Abstract
Native tissues are endowed with a highly organized nanofibrous extracellular matrix (ECM) that directs cellular distribution and function. The objective of this study is to create a purely natural, uniform, and highly aligned nanofibrous ECM scaffold for potential tissue engineering applications. Synthetic nanogratings (130 nm in depth) were used to direct the growth of human dermal fibroblasts for up to 8 weeks, resulting in a uniform 70 μm–thick fibroblast cell sheet with highly aligned cells and ECM nanofibers. A natural ECM scaffold with uniformly aligned nanofibers of 78 ± 9 nm in diameter was generated after removing the cellular components from the detached fibroblast sheet. The elastic modulus of the scaffold was well maintained after the decellularization process because of the preservation of elastin fibers. Reseeding human mesenchymal stem cells (hMSCs) showed the excellent capacity of the scaffold in directing and supporting cell alignment and proliferation along the underlying fibers. The scaffold’s biocompatibility was further examined by an in vitro inflammation assay with seeded macrophages. The aligned ECM scaffold induced a significantly lower immune response compared to its unaligned counterpart, as detected by the pro-inflammatory cytokines secreted from macrophages. The aligned nanofibrous ECM scaffold holds great potential in engineering organized tissues.
Keywords: Nanofibers, Extracellular Matrix, Scaffold, Alignment, Biocompatibility
1. Introduction
Advances in nanotechnology and nanomaterials over the past decade have been applied extensively towards promoting tissue engineering and regenerative medicine. Biomaterials at nanoscale have been highlighted as promising candidates for improving traditional tissue engineering scaffolds due to their ability to effectively mimic surface characteristics of natural tissues, such as surface free energy and topography.[1] In natural tissues, cells directly interact with their surrounding nanobiomaterials, the extracellular matrix (ECM), which plays a critical role in providing mechanical support, directing cell adhesion and growth, as well as regulating development, homeostasis and regeneration.[2] Different electrospun nanofibrous and nanopatterened substrates have been employed to mimic the nanotopography of natural ECM for a wide range of tissue engineering studies including the engineering of neural, bone, and cardiovascular tissues.[3] However, most of these nanoscale scaffolds are fabricated by synthetic materials or a limited set of natural polymer blends, which fail to adequately mimic the complex morphology and composition of natural ECM.[4] In order to more fully replicate the chemical and biological motifs of the ECM found in natural tissues,[5] researchers have adopted the approach of fabricating nanofibrous materials from animal tissues or from cultured cells in vitro.
Compared with animal tissues, cell-derived ECM avoids the problems of pathogen transfer and host immune response.[6, 7] The application of cell-derived ECM as a physiologically functional source of the complex set of naturally occurring bioactive signals has gained increasing interests. For instance, decellularized ECM synthesized by undifferentiated mesenchymal stem cells (MSCs) in vitro has been shown to facilitate cell proliferation, prevent spontaneous differentiation and enhance the chondrogenic and osteogenic potential of freshly reseeded MSCs.[8] Synthetic polymer materials decorated with decellularized ECM were favorable to osteoblastic differentiation of new MSCs.[9] Fibroblast-derived ECM supported the growth of re-seeded fibroblasts and showed low inflammation response when transplanted into rats.[6] However, although all of these natural ECM scaffolds in different forms showed beneficial effects in various applications, none of them presented an organized nanofibrous structure. The development of highly organized engineered ECM scaffolds is crucial to create biomimetic tissues since native tissues are highly organized. It is well accepted that the cell and ECM organization not only dictate the function of a broad range of human body tissues from myocardial to connective tissues,[10] but also influence the in vitro and in vivo inflammatory response.[11, 12] Therefore, mimicking the complexity of ECM and cell organization will make great contributions to effectively study and replicate the biological function of many native tissues in vivo.
Highly organized cell-derived ECM can be generated by using a cell sheet engineering approach in combination with microfabrication technology. Cell sheet engineering is an attractive method to harvest confluent cell cultures as intact, tissue-like cell sheets.[13] The substrate used for cell sheet growth can be microfabricated into micro- or nano-scale gratings in order to organize the cells. Several studies have shown the ability to fabricate cell sheets grown on micrograted substrates.[14–16] At the microscale, gratings as deep as 5 μm appear to produce a non-uniform and a defective cell sheet.[14, 15] The portion of the cell sheet grown on the ridges tends to experience higher tension and secrets less ECM, leading to the formation of holes and tears. Our recent study has shown that synthetic gratings, at nanoscale (240 nm depth), was sufficient to guide cell orientation, without causing undesired cell sheet heterogeneity.[17] Subsequent to ECM synthesis and organization, cellular components of the cell sheet can be removed by decellularization, leaving an intact ECM sheet for an off-the-shelf cell culture scaffold. By utilizing laminar or tubular cellular assemblies, three-dimensional (3D) and tubular tissue structures can be created from these ECM sheets.[16, 18]
Human dermal fibroblasts can be readily isolated and cultured up to passage 14 without a decrease in collagen synthesis or a reduction in their rate of growth.[19] Importantly, dermal fibroblasts are able to synthesize large quantities of ECM biomolecules including elastin, which contributes to the compliance of native tissues.[20] Moreover, the matrices produced by fibroblasts are stronger than reconstituted ECM such as collagen or fibrin gels.[21] Fibroblast-derived cell sheets and the decellularized products have been applied in the fabrication of tissue-engineered blood vessels with seeded smooth muscle cells and endothelial cells, which showed superior mechanical properties.[18] Therefore, an ECM scaffold derived from fibroblast cell sheets holds great potential in serving as a building block to construct strong, complex biological tissues, with highly organized structures. However, few researchers have studied aligned fibroblast cell sheets.
In this work, we produced a uniform and highly aligned nanofibrous natural ECM scaffold from a human dermal fibroblast cell sheet grown on synthetic nanogratings. The fibroblasts were cultured on a nanograted substrate for 8 weeks and formed an ECM-rich cell sheet with well-aligned cells and ECM fibers. A highly organized ECM scaffold with nanofiber diameter of 78 ± 9 nm was obtained by decellularizing the fibroblast cell sheet. The capability of the aligned nanofibrous natural ECM scaffold in directing and supporting cell alignment and growth was tested by re-seeding human MSCs (hMSCs), which showed good alignment and active proliferation. The immune response to the natural ECM scaffolds was tested by an in vitro macrophage culture. Our results indicate that the morphology of nanofibers in the ECM scaffold can mediate the macrophage activities. The alignment of nanofibers significantly improves the biocompatibility of ECM scaffolds.
