Drinking-Water Distribution System: A Field and Pilot Study. Can J Microbiol. 2001;47:782–786. [PubMed] [Google Scholar]
- Chen B, Liang X, Huang X, Zhang T, Li X. Differentiating Anthropogenic Impacts on Args in the Pearl River Estuary by Using Suitable Gene Indicators. Water Res. 2013;47:2811–2820. doi: 10.1016/j.watres.2013.02.042. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline (M45-a) Clinical and Laboratory Standards Institute; Wayne, Pennsylvania: 2006. [Google Scholar]
- Cooper ER, Siewicki TC, Phillips K. Preliminary Risk Assessment Database and Risk Ranking of Pharmaceuticals in the Environment. Sci Total Environ. 2008;398:26–33. doi: 10.1016/j.scitotenv.2008.02.061. [DOI] [PubMed] [Google Scholar]
- Cordova-Kreylos AL, Scow KM. Effects of Ciprofloxacin on Salt Marsh Sediment Microbial Communities. ISME J. 2007;1:585–595. doi: 10.1038/ismej.2007.71. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a Chimera-Checked 16s Rrna Gene Database and Workbench Compatible with Arb. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria C, Vaz-Moreira I, Serapicos E, Nunes OC, Manaia CM. Antibiotic Resistance in Coagulase Negative Staphylococci Isolated from Wastewater and Drinking Water. Sci Total Environ. 2009;407:3876–3882. doi: 10.1016/j.scitotenv.2009.02.034. [DOI] [PubMed] [Google Scholar]
- Fick J, Soderstrom H, Lindberg RH, Phan C, Tysklind M, Larsson DG. Contamination of Surface, Ground, and Drinking Water from Pharmaceutical Production. Environ Toxicol Chem. 2009;28:2522–2527. doi: 10.1897/09-073.1. [DOI] [PubMed] [Google Scholar]
- Figueroa RA, Leonard A, MacKay AA. Modeling Tetracycline Antibiotic Sorption to Clays. Environ Sci Technol. 2004;38:476–483. doi: 10.1021/es0342087. [DOI] [PubMed] [Google Scholar]
- Galloway JM, Haggard BE, Meyer MT, Green WR. Occurrence of Pharmaceuticals and Other Organic Wastewater Constituents in Selected Streams in Northern Arkansas, 2004. USGS Scientific Investigations Report Number 2005-5140; U.S Geological Survey Reston, VA: 2005. [Google Scholar]
- Gavriel AA, Landre JP, Lamb AJ. Incidence of Mesophilic Aeromonas within a Public Drinking Water Supply in North-East Scotland. J Appl Microbiol. 1998;84:383–392. doi: 10.1046/j.1365-2672.1998.00354.x. [DOI] [PubMed] [Google Scholar]
- Goni-Urriza M, Capdepuy M, Arpin C, Raymond N, Caumette P, Quentin C. Impact of an Urban Effluent on Antibiotic Resistance of Riverine Enterobacteriaceae and Aeromonas Spp. Appl Environ Microbiol. 2000;66:125–132. doi: 10.1128/aem.66.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggard BE, Galloway JM, Green WR, Meyer MT. Pharmaceuticals and Other Organic Chemicals in Selected North-Central and Northwestern Arkansas Streams. J Environ Qual. 2006;35:1078–1087. doi: 10.2134/jeq2005.0248. [DOI] [PubMed] [Google Scholar]
- Holmes P, Niccolls LM, Sartory DP. The Ecology of Mesophilic Aeromonas in the Aquatic Environment. In: Austin B, Altwegg M, Gosling PJ, Joseph S, editors. The Genus Aeromonas. John Wiley & Sons; London: 1996. pp. 127–150. [Google Scholar]
