Abstract
Analytical ultracentrifugation has been used to analyze the oligomeric structure of the isolated regulatory domain of phenylalanine hydroxylase. The protein exhibits a monomer–dimer equilibrium with a dissociation constant of ∼46 μM; this value is unaffected by the removal of the 24 N-terminal residues or by phosphorylation of Ser16. In contrast, phenylalanine binding (Kd = 8 μM) stabilizes the dimer. These results suggest that dimerization of the regulatory domain of phenylalanine hydroxylase is linked to allosteric activation of the enzyme.
Phenylalanine hydroxylase (PheH) is an allosteric enzyme that catalyzes the hydroxylation to tyrosine of excess phenylalanine in the diet.1 Incubation with phenylalanine increases the activity up to 100-fold;2 this is accompanied by conformational changes in the interface between the regulatory and catalytic domains.3 Phosphorylation of Ser16 decreases the concentration of phenylalanine required to activate PheH.4 The present understanding of the mechanism of phenylalanine activation is based on a crystal structure of PheH5 in which N-terminal residues 19–29 of the regulatory domain lie across the active site of the catalytic domain. Binding of phenylalanine is proposed to result in a conformational change that displaces the N-terminus of the regulatory domain and opens the active site.5
PheH belongs to the family of aromatic amino acid hydroxylases, along with tyrosine hydroxylase (TyrH) and tryptophan hydroxylase.6 The structures of the catalytic domains of all three are very similar,5,7,8 consistent with a common catalytic mechanism.9 The regulatory domains of PheH and TyrH also have similar structures,5,10 although they display very low levels of sequence identity. However, the solution structure of the isolated regulatory domain of TyrH shows that the protein forms a stable ACT domain dimer,10 whereas the crystal structure of the combined regulatory and catalytic domains of PheH shows no interactions between individual regulatory domains.5 Preliminary characterization of the isolated regulatory domain of PheH, residues 1–117 (RDPheH), revealed that the free domain has an apparent molecular weight between that of a monomer and that of a dimer.11 In part on the basis of this observation, Jaffe et al.12 proposed that the conformational change that occurs upon binding of phenylalanine to PheH involves dimerization of the regulatory domains. Here we describe the use of analytical ultracentrifugation (AUC) to analyze the oligomerization state of RDPheH to determine whether dimerization of this domain is linked to phenylalanine binding and phosphorylation.
Initial sedimentation velocity (SV) experiments were performed at two concentrations of RDPheH; the resulting van Holde–Weischet distributions13 are shown in Figure 1A. The sedimentation distributions show a concentration dependence, in that the sample with a loading concentration of 280 μM has an s value larger than that of the sample with a loading concentration of 7 μM. The weight-average sedimentation coefficients (sw) for both samples are between those calculated for a monomer and a dimer based on the published structures of the regulatory domains of PheH and TyrH, consistent with a monomer–dimer equilibrium for RDPheH.
Figure 1.
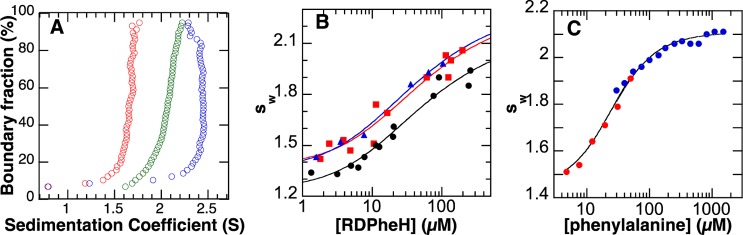
Sedimentation velocity analyses of RDPheH. (A) van Holde–Weischet distribution plot for 7 μM RDPheH (red), 280 μM RDPheH (green), and 7 μM RDPheH with 1 mM phenylalanine (blue). (B) Weight-average sedimentation coefficients (sw) as a function of the concentration of RDPheH (red squares), phosphorylated RDPheH (blue triangles), and RDPheH25–117 (black circles). The lines are from fits of eq 1. (C) Effect of the concentration of phenylalanine on the sw of RDPheH25–117 (7.6 μM, red, or 7.2 μM, blue). The line was generated using the mechanism of Scheme 1 and the values listed in Table 1.
To analyze this equilibrium more quantitatively, a series of SV experiments were conducted at 2.5–206 μM RDPheH. The resulting sw values14 are plotted in Figure 1B as a function of protein concentration. These data were fit well by eq 1, which describes the effect of protein concentration on the average sw value for a monomer–dimer equilibrium
| 1 |
where s1 and s2 are the sedimentation coefficients of the monomer and dimer, respectively, K is the equilibrium constant for dimer formation, and c is the total molar concentration of the protein in terms of monomer. The resulting values are listed in Table 1.