2. Results
2.1 Characterization of aligned fibroblast cell sheet
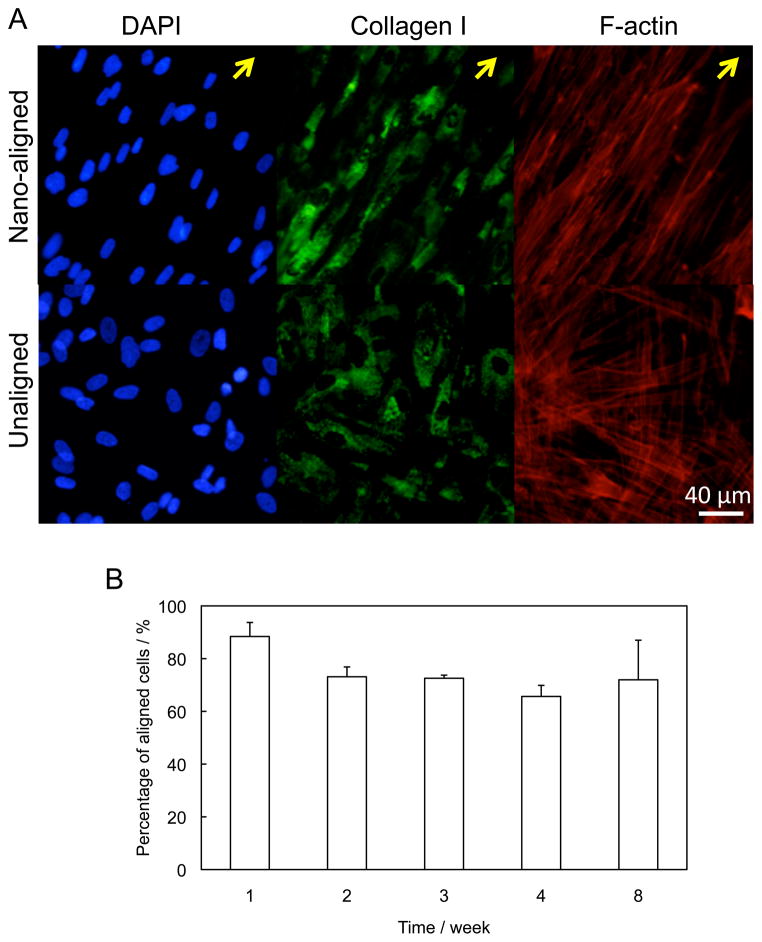
Fibroblasts were continuously cultured for up to 8 weeks, and formed a dense cell sheet composed of cells and ECM biomolecules. Because the interaction between the nanofibers and fibroblasts in the thick cell sheet was stronger than the adhesion force between the cells and the PDMS substrate, the cell sheet was easily peeled off the PDMS substrate as an intact structure (Figure 1). After detachment, the cell sheet incurred less than 5% shrinkage in diameter, with the general shape and structure being well preserved. Inspection of the substrate revealed a negligible amount of retained cells and ECM proteins (Supplemental Figure 1). The cell alignment and ECM organization of fibroblast cell sheets grown on nanograted and flat PDMS substrates were examined every week by staining of cell nuclei, F-actin and ECM proteins, including collagen I, fibronectin, and elastin. After 1 week of culture, both the cells and the ECM proteins demonstrated strong morphological differences in their organization relative to controls. In nanograted samples, both cell nuclei and F-actin were highly aligned in the direction of the nano-patterned grooves (Figure 2A). On the contrary, fibroblasts grown on flat surfaces showed no directionality. The organization of collagen I indicated that the ECM proteins were also highly aligned in the nanopatterned samples, but were randomly distributed in the control specimens.
Figure 1.

Fibroblast cell sheets before and after detachment from PDMS substrate. The cell sheet remained intact tissue organization after detachment.
Figure 2.
Characterization of fibroblast cell sheets. (A) Morphology of nuclei, collagen I and F-actin of fibroblast grown on nanograted and flat surfaces for 7 days. The cells cultured on nanograted surface exhibited well organized F-actin and collagen I. Arrows indicate the direction of the alignment. (B) The percentage of aligned cells (with angles < 15°) in cells sheets. The cells maintained high degree of alignment after the long-term culture.
The alignment of cell sheets with respect to time was quantitatively characterized by measuring cell nucleus alignment, which was defined as the angle between the major axis of each cell nucleus and the main direction of the grating axis.[17] The percentage of aligned cells with angles <15° was calculated for cell sheets grown from 1 to 4 weeks. (Figure 2B). In week 1, 88% cells had less than 15° angles. As the culture time increased, fewer and fewer cells fell within this region, but there was no significant differences (p > 0.05) between samples from week 2 to week 8. After 8 weeks in culture, 72% of cells still displayed good alignment in the direction of the grating axis. Apparently, the drastically increased standard deviation indicated that some cell sheets had much lower percentage of aligned cells.
2.2 Comparison of fibroblast cell sheets before and after decellularization
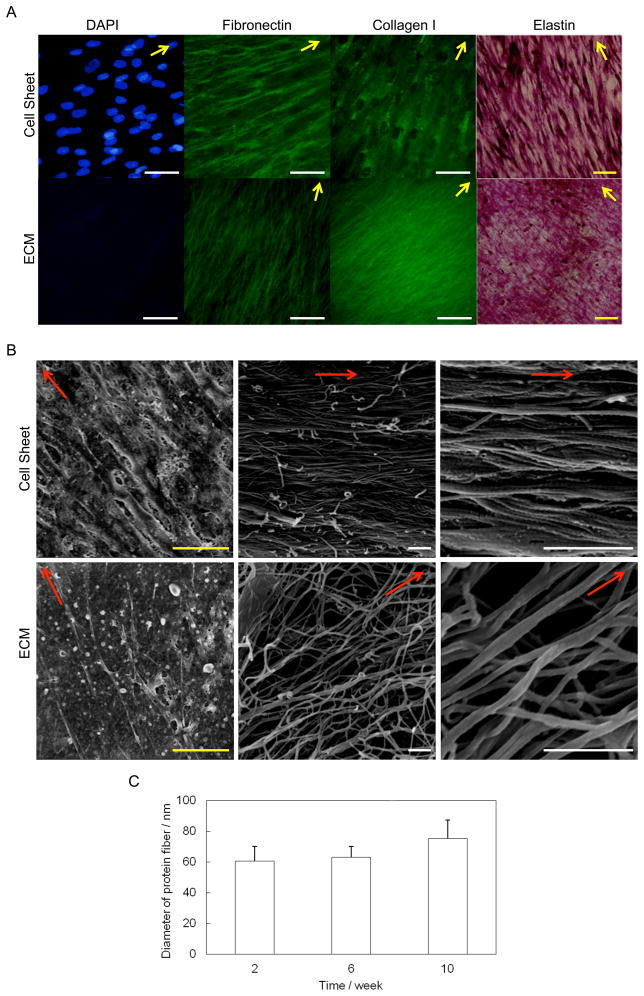
After 4 weeks of culture, cells from the nanopatterned surface still displayed aligned ECM proteins, including collagen I, fibronectin, and elastin (Figure 3A). The ECM proteins were organized into a network, which was well preserved after decellularization. A DNA assay demonstrated that 93% of the DNA content was removed after decellularization. The SEM images of the cell sheets revealed that cells grown on the surface were aligned in a specific direction (Figure 3B). In addition, numerous protein nanofibers embedded in the cell sheets also showed very good alignment along the direction of nanogratings. The diameters of these protein fibers in cell sheet were measured at different growth time points. Figure 3C showed that the average diameter of the fibers slightly increased with time, but there was no significant difference at all the time points (P >0.05). The protein fibers in 8-week old cell sheet had a uniform size of 75 ± 12 nm in diameter. The protein fibers were closely packed together and glued by adhesive molecules, which possess multiple binding domains capable of binding collagen and proteoglycans, as well as the cell surface.[22] After decellularization, cells were lysed and some adhesive molecules and protein were removed. The protein nanofibers were thus clearly exposed. Most fibers were still parallel to each other. Some were interconnected to form a network. There was no significant change in fiber size (78 ± 9 nm in diameter) after the decellularization process (P >0.05).
