Abstract
Effective approaches to neuropsychiatric disorders require detailed understanding of the cellular composition and circuitry of the complex mammalian brain. Here, we present a paradigm for deconstructing the diversity of neurons defined by a specific neurotransmitter, using a microfluidic dynamic array to simultaneously evaluate the expression of 96 genes in single neurons. With this approach, we successfully identified multiple molecularly distinct dopamine neuron subtypes, and localized them in the adult mouse brain. To validate the anatomical and functional correlates of molecular diversity, we provide evidence that one Vip+ subtype, located in the periaqueductal region, has a discrete projection field within the extended amygdala. Another Aldh1a1+ subtype, located in the substantia nigra, is especially vulnerable in the MPTP model of Parkinson’s disease. Overall, this rapid, cost-effective approach enables the identification and classification of multiple dopamine neuron subtypes, with distinct molecular, anatomical, and functional properties.
Introduction
The mammalian central nervous system (CNS) is an extremely heterogeneous organ composed of hundreds of neuronal and non-neuronal cell types (Ng et al., 2009). There is an urgent need to transition from cell groups, traditionally defined by cytology and anatomy, to molecularly defined cell types in order to generate a comprehensive catalog of the mammalian CNS (Bota and Swanson, 2007; Fishell and Heintz, 2013). In this regard, several in situ hybridization and transgene profiling studies have served as an important step in classification of neurons in given brain regions (Shimogori et al., 2010; Siegert et al., 2009).
The next important step towards classifying cellular diversity requires the analysis of a multitude of markers with single-cell resolution. Molecular definition of neurons should rely on a polythetic approach, rather than stochastically expressed markers (Bota and Swanson, 2007). This transition is mandatory for accurately defining neuronal circuitry and its relation to complex neural functions and behaviors. An ideal approach to investigate neuronal diversity would allow a quick, reliable and quantitative assessment of the gene expression profile of single neurons obtained from the mammalian brain. Although recent reports of single-cell transcriptomics have been published, this approach is cost-prohibitive and has yet to reach its maturity before being broadly applicable to the mammalian CNS (Macaulay and Voet, 2014; Tang et al., 2010; Xue et al., 2013). As an alternative technology to simultaneously profile multiple genes from a single cell, we used a microfluidic chip array to investigate neuronal diversity from mouse brain.
We focused on the investigation of the clinically important midbrain dopaminergic (DA) system. Midbrain DA neurons influence a large spectrum of behaviors and DA dysfunction is prominently implicated in a wide variety of disorders including Parkinson’s disease (PD), schizophrenia, attention deficit hyperactivity disorder (ADHD), obsessive-compulsive, addiction and depression, together affecting tens of millions of people. How a small group of neurons, constituting less than 1% of brain neuronal populations, can account for such diverse physiological functions has been a subject of interest since their discovery. We hypothesized that multiple molecularly distinct DA subtypes could be partly responsible for the functional diversity of the DA system.
During embryonic development, midbrain DA neurons are derived from the Shh+ floor plate (FP) (Blaess et al., 2011; Hayes et al., 2011; Joksimovic et al., 2009; Kittappa et al., 2007). FP progenitors already display some heterogeneity, in terms of gene expression and proclivity to contribute to distinct nuclei (Blaess et al., 2011; Hayes et al., 2011; Joksimovic et al., 2009; Wallén et al., 1999). Immature DA neurons migrate from the ventricular zone into the mantle, and express a series of transcription factors and other genes, some of which are heterogeneously expressed in the nascent DA cohort (Ang, 2006; Chung et al., 2010; Di Salvio et al., 2010a; Smidt and Burbach, 2007; Smits et al., 2013). Thus, the earliest aspects of DA neuron heterogeneity appear to be established in the early embryo, although it is likely that subsequent maturation events build on, or refine, DA heterogeneity. This model is akin to that proposed for the generation of cortical interneuron lineages, wherein embryonic cardinal classes are established, and sequentially refined, to generate the full repertoire of interneurons (Kepecs and Fishell, 2014).
When considering the possibility of distinct postnatal DA neuron populations, the overwhelming literature is based on anatomical classification of midbrain DA neurons to three clusters: the substantia nigra pars compacta (SNc; A9), the ventral tegmental area (VTA; A10), and a third smaller cluster, found in the retrorubral area (RR; A8). Accordingly, several studies have addressed the molecular heterogeneity of DA neurons in anatomically defined SNc vs VTA populations with microarray analysis (Chung et al., 2005; Greene et al., 2005; Grimm et al., 2004). Although informative, these studies rely on the premise that the SNc and VTA are composed of molecularly homogeneous populations. Considering the diverse functions of this neurotransmitter system, and recent reports of physiological and functional heterogeneity (Darvas and Palmiter, 2009; Di Salvio et al., 2010b; Ekstrand et al., 2014; Guzman et al., 2010; Lammel et al., 2008; 2012; Margolis et al., 2006a; 2006b; Stamatakis et al., 2013; Tye et al., 2011), we postulated the existence of several molecularly distinct DA neuron subtypes, which may not necessarily be segregated anatomically, but rather may be intermingled with one another. Providing the molecular identity of DA neuron subtypes would have meaningful implications for: (i) targeted therapeutics of the DA system, (ii) understanding the differential vulnerability of DA neurons, (iii) developing more accurate stem cell and induced pluripotent stem cells derived DA neurons, and (iv) a deeper understanding of the DAergic system by providing points of genetic access, thus allowing genetic dissection of this neurotransmitter system.