- Huddleston JR, Zak JC, Jeter RM. Antimicrobial Susceptibilities of Aeromonas Spp. Isolated from Environmental Sources. Appl Environ Microbiol. 2006;72:7036–7042. doi: 10.1128/AEM.00774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CJ, Crawford RL, Knudsen GR, McInerney MJ, Stetzenbach LD. Manual of Environmental Microbiology. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- Janda JM, Abbott SL. 16s Rrna Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clinical Microbiology Reviews. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keevil CW. Encyclopedia of Environmental Microbiology. John Wiley & Sons, Inc.; New York: 2003. Pathogens in Environmental Biofilms. [Google Scholar]
- Kim S, Aga DS. Potential Ecological and Human Health Impacts of Antibiotics and Antibiotic-Resistant Bacteria from Wastewater Treatment Plants. J Toxicol Environ Health B Crit Rev. 2007;10:559–573. doi: 10.1080/15287390600975137. [DOI] [PubMed] [Google Scholar]
- Kim SC, Carlson K. Temporal and Spatial Trends in the Occurrence of Human and Veterinary Antibiotics in Aqueous and River Sediment Matrices. Environ Sci Technol. 2007;41:50–57. doi: 10.1021/es060737+. [DOI] [PubMed] [Google Scholar]
- Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. Pharmaceuticals, Hormones, and Other Organic Wastewater Contaminants in U.S. Streams, 1999-2000: A National Reconnaissance. Environ Sci Technol. 2002;36:1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- Kuhn I, Allestam G, Huys G, Janssen P, Kersters K, Krovacek K, Stenstrom TA. Diversity, Persistence, and Virulence of Aeromonas Strains Isolated from Drinking Water Distribution Systems in Sweden. Appl Environ Microbiol. 1997;63:2708–2715. doi: 10.1128/aem.63.7.2708-2715.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummerer K. Resistance in the Environment. The Journal of Antimicrobial Chemotherapy. 2004;54:311–320. doi: 10.1093/jac/dkh325. [DOI] [PubMed] [Google Scholar]
- Lai HT, Wang TS, Chou CC. Implication of Light Sources and Microbial Activities on Degradation of Sulfonamides in Water and Sediment from a Marine Shrimp Pond. Bioresource technology. 2011;102:5017–5023. doi: 10.1016/j.biortech.2011.01.070. [DOI] [PubMed] [Google Scholar]
- Lee L, Tin S, Kelley ST. Culture-Independent Analysis of Bacterial Diversity in a Child-Care Facility. BMC Microbiol. 2007;7:27. doi: 10.1186/1471-2180-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Pan HY, Liu SM, Lai HT. Effects of Light and Microbial Activity on the Degradation of Two Fluoroquinolone Antibiotics in Pond Water and Sediment. Journal of environmental science and health Part B, Pesticides, food contaminants, and agricultural wastes. 2010;45:456–465. doi: 10.1080/03601231003800222. [DOI] [PubMed] [Google Scholar]
- Lindberg RH, Wennberg P, Johansson MI, Tysklind M, Andersson BA. Screening of Human Antibiotic Substances and Determination of Weekly Mass Flows in Five Sewage Treatment Plants in Sweden. Environ Sci Technol. 2005;39:3421–3429. doi: 10.1021/es048143z. [DOI] [PubMed] [Google Scholar]
- Luo Y, Mao D, Rysz M, Zhou Q, Zhang H, Xu L, P JJA. Trends in Antibiotic Resistance Genes Occurrence in the Haihe River, China. Environ Sci Technol. 2010;44:7220–7225. doi: 10.1021/es100233w. [DOI] [PubMed] [Google Scholar]
- Luo Y, Xu L, Rysz M, Wang Y, Zhang H, Alvarez PJ. Occurrence and Transport of Tetracycline, Sulfonamide, Quinolone, and Macrolide Antibiotics in the Haihe River Basin, China. Environ Sci Technol. 2011;45:1827–1833. doi: 10.1021/es104009s. [DOI] [PubMed] [Google Scholar]