Figure 3.
Comparison of cell sheet and decellularized ECM. (A) ECM staining of fibroblast grown for 4 weeks before and after decellularization. Yellow bar: 100 μm; white bar: 50 μm. ECM fibers maintained the preferential direction after decellularization. Arrows indicate the direction of the alignment. (B) Morphology of cell sheet grown for 8 weeks before and after decellularization. Yellow bar: 25 μm; white bar: 1 μm. Most adhesion molecules were removed after treatment, exposing the aligned nanofibers. Arrows indicate the direction of the alignment. (C) The diameter of protein fibers in cell sheet at different growth time points. The 8-week cell sheet had a uniform size of 75 ± 12 nm in fiber diameter. There was no significant difference in the size of fiber diameters over time.
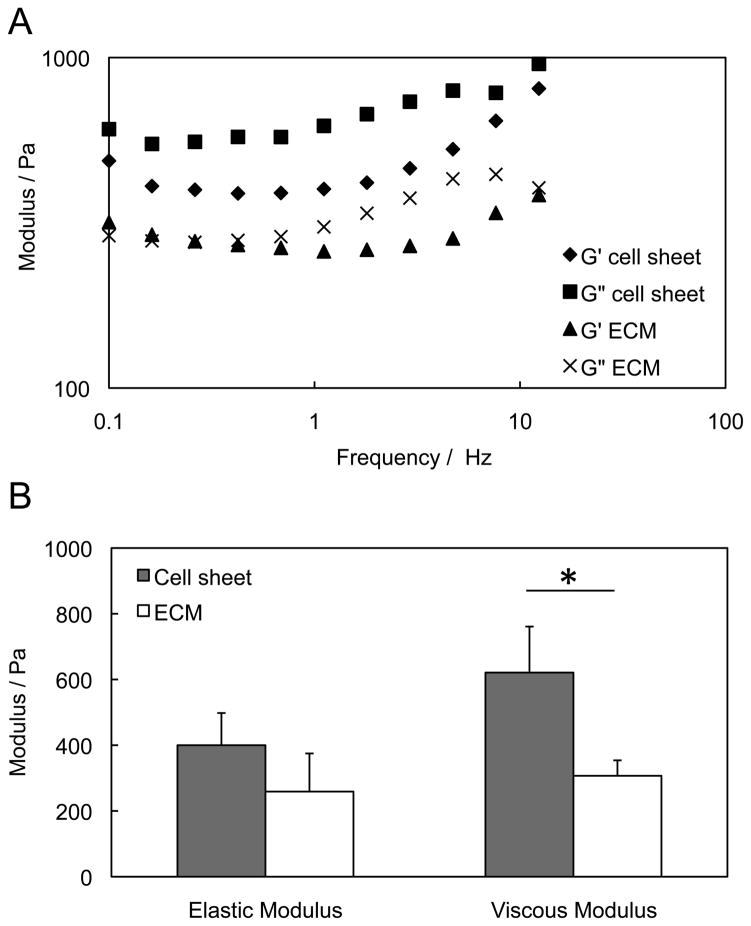
Mechanical properties of the cell sheet and ECM after decellularization are shown in Figure 4. Due to the high content of free water in the cell sheet and ECM, the viscous modulus (G”) is higher than the elastic modulus (G'), as shown in Figure 4A. After decellularization, both elastic and viscous modulus decreased. There was no significant decline (P > 0.05) for elastic modulus, but around 50% of viscous modulus was lost. The significant reduction (P < 0.05) of the mechanical strength of the cell sheets may be caused by the loss of “adhesive molecules” such as proteoglycans that keep the tissue hydrated, as shown in the Figure 4B.
Figure 4.
Mechanical strength of 8-week old cell sheet before and after decellularization. (A) Frequency sweeps of cell sheets and ECM sheet after decellularization. The elastic modulus (G') and viscous modulus (G”) remained constant over all frequencies. (B) Comparison of the average modulus at frequency 1 Hz. The ECM scaffold maintained elastic modulus, but had significant lower viscous modulus than the cell sheet.