Results
Comprehensive single-cell gene expression profile of midbrain dopamine neurons
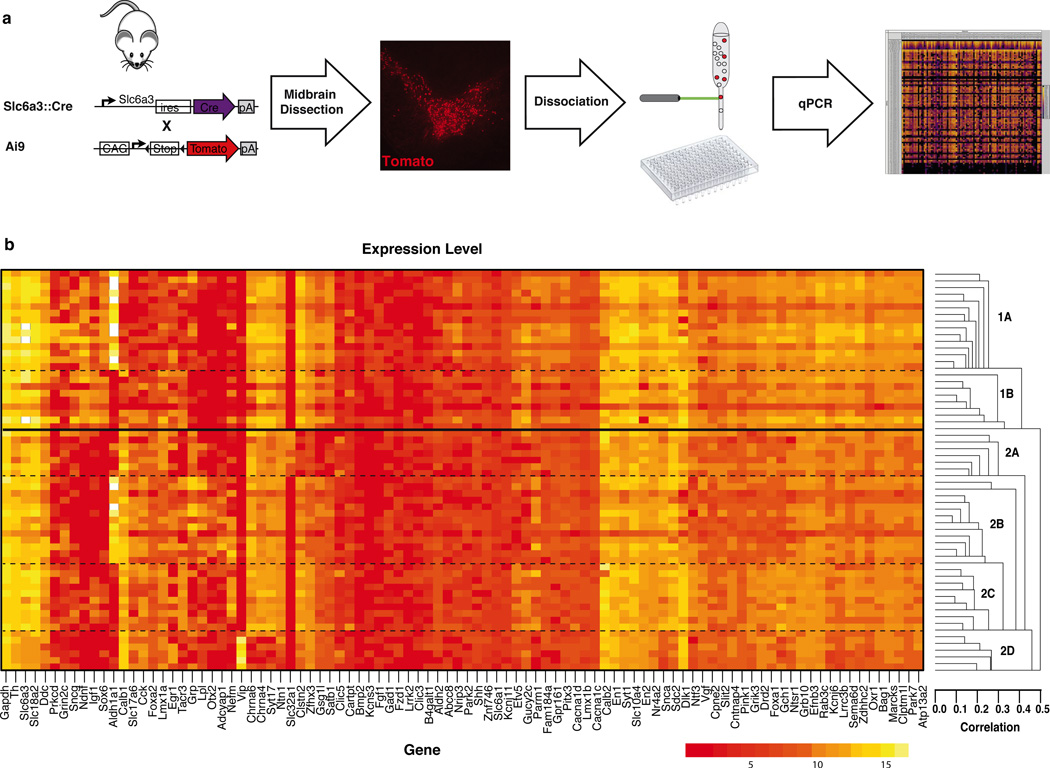
To redefine the DA system on a molecular basis we devised a novel, neuroanatomically unbiased, systematic approach to profile single midbrain DA neurons. We opted for a 96.96 dynamic array to evaluate 96 cells, each for the expression of 96 key genes, an approach used previously to discriminate stem cell identities (Buganim et al., 2012; Guo et al., 2010). We selected 96 gene candidates based on reported differential expression between SNc and VTA (Chung et al., 2005; Greene et al., 2005; Grimm et al., 2004), with a positive bias towards genes with validated midbrain mRNA expression as shown by in situ hybridization in public databases (Table S1). In addition, we evaluated the expression of housekeeping genes (Actb, Gapdh, Hprt), genes linked to PD (Atp13a2, Lrrk2, Park2, Park7, Pink1, Snca,), as well as validated DA neuronal markers (Ddc, Th, Slc6a3, Slc18a2). All primers used were pre-validated using serial dilution of commercial reference samples (see online Material and Method). To isolate single DA neurons, we grossly dissected and dissociated the midbrain of postnatal day four (P4) Slc6a3::Cre, Ai9 mouse. These mice have been described previously (Bäckman et al., 2006; Madisen et al., 2010), and in our hands, resulted in more than 95% of Th+ midbrain DA neurons expressing tdTomato following Cre dependent recombination. Fluorescently labeled single cell expressing tdTomato were FACS sorted into 96-well plates (Fig. 1a). P4 was selected as a specific time point, in part because of the technical ease of FACS sorting at this stage (Arlotta et al., 2005). Supporting this choice, it has been argued that neuronal molecular identity is established shortly after cells exit their last mitotic cycle (Fishell and Heintz, 2013). Indeed, the vast majority of markers that we selected are expressed in the developing embryo or in the neonate supporting P4 as a meaningful time point (Hoekstra et al., 2013; Veenvliet et al., 2013).
Figure 1. High throughput single-cell gene expression analysis of midbrain dopamine neurons.
(a) The midbrain of postnatal day four Slc6a3::Cre; Ai9 midbrain was dissected and dissociated. Fluorescent (tdTomato+) single cells were FACS sorted into 96-well plate and analyzed for the expression of 96 genes using Fluidigm Biomark system. (b) A sample array in which the expression level of each gene is depicted on a logarithmic scale with red indicating low expression and yellow representing a high expression. Unbiased clustering analysis revealed the presence of two main clusters that could be further divided into a total of six molecularly distinct dopamine (DA) neuron subtypes.