- Martone-Rocha S, Piveli RP, Matte GR, Doria MC, Dropa M, Morita M, Peternella FA, Matte MH. Dynamics of Aeromonas Species Isolated from Wastewater Treatment System. Journal of Water and Health. 2010;8:703–711. doi: 10.2166/wh.2010.140. [DOI] [PubMed] [Google Scholar]
- Massey LB, Haggard BE, Galloway JM, Loftin KA, Meyer MT, Green WR. Antibiotic Fate and Transport in Three Effluent-Dominated Ozark Streams. Ecol Eng. 2010;36:930–938. [Google Scholar]
- McGowan JE., Jr Economic Impact of Antimicrobial Resistance. Emerging Infectious Diseases. 2001;7:286–292. doi: 10.3201/eid0702.010228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao XS, Bishay F, Chen M, Metcalfe CD. Occurrence of Antimicrobials in the Final Effluents of Wastewater Treatment Plants in Canada. Environ Sci Technol. 2004;38:3533–3541. doi: 10.1021/es030653q. [DOI] [PubMed] [Google Scholar]
- Mispagel H, Gray JT. Antibiotic Resistance from Wastewater Oxidation Ponds. Water Environment Research. 2005;77:2996–3002. doi: 10.2175/106143005x73875. [DOI] [PubMed] [Google Scholar]
- Monfort P, Baleux B. Distribution and Survival of Motile Aeromonas Spp. In Brackish Water Receiving Sewage Treatment Effluent. Appl Environ Microbiol. 1991;57:2459–2467. doi: 10.1128/aem.57.9.2459-2467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Shaw JG. Aeromonas Spp. Clinical Microbiology and Disease. The Journal of Infection. 2011;62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Peduzzi R, Demarta A, Tonolla M. Seasonal Changes of Microbial Populations in the Sediments of the Basins of Lugano and Agno. Aquatic Sci. 1992;54:331–337. [Google Scholar]
- Pei R, Kim SC, Carlson KH, Pruden A. Effect of River Landscape on theSediment Concentrations of Antibiotics and Corresponding Antibiotic Resistance Genes (Arg) Water Res. 2006;40:2427–2435. doi: 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- Poffe R, Op de Beeck E. Enumeration of Aeromonas Hydrophila from Domestic Wastewater Treatment Plants and Surface Waters. The Journal of Applied Bacteriology. 1991;71:366–370. doi: 10.1111/j.1365-2672.1991.tb03802.x. [DOI] [PubMed] [Google Scholar]
- Pruden A, Pei R, Storteboom H, Carlson KH. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ Sci Technol. 2006;40:7445–7450. doi: 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- Rahman M, Huys G, Rahman M, Albert MJ, Kuhn I, Mollby R. Persistence, Transmission, and Virulence Characteristics of Aeromonas Strains in a Duckweed Aquaculture-Based Hospital Sewage Water Recycling Plant in Bangladesh. Appl Environ Microbiol. 2007;73:1444–1451. doi: 10.1128/AEM.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes G, Huys G, Swings J, McGann P, Hiney M, Smith P, Pickup RW. Distribution of Oxytetracycline Resistance Plasmids between Aeromonads in Hospital and Aquaculture Environments: Implication of Tn1721 in Dissemination of the Tetracycline Resistance Determinant Tet A. Appl Environ Microbiol. 2000;66:3883–3890. doi: 10.1128/aem.66.9.3883-3890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S, Weinberg HS. Pharmaceuticals in Water - an Interdisciplinary Approach to a Public Health Challenge. Environ Health Perspect. 2010;118:1016–1020. doi: 10.1289/ehp.0901532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt-Farrell N. The Landscape of Antibiotic Resistance. Environ Health Perspect. 2009;117:A244–250. doi: 10.1289/ehp.117-a244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossolini GM, Walsh T, Amicosante G. The Aeromonas Metallo-Beta-Lactamases: Genetics, Enzymology, and Contribution to Drug Resistance. Microb Drug Resist. 1996;2:245–252. doi: 10.1089/mdr.1996.2.245. [DOI] [PubMed] [Google Scholar]