2.3 Aligned nanofibrous ECM scaffold in guiding cell growth
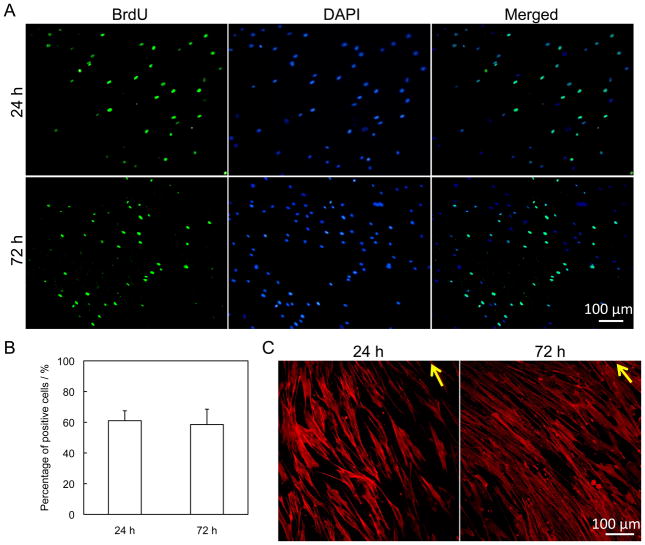
The supporting and morphological effects of the ECM scaffold were examined by growing hMSCs in the scaffold. The proliferation of hMSCs was analyzed by a BrdU assay. BrdU-positive cells were stained, counted, and compared to the total number of cells that were stained with DAPI. An arbitrary view field is shown in Figure 6A. The average percentage of cells incorporating BrdU into their DNA after 24 h and 72 h culture was 61% and 58% respectively. There was no significant difference between these two samples as demonstrated in Figure 6B. During 72 h of culture on the ECM scaffold, hMSCs maintained a high proliferation rate, suggesting that the fibroblast-derived ECM scaffold could provide a suitable environment for cell adhesion and growth. The cytoskeleton protein F-actin was stained for hMSCs after 24 h and 72 h, shown in Figure 6C. The organization of F-actin filaments indicated that the cells were well aligned under the guidance of underlying ECM fibers.
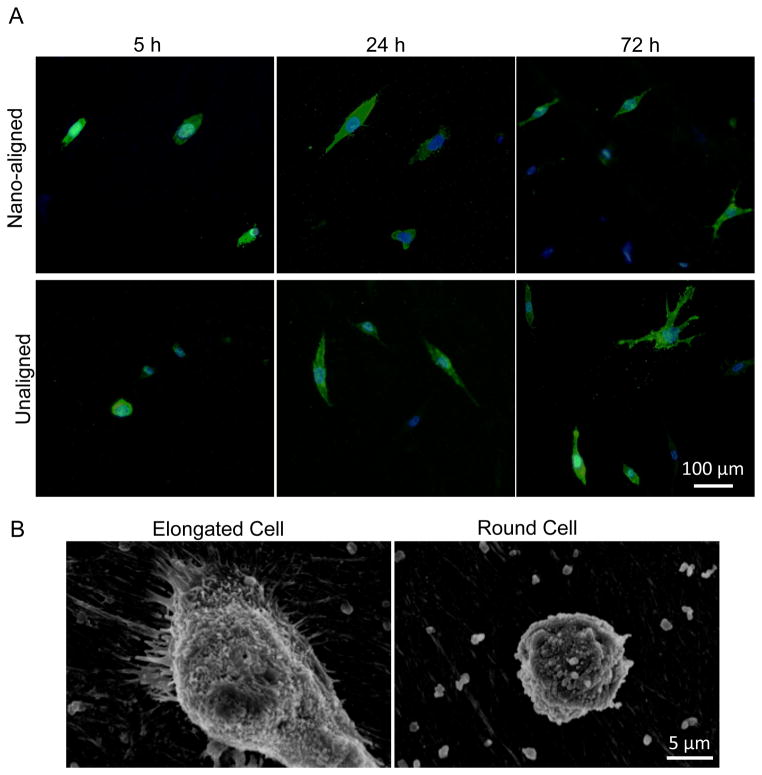
Figure 6.
Morphology of macrophages on ECM scaffolds. (A) CD 14 immunofluorescent staining of cells grown on nano-aligned and unaligned ECM scaffolds for 5, 24, and 72 h. Scale bar: 100 μm. (B) SEM images of an elongated macrophage and a round macrophage.
2.4 In vitro evaluation of inflammatory response to ECM scaffold
The inflammatory response of the decellularized ECM was investigated by analyzing the morphology and cytokine secretion of differentiated macrophages cultured on both aligned and unaligned nanofibrous ECM. The macrophages cultured on a plastic 12-well plate were used as the control. Differentiated macrophages were visualized by immunostaining of CD14 and SEM (Figure 6). The aspect ratio, R-ratio = length of major axis / length of minor axis, was measured for each cell in three different views. The elongated cells were defined as those with R ≥ 2.5. Both round and elongated cells were observed on all the samples. More macrophages examined at 5 h exhibited relatively round shape than at 24 and 72 h. Elongated cells exhibited higher filopodium interactions with surrounding ECM nanofibers than round cells that have few cytoplasmic projections and low spreading (Figure 6B). The percentage of elongated cells and the average aspect ratio of elongated cells on aligned ECM were not significantly different than on unaligned ECM (Supplemental Figure 2).
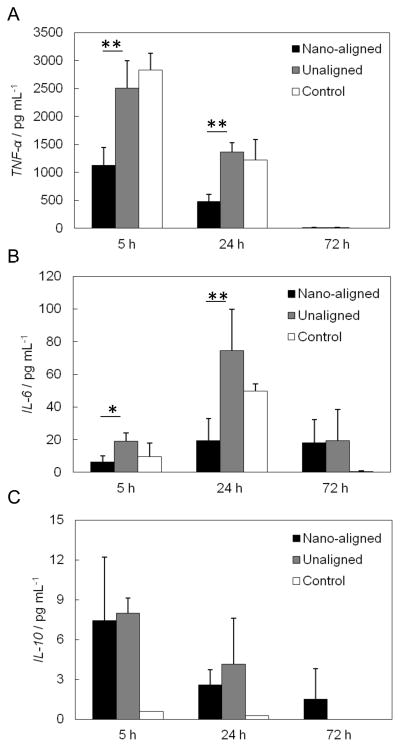
Macrophages can secret different cytokines and chemokines in order to modulate the immune response to the presence of biomaterials. Evaluation of the cytokine secretion level can therefore help determine whether a material is suitable for implantation. The cytokines examined were TNF-α, IL-6, and IL-10 (Figure 8). At 5 h, the average TNF-α secretion levels from aligned and unaligned nanofibrous ECM were lower than the corresponding control. Although unaligned samples showed no significant difference relative to the control (P > 0.05), the aligned samples showed a 45% lower secretion of TNF-α than the unaligned samples (P < 0.01) and a 60% lower secretion relative to the control (P < 0.01). All the 24 h samples had significantly lower TNF-α secretion than 5 h samples. The average secretion from aligned samples was still significantly lower than the unaligned samples (P < 0.01) and the corresponding control (P < 0.05). The secretion from the decellularized unaligned samples still showed no statistical difference from the control (P > 0.05). At 72 h, TNF-α secretion from all the samples dramatically decreased. The average IL-6 secretion followed a similar trend as TNF-α. The nano-aligned samples showed significantly lower IL-6 expression than unaligned samples at both 5 h (P < 0.05) and 24 h (P < 0.01). At 72 h, the nano-aligned samples and unaligned samples secreted similar levels of IL-6. The unaligned samples had comparable IL-6 secretion to the controls at all time points. There was a decreasing trend of IL-10 secretion in macrophages with time. At 5 h and 24 h, the nano-aligned and unaligned samples expressed similar levels of IL-10 (P > 0.05), but both were significantly higher than the control (P < 0.05).