In the initial experiment, 87 single cells were sorted in 96-well plate for gene expression analysis (Fig. 1b). The remaining 9 wells included a brain cDNA dilution series to generate a standard curve. The Biomark microfluidics chip allow the combination of samples and primer-probe sets for 9,216 quantitative RT-PCR (RT-qPCR) reactions with cycle threshold (Ct) values as output (Fig. S1). For quality control purposes, the melting curve of every reaction was determined, and every reaction with a peak falling outside the expected value was removed from the analysis (example of standard curve and melting curve are provided in Fig. S2). This experiment was replicated by two other arrays filled with single cells sorted from two additional mice. Using a cutoff criterion of two standard deviations away from the median of the housekeeping gene Gapdh, and key DA neurons defining genes Th, Slc6a3 (Dat), Slc18a2 (Vmat2), and Ddc (Aaad), we removed all cells exhibiting a weak and unreliable expression. Altogether, we were left with 159 DA neurons for further analysis and the complete expression profile of these cells is provided in Fig. S3. All these neurons had expression of key transcription factors such as Foxa1, En1/2, Lmx1b, Pitx3 and Nr4a2; since the combinatorial expression of these transcription factors is unique to midbrain DA neurons, their expression confirms the absence of hypothalamic DA neurons in the FACS sorted cells. Additionally, all samples had virtually undetectable levels of Gad1, confirming the absence of GABAergic neurons.
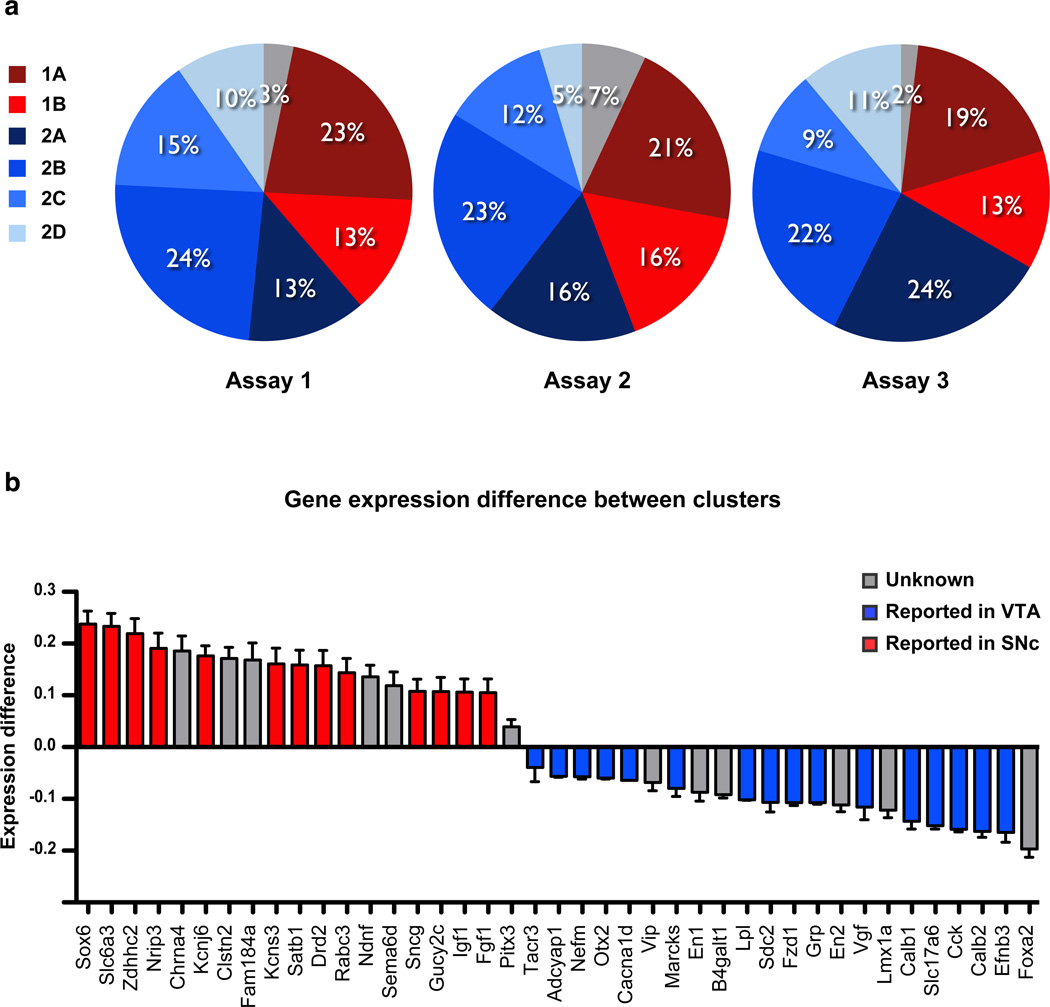
From gene expression data (Fig. 1b and S3), it is apparent that a large number of genes are expressed in all DA neurons. The genes expressed in most DA neurons include PD-linked genes Atp13a2, Lrrk2, Park2, Park7, and Pink1, as well as the transcription factors Foxa1, En1/2, Lmx1b, Pitx3 and Nr4a2. In contrast to these uniformly expressed genes, several genes are differentially expressed between individual DA neurons. As a first approach to investigate molecular heterogeneity of DA populations, we used an unbiased coefficient similarity hierarchical clustering analysis to parcel these cells according to their gene expression profile (Fig. 1b and S4). This analysis revealed the presence of two main clusters that could be further divided into a total of six molecularly distinct DA subtypes (Fig. 2a). Out of 159 cells, 55 cells (35%) belong to the first cluster, 98 cells (61%) were assigned to the second cluster, whereas six cells (4%) could not be assigned. The first cluster can be subdivided into two subtypes, here referred to as DA1A and DA1B, which are composed of 33 cells (21%) and 22 cells (14%) respectively. The second cluster can be subdivided into four molecularly distinct subtypes labeled DA2A (18%), DA2B (23%), DA2C (12%), and DA2D (9%). Interestingly, these six molecular subtypes were represented in a roughly similar proportion in all three microfluidic arrays (Fig. 2a).