Table 1. Sedimentation Coefficients and Dissociation Constants for RDPheH.
| s1 (S) | s2 (S) | K1 (μM)a | |
|---|---|---|---|
| RDPheHb | 1.38 ± 0.08 | 2.31 ± 0.13 | 46 ± 35 |
| RDPheH25–117b | 1.24 ± 0.05 | 2.17 ± 0.07 | 46 ± 19 |
| phosphorylated RDPheHb | 1.36 ± 0.05 | 2.32 ± 0.10 | 37 ± 18 |
| RDPheH25–117c | 1.22 (1.14–1.29) | 2.11 (2.11–2.12) | 34 (22–56) |
Dissociation constant for dimerization.
Based on eq 1.
From global analyses using KinTek Explorer.
The structure of PheH containing both the catalytic and regulatory domains5 shows the ∼20 N-terminal residues to be unstructured. To determine the effect of this N-terminal region on dimerization, a series of mutants of the RDPheH lacking ∼20 N-terminal residues were constructed. The protein lacking the 24 N-terminal residues (RDPheH25–117) was the best-behaved in solution. The results of an SV analysis of the solution behavior of RDPheH25–117 are shown Figure 1B. The dissociation constant of RDPheH25–117 obtained from this analysis is very close to that of RDPheH (Table 1), establishing that the 24 N-terminal residues have no significant effect on dimerization. The 1H–15N heteronuclear single-quantum coherence nuclear magnetic resonance spectrum of RDPheH25–117 overlaps with that of RDPheH (data not shown), establishing that the deletion did not alter the structure.
Next, AUC was used to determine whether phosphorylation alters the monomer–dimer equilibrium. As shown in Figure 1B (green) and Table 1, the effect of protein concentration on the sedimentation behavior of RDPheH is unaffected by phosphorylation of the protein.
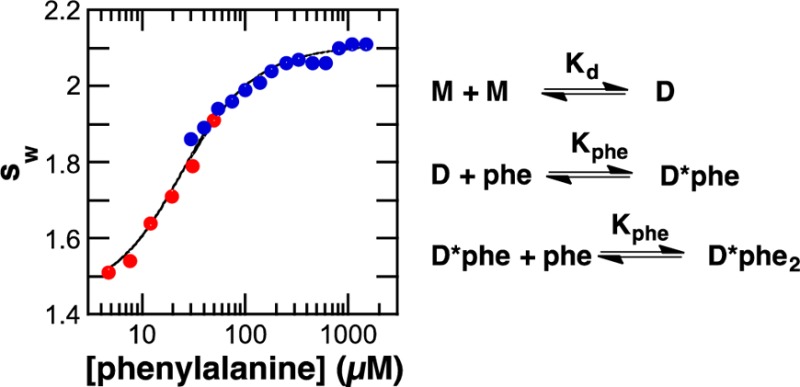
In contrast to the lack of an effect of phosphorylation, phenylalanine has a substantial effect on the sedimentation behavior. Addition of 1 mM phenylalanine to 7 μM RDPheH results in an increase in the s value of the protein to a value consistent with a dimer (Figure 1A). SV analyses were conducted with a fixed concentration of RDPheH25–117 (∼7 μM total monomer) and a range of phenylalanine concentrations (4.8–1500 μM). The resulting sw values are shown in Figure 1C as a function of phenylalanine concentration. There is a clear increase in the average sw value as the concentration of phenylalanine increases.
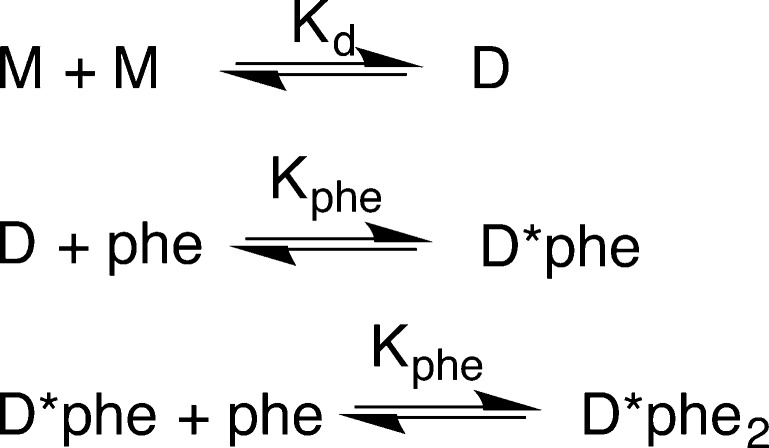
The data in panels B and C of Figure 1 for RDPheH25–117 were analyzed globally15 to evaluate individual dimerization models and calculate values for the equilibrium constants for phenylalanine binding and dimerization. The model shown in Scheme 1, which involves binding of two molecules of phenylalanine per dimer with equal affinity, gave the best fit (χ2 = 0.339) with a Kd value for phenylalanine of 8.3 μM (6.4–13 μM) and the values listed in Table 1. A model in which the affinities for the first and second molecules of phenylalanine differ gave values for Kphe-1 and Kphe-2 of 13 and 4.6 μM, respectively, but did not yield an improvement in the χ2 value (0.335). A model in which only one phenylalanine binds to the dimer resulted a much higher χ2 value (0.561).