3. Discussion
Three-dimensional (3D) engineered scaffolds that mimic the structure and composition of native tissues possess elevated promise in regenerating more functional tissues. Cells are profoundly influenced by the surrounding nanofibrous environment, which includes topography, rigidity, and extracellular biochemical cues.[23] Current 3D nanofibrous scaffold fabrication in the field is dominated by an electrospinning technique that can control the mechanical and chemical properties of the nanofibers.[24] Nevertheless, only natural ECM derived from cells or tissues can potentially provide suitable nanofibrous topography, mechanical support, and chemical and biological stimuli at the same time. For this reason, it is critical to create a completely natural 3D nanofibrous scaffold that mimics ECM in native tissues, including both morphology and biochemical constituents. Cultured cells can be screened for pathogens and then maintained in a pathogen-free condition for ECM harvesting. Therefore, cell-derived ECM offers a promising alternative to the use of ECM matrices derived from natural tissues. In addition, the cell-derived ECM scaffolds eliminate the provocation of undesirable inflammatory and immunological reactions from foreign natural tissues and organs.[6, 7] They also provide the desired geometry and porosity without the limitation of poor cell penetration that can occur during the repopulation of decellularized native tissues. In this study we report the development of a uniform, highly aligned, and completely biological 3D nanofibrous ECM scaffold derived from human dermal fibroblasts. The alignment of the nanofibers not only holds great potential to engineer highly organized tissues, but also contributes to the improved biocompatibility of the scaffolds.
3.1 Uniformity and alignment
The fibroblasts were grown for up to 8 weeks to obtain a 70 μm-thick cell sheet. The culture period could be shortened to 3 weeks using high cell seeding density and supplementation of growth factors, including epidermal growth factor, basic fibroblast growth factor, as well as dexamethasone and insulin into cell culture medium.[21] This chemically enhanced medium could promote cell growth and ECM protein synthesis. The long-term cultured cell sheet can be manually detached from the PDMS substrate without damaging the whole structure. Almost all other cell sheet engineering techniques require a thermoresponsive polymer coating on substrates to facilitate the detachment of cell sheet. Our results showed that use of a long-term cultured thick fibroblast cell sheet could significantly simplify the procedure.
Cells can sense the topography of the underlying substrate and respond to the physical cues by adjusting their alignment and migration.[17] We found that as cell layers became thicker and thicker due to the continuous cell proliferation and ECM deposition, each newly formed layer of cells and ECM still followed the orientation guidance of the underlying layer. After 1 week culture, an average of 88% of the cells were highly aligned, which demonstrated the effectiveness of the nanograted substrate in directing the cell alignment. In the following 7 weeks, at least 65% of the cells remained organized. This result suggests that the cell alignment and matrix deposition in the growing upper layer could be guided by the immediate underlying aligned cell layer without direct contact with the PDMS substrate. The nanogratings are only 130 nm in depth, which is negligible comparing to the thickness of a single cell layer (around 5–10 μm). So the nanogratings only direct the alignment of the cells, but not cause any formation of tears and gratings as shown in those harvested from large arrays of alternating ridges and grooves in micropatterns.[14, 15] Therefore, the cell sheet obtained from these nanogratings is much more uniform.
3.2 Elastic and viscous moduli
Collagen and elastin are the two major structural proteins in the ECM that impart mechanical properties to the natural tissues.[25] Elastin is a highly elastic rubberlike protein that has the ability to stretch 2–3 times its initial length and recoil back to the original state with little energy loss.[26] The cross-linked elastic network in engineered arteries is responsible for preventing vascular dilation in response to the continuous pressure exerted by blood flow in vivo[27]. Black et al. proved that loss of elastin significantly decreased the elastic modulus.[28] The elastic modulus after our decellularization treatment decreased; however, the lack of significant change may indicate that most elastic fibers in the ECM scaffolds were preserved. Collagen is nearly 100 times stiffer than elastin and nearly inextensible. It has been demonstrated that collagen is more hysteretic than elastin,[29] and thus this molecule contributes more to the cell sheet's ability to dissipate energy with strain. Proteoglycans are proteins that have covalently attached to highly anionic glycosaminoglycans (GAG) and fill the extracellular space.[30] They can form large complexes with fibrous matrix protein such as collagen[31] and may serve as one of the “adhesive molecules” to glue collagen fibrils together. It was hypothesized that the proteoglycan bridging between collagen fibrils played a role in transmitting and resisting tensile stresses and contributed to the strength of the tissues.[32] Another work showed that one of the major functions of GAGs in the tissue was water retention. Removal of GAGs had a negative effect on the viscoelastic properties of the scaffold.[33] Decellularization processes can cause a significant effect on the content of ECM biomacromolecules. The amount of collagens, elastin, fibronectin and GAGs generally decrease but are still present in different decellularized tissues. For example, SDS treatment can significantly decrease GAG but retain collagen relative amount in native rat liver.[34] Acellular pig pleura retained gross configuration of elastin and collagen layers, but did not retain collagen IV or laminin.[35] Based on the above studies, the significant decrease in viscous modulus after decellularization treatment could be due to the loss of the “adhesion molecules” including GAGs, which decreased the friction between adjacent collagen fibers or water content in the tissue.[36] The SEM images in Figure 3B clearly show that the filler substance between fibers was significantly removed after decellularization, but the nanofiberous microstructure was well retained.