Figure 2. Dopamine neurons cluster into distinct subtypes.
(a) Out of 159 cells, 55 cells (35%) belong to the first cluster, 98 cells (62%) were assigned to the second cluster, whereas six cells (3%) could not be assigned. Overall, clustering analysis identified six DA subtypes, represented in a roughly similar proportion in all three microfluidic arrays. The first cluster can be subdivided into two subtypes, referred to as DA1A and DA1B, which are composed of 33 cells (21%) and 22 cells (14%) respectively. The second cluster can be subdivided into four molecularly distinct subtypes, labeled DA2A (18%), DA2B (23%), DA2C (12%), and DA2D (9%). (b) We compared the gene expression level normalized to Gapdh between the two identified clusters. Genes previously reported to be overexpressed in the VTA are illustrated in blue and genes overexpressed in the SNc illustrated in red (see Table S1 for references). This quantitative analysis demonstrates the similarity between the gene expression profile of Cluster 1 and previously reported profile of SNc, as well as the similarity between Cluster 2 and previously reported profile of the VTA.
We compared the relative gene expression level, normalized to Gapdh, of the two identified main clusters by subtracting the average value of cells assigned to the second cluster to the average value of cells assigned to the first cluster (Fig. 2b). Gapdh proved to be a reliable housekeeping gene since there were significant correlations between this gene and other known housekeeping genes, Actb (r = 0.939, n = 105, p < 0.0001) and Hprt (r = 0.922, n = 105, p < 0.0001). This analysis revealed significant differences between these clusters in the expression of some key genes. For instance, Sox6, Slc6a3, Zdhhc2, and Nrip3 have a higher expression in the first, compared to the second cluster. Interestingly, those four genes were previously reported to have a higher expression in pooled cells of the SNc compared to the VTA (illustrated in red in Fig. 2b; see Table S1 for references). In comparison, we found that Calb1, Slc17a6, Cck, Calb2, and Efnb3 are five genes found to have a higher expression in the second cluster. These five genes, like the majority of genes overexpressed in the second cluster, were previously associated with cells of the VTA (illustrated in blue in Fig. 2b; see Table S1 for references). It is important to note that cells were not identified based on their neuroanatomical position, since the anatomical origin of each FACS sorted cell is undetermined. However, we can conclude that the expression profile of cells from cluster 1 has more similarity with the molecular signature of cells of the SNc, whereas cells from cluster 2 have an expression profile related to the VTA. This prediction is indeed borne out in subsequent anatomical analysis, which show that DA1A and DA1B (cluster 1) are more localized to the SNc, whereas DA2A–D (cluster 2) are preferentially localized to the VTA.
Molecular signature of dopamine neuron subtypes
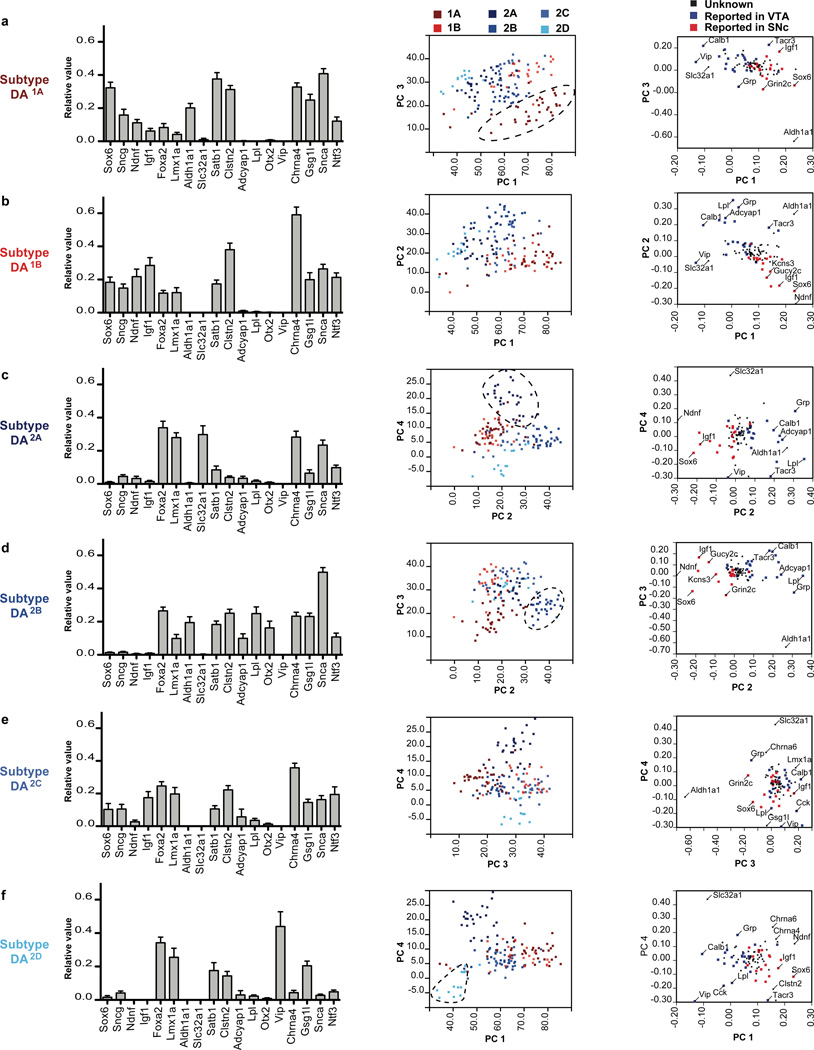