Scheme 1. RDPheH Dimerization Model.
The data presented here support a model in which the isolated RDPheH is present in a monomer–dimer equilibrium, with phenylalanine binding stabilizing the dimer. The data support the model of Jaffe et al.12 in which the structural change associated with phenylalanine activation involves dimerization of regulatory domains within the PheH tetramer. While these results do not address the structure of the dimer, the finding that the isolated regulatory domain of TyrH is an ACT domain dimer10,16 and the similar structures of the regulatory domains of the two proteins suggest that the RDPheH dimer is also an ACT domain dimer.
The lack of an effect of phosphorylation at Ser16 on dimerization suggests that phosphorylation of this residue disrupts interactions of the ∼20 N-terminal residues with the catalytic domain of the enzyme. Weakening such an interaction would make formation of the proposed regulatory domain dimer more favorable. The lack of an effect of deleting the 24 N-terminal residues on dimerization is also consistent with these residues interacting with the catalytic domain.
Acknowledgments
We thank Mr. Virgil Schirf for assistance with the AUC experiments and Dr. Borries Demeler for helpful discussion. Support for the Center for Macromolecular Interactions from the Office of the Vice President for Research at University of Texas Health Science Center at San Antonio is gratefully acknowledged.
Supporting Information Available
Experimental procedures, including details of data analyses. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported in part by National Institutes of Health Grant R01 GM098140.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fitzpatrick P. F. (2012) Arch. Biochem. Biophys. 519, 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiman R.; Jones S. H.; Gray D. W. (1990) J. Biol. Chem. 265, 11633–11642. [PubMed] [Google Scholar]
- Li J.; Dangott L. J.; Fitzpatrick P. F. (2010) Biochemistry 49, 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doskeland A. P.; Martinez A.; Knappskog P. M.; Flatmark T. (1996) Biochem. J. 313, 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B.; Jennings I. G.; House C. M.; Michell B. J.; Goodwill K. E.; Santarsiero B. D.; Stevens R. C.; Cotton R. G. H.; Kemp B. E. (1999) Nat. Struct. Biol. 6, 442–448. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick P. F. (1999) Annu. Rev. Biochem. 68, 355–381. [DOI] [PubMed] [Google Scholar]
- Wang L.; Erlandsen H.; Haavik J.; Knappskog P. M.; Stevens R. C. (2002) Biochemistry 41, 12569–12574. [DOI] [PubMed] [Google Scholar]
- Goodwill K. E.; Sabatier C.; Marks C.; Raag R.; Fitzpatrick P. F.; Stevens R. C. (1997) Nat. Struct. Biol. 4, 578–585. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick P. F. (2003) Biochemistry 42, 14083–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Huang T.; Ilangovan U.; Hinck A. P.; Fitzpatrick P. F. (2014) J. Mol. Biol. 426, 1483–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Ilangovan U.; Daubner S. C.; Hinck A. P.; Fitzpatrick P. F. (2011) Arch. Biochem. Biophys. 505, 250–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe E. K.; Stith L.; Lawrence S. H.; Andrake M.; Dunbrack R. L. Jr. (2013) Arch. Biochem. Biophys. 530, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., and Rowe A. J., Eds.) pp 210–229, Royal Society of Chemistry, London. [Google Scholar]
- Gabrielson J. P.; Randolph T. W.; Kendrick B. S.; Stoner M. R. (2007) Anal. Biochem. 361, 24–30. [DOI] [PubMed] [Google Scholar]
- Johnson K. A.; Simpson Z. B.; Blom T. (2009) Anal. Biochem. 387, 20–29. [DOI] [PubMed] [Google Scholar]
- Grant G. A. (2006) J. Biol. Chem. 281, 33825–33829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.