3.3 Biocompatibility
Although some studies have confirmed that human fibroblasts possess similar immunosuppresive functions as hMSCs, the immunomodulatory properties of the ECM obtained from decellularized fibroblast cell sheet have not been tested. In the absence of fibroblasts, the nano-aligned ECM still showed improved biocompatiblity compared with unaligned ECM and plastic controls. The classical activation of macrophage cells in M1 phase leads to production of pro-inflammatory cytokines such as TNF-α, IL-6 and IL-1β. These cytokines are typically associated with inflammation, tumor resistance, and graft rejection. The alternative M2 activation of macrophage cells stimulates the production of anti-inflammatory cytokines such as IL-10, TGF-β and vascular endothelial growth factor (VEGF), which are involved in immunoregulation, matrix deposition, and remodeling. The expression level of TNF-α and IL-6 significantly downregulated in aligned nanofibrous ECM scaffolds compared with their unaligned counterparts, indicating a lower inflammatory response. It has been suggested that TNF-α cytokine production from macrophages is surface texture dependent, especially at the nanometric scale.[37] Several studies on nanograted substrates including Ti-coated Si wafer (33 – 158 nm in depth), polycaprolactone (PCL) (114 – 1,972 nm) and PDMS (350 nm in depth) showed that the in vitro immunological response to a biomaterial surface can be altered by introducing the nanoscale features.[12, 37] However, these substrates cannot recapitulate the 3D fibrous characteristics of natural ECM. Electrospun nanofibers can provide a 3D nanofibrous environment for cells. Several studies demonstrated that the aligned nanofibrous scaffold had lower in vitro TNF-α secretion at 24 h and induced less foreign body response in vivo than a unaligned fiber scaffold.[11, 38] However, all of these tested nanofibers are synthetic materials that can not mimic the real protein-rich environment in the exudates and might cause the inconsistency between in vitro and in vivo results.[37] In addition, the diameter of these nanofibers ranges from 313 to 1,600 nm, a submicron to micro scale that is much larger than the ECM fibers in native tissues. The fiber size (average diameter 78 ± 9 nm) of our scaffold is comparable to the natural ECM fibers such as organized collagen I fibrils (fiber diameter around 80 – 100 nm).[39] Therefore, by comparing the macrophages-mediated immune response to aligned and randomly organized nanofibrous ECM scaffolds, we definitively proved that the alignment of fibers is a crucial parameter that positively influences the biocompatibility of biomaterials.
Cells, including macrophages, can attach to the ECM via various surface receptors to trigger a series of cellular signals to direct the cell adhesion, migration, proliferation, protein synthesis and secretion. Our natural ECM scaffold is composed of a complex of proteins and polysaccharides. All of these components may contribute to the biocompatibility of the scaffold. It has been demonstrated that elastin, as an ECM protein, can effectively enhance the immunosuppressive function of polydioxanone electrospun nanofibers.[40] The hyaluronic acid, a polysaccharide in natural ECM, possesses immunomodulatory properties and can dampen inflammatory macrophage activities.[41] Therefore, the rich extracellular matrices constituting the scaffold may play roles in improving the scaffold biocompatibility. Moreover, the signaling molecules including growth factors, cytokines and chemokines trapped in the ECM also regulate cellular functions. After tissue decellularization, the amount of growth factors such as basic fibroblast grow factor (bFGF), transforming growth factor beta 1 (TGF-β1), and VEGF decreased but were still present in the tissues.[42] Aligned fibroblast cell sheets have been shown to secrete significantly higher amount of VEGF than the cell sheets with random cell orientation.[14] It is possible that the aligned and unaligned fibroblast cell sheets retain different quantities of signaling factors after decellularization, and these preserved signaling molecules further contribute to the immunomodulatory property of aligned ECM nanofibers. The exact mechanism of these possible parameters will be investigated in the future.
4. Conclusions
We constructed a uniform and highly aligned scaffold solely composed of natural ECM molecules, which possessed a nanofibrous structure with the fiber diameter (78 ± 9 nm). This fiber size is comparable to natural ECM protein fibers. The scaffold supported cell proliferation and provided directional cues for cell alignment. The nano-aligned ECM scaffold showed superior in vitro immunosuppression compared with unaligned ECM and conventional tissue culture plastics. The natural, highly aligned, nanofibrous ECM scaffold holds great potential in providing a biomimetic cell delivery platform for various tissue engineering applications.
5. Experimental Section
Fibroblast cell sheet culture
A nanopattern was produced on a PDMS substrate surface using soft lithography following our previous publication.[17] The gratings on the substrate were 130 nm in depth and 350 nm in width (Supplemental Figure 3). The nano-patterned PDMS was coated with bovine collagen I to facilitate cell adhesion.[17] A PDMS substrate without any patterns was used to prepare unaligned or randomly organized cell sheets and ECM scaffolds. Human dermal fibroblasts (ATCC, Manasass, VA) at the passage between 3 to 5 were seeded on the PDMS at a density of 6,000 cells cm−2. The cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 20% fetal bovine serum (FBS), 20% Ham F12, 500 μM sodium ascorbate, and 1% penicillin/streptomycin (Life Technologies, Rockville, MD). The culture was maintained by changing medium twice per week and cells were allowed to proliferate for up to 8 weeks. The cell sheet was harvested by gently pulling the cell layers off the PDMS substrate.
Cell sheet decellularization
The fibroblast cell sheet was placed into the first decellularization solution, which contained 1 M NaCl, 10 mM Tris, and 5 mM EDTA (Sigma, St Louis, MO). The cell sheet was shaken for 1 h at room temperature and rinsed thoroughly with phosphate buffered saline (PBS). The cell sheet was then placed in a second decellularization solution containing 0.5% SDS, 10 mM Tris, and 25 mM EDTA (Sigma, St Louis, MO), and shaken for 0.5 h at room temperature. After a PBS wash, the sample was rinsed in DMEM medium with 20% FBS for 48 h at room temperature and rinsed again with PBS.
Cell and ECM protein characterization
The expression of the ECM proteins collagen I and fibronectin were examined by immunofluorescent staining. Samples were fixed, blocked, and incubated with the primary antibody against fibronectin/collagen I (Abcam, Cambridge, MA). Samples were washed and incubated with a mixture of secondary antibodies conjugated to Alexa Fluor 488 and phalloidin conjugated to Alexa Fluor 594 (Life Technologies, Rockville, MD). The samples were then washed and incubated in 4′,6-diamidino-2-phenylindole (DAPI) solution to counter stain the cell nuclei. Finally the samples were mounted and viewed using an Olympus BX-51 fluorescent microscope. The Elastin was stained using a Verhoeff Van Gieson Elastin Stain Kit (Polysciences, Warrington, PA).
Scanning electron microscopy (SEM) imaging
Samples were fixed with 2% glutaraldehyde, washed with PBS, and then dehydrated through a graded series of ethanol. Finally the samples were dried in Hexamethyldisilazane (Sigma, St Louis, MO) and viewed using a Hitachi S-4700 field emission scanning electron microscope. The SEM images were analyzed using Photoshop software to calculate the average diameter of protein fibers. Three digital images of each time point were measured with 30 fibers in every image.