The DA neuron subtypes defined by clustering analysis harbor an organized gene expression profile, which can be used to derive a molecular bar code unique to each subtype (Fig. 3). To gain further insight into the data, we used principal component analysis (PCA) to identify components responsible for the variance observed. Applied to the expression data of the 159 cells, we extracted four principal components (PC) and found that the first principal component (PC1) underlies 22.1% of the observed variance, PC2 contributes 13.8% to the variance, whereas PC3 and PC4 contributes to 8.7% and 6.7% of the variance respectively. Together, these four PCs account for more than 50% of the variation of the data set. The first cluster is divided into DA1A and DA1B, and both express high levels of Ndnf, Sox6, and Sncg mRNA and the loading of these genes contribution to PC1 are 0.237 (Ndnf), 0.232 (Sox6) and 0.145 (Sncg) (Fig. 3a–b; Fig. S5a). However, the high expression of Aldh1a1 in DA1A, a gene absent from DA1B, as well as the higher expression of Snca gene, allowed us to differentiate these two populations (Fig. 3a–b). In contrast, Igf1 is more highly expressed in DA1B compared to DA1A. Interestingly, all these genes contribute significantly to PC3, and thus allowed us to segregate those two populations by PCA (Fig. 3a).
Figure 3. The distinctive molecular signatures of the six dopamine neuron subtypes.
The normalized expression profile of the six dopamine (DA) neuron subtypes is illustrated using 18 key genes. Plotting cells on the four extracted components of the principal component analysis allow to differentiate most DA subtypes. (a) DA1A is the only subtype with high expression of Aldh1a1. Sox6, Ndnf and low/absent expression of Foxa2 and Lmx1a. (b) DA1B depicts an expression profile similar to DA1A, but lacks Aldh1a1 expression. (c) DA2A is characterized by the high expression of Slc32a1, but low expression of Satb1, Gsg1l, and Clstn2. (d) DA2B is defined by the unique co-expression of Lpl, Adcyap1, Otx2 and Aldh1a1. (e) No distinctive marker could be identified for subtype DA2C, which also possesses some similarity with DA1B, if it was not for high levels of Calb1, Cck and Slc17a6. (f) DA2D can be identified by the unique expression of Vip, combined with low expression of Snca, Chrna4, and Ntf3.
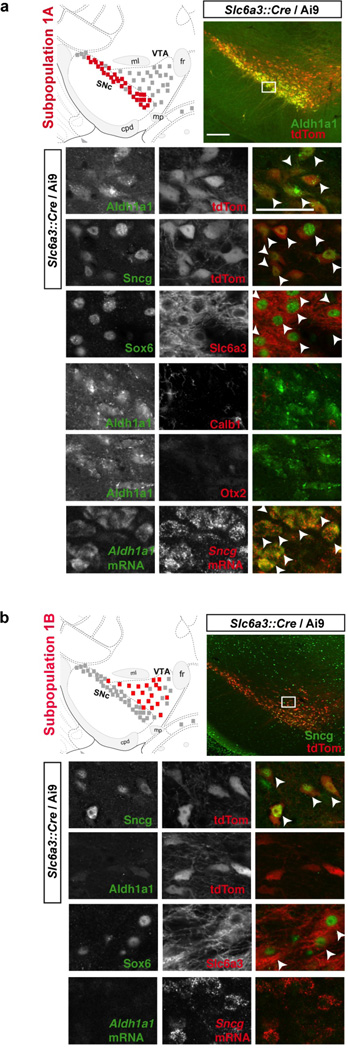
In order to validate these results and anatomically localize these DA subtypes in the adult midbrain, we used a combination of immunofluorescence, fluorescent in situ hybridization, and transgenic mice. The majority of DA1A neurons are located in the ventral tier of the SNc (Fig. 4a), with a few cells observed in RR. In those two regions, Aldh1a1 labels 36.7 ± 1.8% and 12.1± 2.3% of DA neurons respectively, overall accounting for approximately 15.4 ± 1.3% of all midbrain DA neurons. Within the DAergic field, Aldh1a1 antibody overwhelmingly labeled DA neurons (Fig. S6), with occasionally one cell per section appearing negative for Th. Within the SNc, Aldh1a1+ neurons are located in the ventral tier and are also immunolabeled by Sncg and Sox6, but are not labeled with Calb1 and Otx2 antibodies (Fig. 4a). These neurons also co-expressed Aldh1a1 and Sncg mRNA, which is also in complete agreement with our single-cell expression profile of DA1A cells (Fig. 3a). Indeed according to the array, only the DA1A subtype expresses a combination of Aldh1a1, Sox6, Sncg, but do not express Otx2 and Calb1. In contrast to DA1A, DA1B neurons are located in the dorsal part of the SNc, and intermingled with other DA subtypes in the rostro-dorsal VTA (Fig. 4b). These neurons are Sox6+ and Sncg+, but negative for Aldh1a1. Moreover, Calb1 was variably expressed in Sox6 positive cells of DA1B, which also appear to mirror the single cell array data whereby varying levels of Calb1 mRNA were detected in this population.