- Schwartz T, Kohnen W, Jansen B, Obst U. Detection of Antibiotic-Resistant Bacteria and Their Resistance Genes in Wastewater, Surface Water, and Drinking Water Biofilms. FEMS Microbiol Ecol. 2003;43:325–335. doi: 10.1111/j.1574-6941.2003.tb01073.x. [DOI] [PubMed] [Google Scholar]
- Storteboom H, Arabi M, Davis JG, Crimi B, Pruden A. Identification of Antibiotic-Resistance-Gene Molecular Signatures Suitable as Tracers of Pristine River, Urban, and Agricultural Sources. Environ Sci Technol. 2010;44:1947–1953. doi: 10.1021/es902893f. [DOI] [PubMed] [Google Scholar]
- Szabo G, Khayer B, Rusznyak A, Tatrai I, Devai G, Marialigeti K, Borsodi AK. Seasonal and Spatial Variability of Sediment Bacterial Communities Inhabiting the Large Shallow Lake Balaton. Hydrobiologia. 2011;663:217–232. [Google Scholar]
- ter Laak TL, Gebbink WA, Tolls J. Estimation of Soil Sorption Coefficients of Veterinary Pharmaceuticals from Soil Properties. Environ Toxicol Chem. 2006;25:933–941. doi: 10.1897/05-229r.1. [DOI] [PubMed] [Google Scholar]
- Villarruel-Lopez A, Fernandez-Rendon E, Mota-de-la-Garza L, Ortigoza-Ferado J. Presence of Aeromonas Spp in Water from Drinking-Water- and Wastewater-Treatment Plants in Mexico City. Water Environment Research. 2005;77:3074–3079. doi: 10.2175/106143005x73974. [DOI] [PubMed] [Google Scholar]
- Warren WJ, Jeter RM, Kimbrough RC, Zak JC. Population Patterns and Antimicrobial Resistance of Aeromonas in Urban Playa Lakes. Can J Microbiol. 2004;50:397–404. doi: 10.1139/w04-029. [DOI] [PubMed] [Google Scholar]
- Watkinson AJ, Micalizzi GB, Graham GM, Bates JB, Costanzo SD. Antibiotic-Resistant Escherichia Coli in Wastewaters, Surface Waters, and Oysters from an Urban Riverine System. Appl Environ Microbiol. 2007;73:5667–5670. doi: 10.1128/AEM.00763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte W. Medical Consequences of Antibiotic Use in Agriculture. Science. 1998;279:996–997. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- Zalmum AA, Marialegite K, Ghenghesh KS. Bacterial Composition of the Biofilm on the Surface of Course Sediment of the Danube: With Special Reference to the Clinically Important Bacteria. Arch Inst Pasteur Tunis. 1998;75:205–209. [PubMed] [Google Scholar]
- Zhang XX, Zhang T, Fang HH. Antibiotic Resistance Genes in Water Environment. Applied Microbiology and Biotechnology. 2009a;82:397–414. doi: 10.1007/s00253-008-1829-z. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Marrs CF, Simon C, Xi C. Wastewater Treatment Contributes to Selective Increase of Antibiotic Resistance among Acinetobacter Spp. Sci Total Environ. 2009b;407:3702–3706. doi: 10.1016/j.scitotenv.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Zhou LJ, Ying GG, Zhao JL, Yang JF, Wang L, Yang B, Liu S. Trends in the Occurrence of Human and Veterinary Antibiotics in the Sediments of the Yellow River, Hai River and Liao River in Northern China. Environ Pollut. 2011;159:1877–1885. doi: 10.1016/j.envpol.2011.03.034. [DOI] [PubMed] [Google Scholar]