Mechanical testing
The mechanical strength measurement was performed in Bohlin CVOR rheometer (Malvern Instruments, UK) using a parallel-plate of 25 mm in diameter. The measurement went through frequency scanning at room temperature in the range of 0.1–2 Hz. The elastic modulus and viscous modulus were recorded as a function of frequency. Each measurement was performed at least three times on three different samples.
hMSCs proliferation and morphology assay
Bone marrow–derived hMSCs were provided by Tulane University Health Sciences Center.[17] The decellularized fibroblast cell sheet was sterilized with 70% ethanol and rinsed with PBS. Passage 5 hMSCs were seeded at the density of 7,000 cells cm−2 and cultured in alpha-MEM supplemented with 20% FBS, 1% L-glutamine, and 1% penicillin/streptomycin (Life Technologies, Rockville, MD). At specific time points, samples were fed with BrdU (Sigma, St Louis, MO) for 15 h, and then fixed stained with anti-BrdU conjugated to fluorescein isothiocyanate (FITC) (Life Technologies, Rockville, MD) and DAPI. The percentage of BrdU-positive cells was calculated as the number of BrdU-positive cells divided by the total number of cells obtained from Dapi staining, and the data were pooled for statistical analysis. n=3 for each sample. The F-actin was stained with rodamine phalloidin (Life Technologies, Rockville, MD).
Inflammatory response test
The human acute monocytic leukemia THP1 cells were obtained from ATCC, and maintained in RPMI 1640 medium supplemented with 10% FBS, 0.1% beta-mercaptoethanol and 1% pen-strep. To induce the differentiation of THP1 cells into monocytes-derived macrophages, THP1 cells (6.8 x 105 mL−1) were incubated in the cell culture media added with 200 nM phorbol 12-myristate 13-acetate (PMA) (Sigma, St Louis, MO) for 3 days. The expression of CD14 in the differentiated cells was examined by immunofluorescent staining (Supplemental Figure 2). The differentiated cells were collected and seeded on decellularized cell sheets at the seeding density of 1.1 x 104 cm−2. After 3 days, cell culture medium was changed to serum-free medium containing 1 μg/ml lipopolysaccharide (LPS) (Sigma, St Louis, MO). The medium was collected at 5, 24 and 72 h to quantify the secretion of TNF-α, IL-6 and IL-10 using enzyme-linked immunosorbent assay (ELISA) kit (Abcam, Cambridge, MA). Differentiated macrophages were also seeded on plastic 12-well plate as control. Immunofluorescence staining of macrophage marker CD 14 and SEM were performed to observe the macrophage morphology.
Statistics/data analysis
Experiment results were expressed as means ± standard deviation (SD) of the means of the samples. Student's t-test (Microsoft Excel) was used for comparisons, and statistical significance was accepted at P < 0.05.
Supplementary Material
Figure 5.
Proliferation and morphology of hMSCs seeded on ECM derived from nanopatterned fibroblast cell sheets. (A) BrdU immunofluorescent staining of hMSCs grown for 24 h and 72 h. (B) Percentage of BrdU-positive cells, obtained from arbitrary fields at 24 h and 72 h in culture. Results from arbitrary view fields (n=5) were collected. (C) Immunofluorescent staining of F-actin of hMSCs grown for 24 h and 72 h. Arrows indicate the direction of the alignment.
Figure 7.
Cytokine secretion by differentiated macrophages seeded on the top of the nano-aligned ECM, unaligned ECM and plastic plates (control). (A) TNF-α, (B) IL-6, (C) IL-10. * P< 0.5; ** P<0.01.
Acknowledgments
This study was supported by the National Instiutues of Health (1R15HL115521-01A1 to F.Z., EB015300 to K.W.L), Young Clinical Scientist Award from Flight Attendance Medical Research Institute (062518-YCSA) to F.Z., and the Research Excellence Fund-Research Seed Grant (REF-RS) from Michigan Technological University to F.Z. We also acknowledge the Summer Undergraduate Research Fellowship (SURF) from Michigan Technological University to C.V.
References
- 1.Zhang LJ, Webster TJ. Nano Today. 2009;4:66. [Google Scholar]
- 2.a) Keely PJ, Fong AM, Zutter MM, Santoro SA. J Cell Sci. 1995;108:595. doi: 10.1242/jcs.108.2.595. [DOI] [PubMed] [Google Scholar]; b) Mostafavi-Pour Z, Askari JA, Parkinson SJ, Parker PJ, Ng TTC, Humphries MJ. J Cell Biol. 2003;161:155. doi: 10.1083/jcb.200210176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Koutsopoulos S, Zhang SG. Acta Biomater. 2013;9:5162. doi: 10.1016/j.actbio.2012.09.010. [DOI] [PubMed] [Google Scholar]; b) Subramanian A, Krishnan UM, Sethuraman S. Ann Biomed Eng. 2012;40:2098. doi: 10.1007/s10439-012-0592-6. [DOI] [PubMed] [Google Scholar]; c) Lin YD, Luo CY, Hu YN, Yeh ML, Hsueh YC, Chang MY, Tsai DC, Wang JN, Tang MJ, Wei EIH, Springer ML, Hsieh PCH. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003841. [DOI] [PubMed] [Google Scholar]
- 4.Badylak SF, Freytes DO, Gilbert TW. Acta Biomater. 2009;5:1. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Reing JE, Zhang L, Myers-Irvin J, Cordero KE, Freytes DO, Heber-Katz E, Bedelbaeva K, McIntosh D, Dewilde A, Braunhut SJ, Badylak SF. Tissue Eng Part A. 2009;15:605. doi: 10.1089/ten.tea.2007.0425. [DOI] [PubMed] [Google Scholar]
- 6.Lu HX, Hoshiba T, Kawazoe N, Koda I, Song MH, Chen GP. Biomaterials. 2011;32:9658. doi: 10.1016/j.biomaterials.2011.08.091. [DOI] [PubMed] [Google Scholar]
- 7.Lu HX, Hoshiba T, Kawazoe N, Chen GP. Biomaterials. 2011;32:2489. doi: 10.1016/j.biomaterials.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Pei M, He F, Kish VL. Tissue Eng Part A. 2011;17:3067. doi: 10.1089/ten.tea.2011.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadr N, Pippenger BE, Scherberich A, Wendt D, Mantero S, Martin I, Papadimitropoulos A. Biomaterials. 2012;33:5085. doi: 10.1016/j.biomaterials.2012.03.082. [DOI] [PubMed] [Google Scholar]