Figure 4. Molecular and neuroanatomical characterization of dopamine neuron subtypes of the first cluster in the adult brain.
(a) Subtype DA1A is mainly located in the SNc (red square), with a few cells observable in the RR (not shown). These neurons are positive for Aldh1a1, Sncg, and Sox6, but do not show the expression of Calb1 and Otx2. (b) DA1B subtype neurons are generally located just dorsally to DA1A, encompassing the dorsal portion of the SNc and dorso-rostral VTA, as well as RR (not shown). DA1B neurons can be labeled for Sncg, Sox6, but Aldh1a1 is absent in this cell population. Scale bar: low magnification = 200µm, high magnification = 50µm. Colored square = 5 neurons mentioned population, black square = 5 DA neurons.
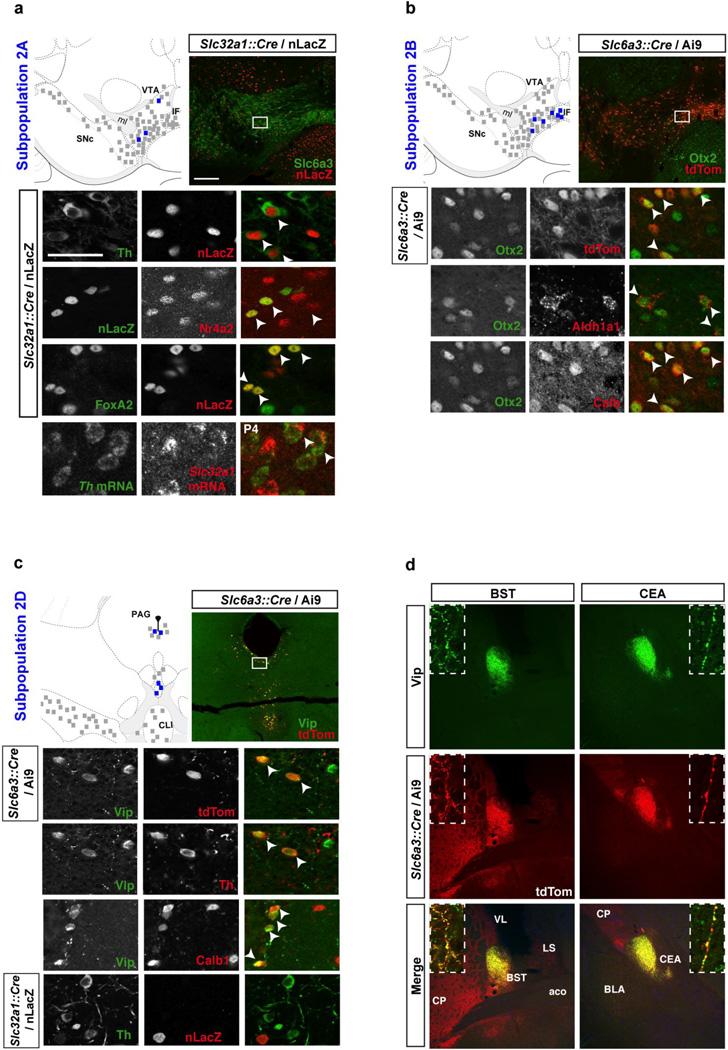
The second cluster can be defined by a particularly high expression of Calb1, Slc17a6, and Cck among others, and can be further divided into four DA subtypes (DA2A–D), each defined by a unique molecular signature (Fig. 3c–f). Subtype DA2A is specifically defined by the expression of the GABA vesicular transporter Slc32a1 (also known as Vgat), since this gene is not significantly expressed by any other subpopulation (Fig. 3c and Fig. S5a). This opens the intriguing possibility that DA2A may co-release GABA and it is conceivable that this subtype matches a described DA population sending an inhibitory projection to lateral habenula (Stamatakis et al., 2013). Notably, these cells did not reliably express Gad1, a key GABA synthesis gene, ruling out the possibility of GABAergic neuron contamination. This DA subtype is also uniquely defined by low expression of a list of genes, which includes Th, Satb1, Clstn2, Gsg1l, Zfhx3, and Tacr3 (Fig. S5). Interestingly, all these genes contribute significantly to PC4, which allowed us to segregate DA2A from the other populations (Fig. 3c). In order to validate the molecular bar code obtained from our single cell approach, we aimed at localizing DA2A neurons in the adult brain. Since Slc32a1 immunoreactivity is localized in dendrites, and is also present in GABAergic neurons, co-immunolabeling analyses proved challenging. Therefore, we crossed Slc32a1::Cre knock-in line to a nuclear LacZ (nLacZ) reporter (Fig. 5a; see methods for details). Although nLacZ was present in a large number of neurons, presumably GABAergic, confocal analysis revealed in some cells, the colocalization of nLacZ with several markers of midbrain DA neurons including Th, Nr4a2, and FoxA2 as shown in Fig. 5a. These neurons were observed intermingled with other cell types within the VTA, IF and CLI regions. Quantification of nLacZ and Nr4a2 co-labeled cells shows that subtype DA2A might comprise approximately 14.4 ± 3.8% of DA neurons of the VTA, but only about 10% of the IF/CLI. We also observed the colocalization of Slc32a1 and Th mRNA supporting our single cell qPCR and Slc32a1::Cre recombination data (Fig. 5a)..