- 10.a) Papadaki M, Bursac N, Langer R, Merok J, Vunjak-Novakovic G, Freed LE. Am J Physiol-heart C. 2001;280:H168. doi: 10.1152/ajpheart.2001.280.1.H168. [DOI] [PubMed] [Google Scholar]; b) Vunjak-Novakovic G, Altman G, Horan R, Kaplan DL. Annu Rev Biomed Eng. 2004;6:131. doi: 10.1146/annurev.bioeng.6.040803.140037. [DOI] [PubMed] [Google Scholar]
- 11.Saino E, Focarete ML, Gualandi C, Emanuele E, Cornaglia AI, Imbriani M, Visai L. Biomacromolecules. 2011;12:1900. doi: 10.1021/bm200248h. [DOI] [PubMed] [Google Scholar]
- 12.Chen SL, Jones JA, Xu YG, Low HY, Anderson JM, Leong KW. Biomaterials. 2010;31:3479. doi: 10.1016/j.biomaterials.2010.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda N, Shimizu T, Yamato M, Okano T. Adv Mater. 2007;19:3089. [Google Scholar]
- 14.Isenberg BC, Backman DE, Kinahan ME, Jesudason R, Suki B, Stone PJ, Davis EC, Wong JY. J Biomech. 2012;45:756. doi: 10.1016/j.jbiomech.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Isenberg BC, Tsuda Y, Williams C, Shimizu T, Yamato M, Okano T, Wong JY. Biomaterials. 2008;29:2565. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi H, Nakayama M, Shimizu T, Yamato M, Okano T. Biomaterials. 2011;32:8830. doi: 10.1016/j.biomaterials.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhao F, Veldhuis JJ, Duan YJ, Yang Y, Christoforou N, Ma T, Leong KW. Mol Ther. 2010;18:1010. doi: 10.1038/mt.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L'Heureux N, Paquet S, Labbe R, Germain L, Auger FA. Faseb J. 1998;12:47. doi: 10.1096/fasebj.12.1.47. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa O, Kondo A, Okada K, Miyachi Y, Furumura M. Brit J Dermatol. 1997;136:6. [PubMed] [Google Scholar]
- 20.Mitchell SL, Niklason LE. Cardiovasc Pathol. 2003;12:59. doi: 10.1016/s1054-8807(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 21.Ahlfors JEW, Billiar KL. Biomaterials. 2007;28:2183. doi: 10.1016/j.biomaterials.2006.12.030. [DOI] [PubMed] [Google Scholar]
- 22.Friedl P, Brocker EB. Cell Mol Life Sci. 2000;57:41. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toh YC, Ng S, Khong YM, Zhang X, Zhu YJ, Lin PC, Te CM, Sun WX, Yu HR. Nano Today. 2006;1:34. [Google Scholar]
- 24.Nisbet DR, Forsythe JS, Shen W, Finkelstein DI, Horne MK. J Biomater Appl. 2009;24:7. doi: 10.1177/0885328208099086. [DOI] [PubMed] [Google Scholar]
- 25.Lu QJ, Ganesan K, Simionescu DT, Vyavahare NR. Biomaterials. 2004;25:5227. doi: 10.1016/j.biomaterials.2003.12.019. [DOI] [PubMed] [Google Scholar]; Uchimura E, Sawa Y, Taketani S, Yamanaka Y, Hara M, Matsuda H, Miyake J. J Biomed Mater Res Part A. 2003;67A:834. doi: 10.1002/jbm.a.10097. [DOI] [PubMed] [Google Scholar]
- 26.Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T. Phil Trans R Soc B. 2002;357:169. doi: 10.1098/rstb.2001.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell SL, Niklason LE. Cardiovasc Pathol. 2003;12:59. doi: 10.1016/s1054-8807(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 28.Black LD, Allen PG, Morris SM, Stone PJ, Suki B. Biophys J. 2008;94:1916. doi: 10.1529/biophysj.107.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mijailovich SM, Stamenovic D, Brown R, Leith DE, Fredberg JJ. J Appl Physiol. 1994;76:773. doi: 10.1152/jappl.1994.76.2.773. [DOI] [PubMed] [Google Scholar]
- 30.Perrimon N, Bernfield M. Semin Cell Dev Biol. 2001;12:65. doi: 10.1006/scdb.2000.0237. [DOI] [PubMed] [Google Scholar]
- 31.a) Lewis PN, Pinali C, Young RD, Meek KM, Quantock AJ, Knupp C. Structure. 2010;18:239. doi: 10.1016/j.str.2009.11.013. [DOI] [PubMed] [Google Scholar]; b) Scott JE. Ciba Found Symp. 1986;124:104. doi: 10.1002/9780470513385.ch7. [DOI] [PubMed] [Google Scholar]
- 32.Cribb AM, Scott JE. J Anat. 1995;187:423. [PMC free article] [PubMed] [Google Scholar]
- 33.Lovekamp JJ, Simionescu DT, Mercuri JJ, Zubiate B, Sacks MS, Vyavahare NR. Biomaterials. 2006;27:1507. doi: 10.1016/j.biomaterials.2005.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren HZ, Shi XL, Tao L, Xiao JQ, Han B, Zhang Y, Yuan XW, Ding YT. Liver Int. 2013;33:448. doi: 10.1111/liv.12088. [DOI] [PubMed] [Google Scholar]
- 35.Nichols JE, Niles J, Riddle M, Vargas G, Schilagard T, Ma L, Edward K, La Francesca S, Sakamoto J, Vega S, Ogadegbe M, Mlcak R, Deyo D, Woodson L, McQuitty C, Lick S, Beckles D, Melo E, Cortiella J. Tissue Eng Part A. 2013;19:2045. doi: 10.1089/ten.tea.2012.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mok MT, Ilic MZ, Handley CJ, Robinson HC. Arch Biochem Biophys. 1992;292:442. doi: 10.1016/0003-9861(92)90014-n. [DOI] [PubMed] [Google Scholar]
- 37.Lamers E, Walboomers XF, Domanski M, Prodanov L, Melis J, Luttge R, Winnubst L, Anderson JM, Gardeniers H, Jansen JA. Nanomed Nanotech Biol Med. 2012;8:308. doi: 10.1016/j.nano.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Cao HQ, McHugh K, Chew SY, Anderson JM. J Biomed Mater Res Part A. 2010;93A:1151. doi: 10.1002/jbm.a.32609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner S, Wagner HD. Annu Rev Mater Sci. 1998;28:271. [Google Scholar]
- 40.Sell S, Barnes C, Smith M, McClure M, Madurantakam P, Grant J, McManus M, Bowlin G. Polym Int. 2007;56:1349. [Google Scholar]
- 41.Franz S, Allenstein F, Kajahn J, Forstreuter I, Hintze V, Moller S, Simon JC. Acta Biomater. 2013;9:5621. doi: 10.1016/j.actbio.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Reing JE, Brown BN, Daly KA, Freund JM, Gilbert TW, Hsiong SX, Huber A, Kullas KE, Tottey S, Wolf MT, Badylak SF. Biomaterials. 2010;31:8626. doi: 10.1016/j.biomaterials.2010.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.