Figure 5. Molecular and neuroanatomical characterization of dopamine neuron subtypes of the second cluster in the adult brain.
(a) Subtype DA2A is uniquely defined by the expression of Slc32a1, here mapped using a Slc32a1::Cre mouse with a nuclear LacZ reporter (nLacZ). These cells are bona fide DA neurons as they can be labeled for Th, Nr4a2 (Nurr1), FoxA2, and Slc6a3 (Dat). These cells are intermingled with other DA subtypes throughout the VTA. (b) DA2B neurons can be labeled with Otx2, Aldh1a1 and Calb1, but not Sox6 (not shown). These cells are positioned mainly in the ventromedial VTA and IF. (c) Subpopulation DA2D is located in the periaqueductal gray (PAG) and dorsal raphe (DR) region, and can be labeled with Vip antibody. These DA cells express Th and FoxA2, and are labeled by Slc6a3::Cre driver. These cells are also Calb1 positive, but negative for Otx2, Aldh1a1, and Slc32a1::Cre. (d) Only subset of midbrain DA neurons axonal projections, as labeled with tdTomato in Slc6a3::Cre / Ai9, colocalized with Vip. These fibers, which originate from subtype DA2D form discrete projections to the oval part of the bed nucleus of the stria terminalis (BSTov) and the lateral part of the central amygdala (CEAl), with apparent boutons colocalizing Vip (green) and tdTomato (red; inset). Scale bar: low magnification = 200µm, high magnification = 50µm. Colored square = 5 neurons mentioned population, black square = 5 DA neurons.
Subpopulation DA2B is uniquely defined by high/exclusive expression of Lpl, Otx2, Adcyap1 as well as Grp (Fig. 3d; Fig. S5). These genes contribute strongly to PC2 to the order of 0.355 (Lpl), 0.221 (Otx2), 0.242 (Adcyap1), and 0.309 (Grp). This population displays also strong expression of Aldh1a1, but contrary to DA1A, which also expresses Aldh1a1, DA2B depicts high levels of Calb1 and other molecular determinant of the second cluster. Subtype DA2B can be isolated by plotting cells according to PC3 and PC2 (Fig. 3d). We validated the localization of some of those markers in the adult mouse brain. We could locate subtype DA2B neurons by co-labeling of Aldh1a1 and Otx2 (Fig. 5b), a combinatorial code unique to this DA subtype. Aldh1a1+ neurons composed approximately 18.7 ± 2.4% of the DA neurons of the VTA, 52.3 ± 5.1% of the interfacsicular nucleus (IF), and 17.5 ± 2.3% of the caudal linear nucleus (CLI). Together, Aldh1a1+ neurons in these regions account for 11.3 ± 1.3% of all midbrain DA neurons. The marker Otx2 is less specific to DA neurons with numerous Th negative cells observed within the VTA and neighboring regions (i.g. interpeduncular nucleus; Fig 5b). Within the VTA, IF and CLI, more than 60% of Aldh1a1-labeled DA neurons were also positive for Otx2. These cells contain high levels of Calb1, but are negative for Sox6 and Sncg (not shown), which confirms the single cell expression profile obtained with the array. This Grp+ DA2B population matches a recently described DA population that projects to the nucleus accumbens (Ekstrand et al., 2014).
In comparison to other DA subtypes, DA2C is harder to define molecularly since it is not uniquely distinguished by expression of a single gene like other subtypes of the second cluster. All genes characterizing the second cluster are strongly expressed in this population (i.g. Calb1, Cck, Slc17a6), but none of the markers identifying other subpopulations are present Slc32a1 (DA2A), Adcyap1 (DA2B), and Vip (DA2D) (Fig 3e). Interestingly, genes characterizing cluster 1 are also expressed in this subpopulation, albeit at lower levels (i.g. Sox6, Igf1, but not Ndnf). Not surprisingly, this subtype could not be isolated by PCA (Fig 3e). It is also possible that DA2C could be comprised of multiple DA subtypes, that are indistinguishable by our analyses.
The final DA subtype identified, DA2D, depicts a unique and well-defined gene expression profile with the exclusive expression of Vip (Fig. 3f) combined to low/absent expression of Snca, Chrna4, and Ntf3 (Fig. 3f and Fig. S5a). Like most cells belonging to cluster 2, these cells express Calb1, Cck, and Foxa2, but do not express Sox6 and Ndnf. In the adult mouse brain, these neurons are located in the periaqueductal gray (PAG) and dorsal raphe nucleus (Fig. 5c). Within the PAG, we observed that in the Slc6a3::Cre/Ai9 mouse, about 48.7 ± 1.2% of tdTomato positive cells expressed Vip. On the other hand, 50.0 ± 1.3% of Vip+ cells were labeled with tdTomato, suggesting that some cells of this population could have evaded recombination due to very low Slc6a3 expression level. Vip-labeled DA neurons co-express most DAergic markers including Th and FoxA2, but little to no expression of Sox6, Sncg, Aldh1a1 and Otx2 (Fig. 5c and data not shown) was detected by immunofluorescence in the adult brain. Interestingly, for subtype DA2D, we were able to take advantage of strong Vip immunolabeling to visualize its projections in Slc6a3::Cre; Ai9 mice (Fig. 5d). We find that Vip immunoreactivity is found only in a subset of the DA projections. While DA projections are extensive in the extended amygdala, Vip immunolabeling is only observed in the oval portion of the bed nucleus of the stria terminalis (BSTov), and the lateral portion of central nucleus of the amygdala (CEAl). High-magnification images of fibers depict the colocalization of Vip and tdTomato in axonal boutons of DA neurons terminating in the BSTov and CEAl (Fig. 5d, inset). Vip labeling is excluded from other DA fields including the lateral septum (Fig. 5d; LS), striatum, accumbens, and cortex. This labeling is consistent with retrograde tracer analysis showing a dopaminergic projection from the PAG/DR directed toward the extended amygdala in the rat (Hasue and Shammah-Lagnado, 2002; Meloni et al., 2006). Overall, the presence and absence of these markers by co-labeling confirms the finding of our high-throughput single cell qPCR study, as well as validates the existence of this DA subtype with distinctive anatomical location in the PAG/DR and specific projection within the extended amygdala in the adult brain.
Finally, we found it useful to examine the expression of various classes of genes across DA subtypes (Fig. S5). We found that DA subtypes exhibit distinct transcription factor profiles that could in part underlie their molecular phenotype. Additionally, neuropeptides, channels and neurotrophic factors were substantially different between DA subtypes, suggestive of functional heterogeneity between these DA subtypes. With respect to PD related genes, we found that these are expressed in all DA subtypes; however, Snca levels are increased in DA1A and DA2B (Fig. S5d).
Relevance of dopamine neuron identity to Parkinson’s disease
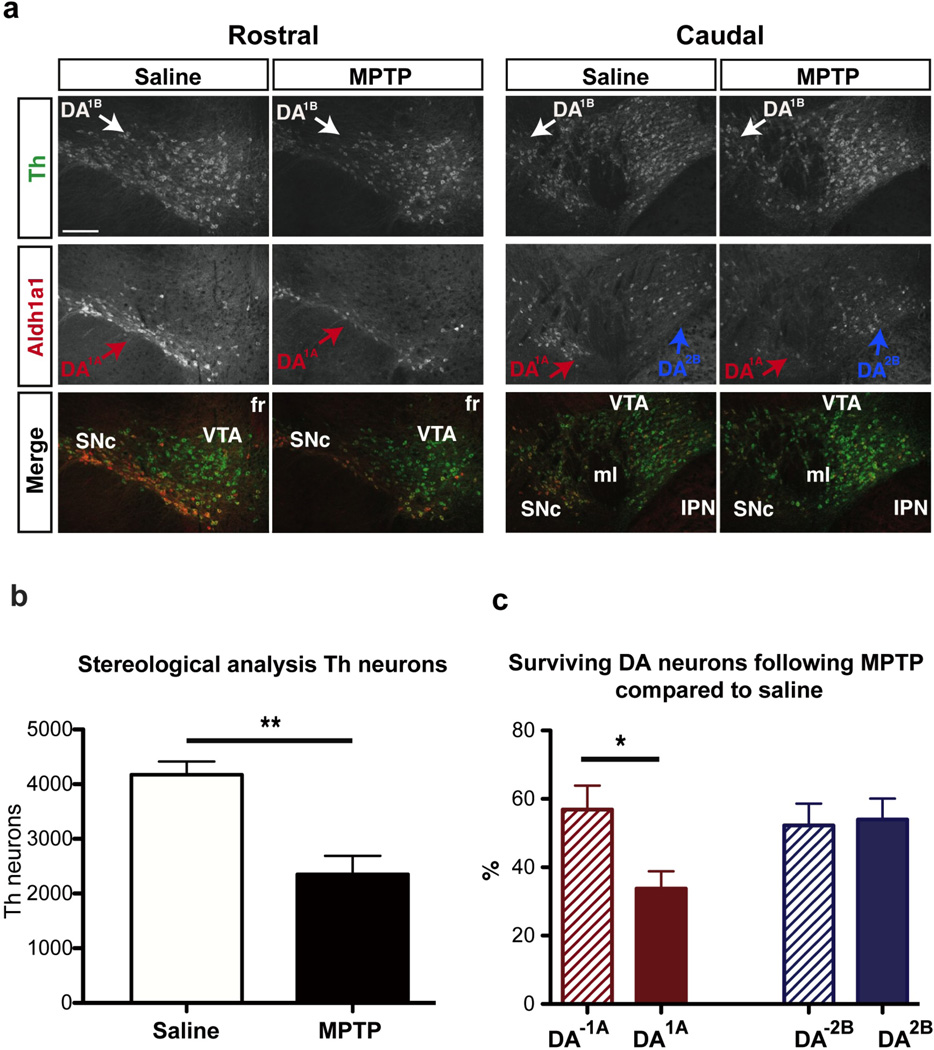
In Parkinson’s disease, DA neurons located in the ventral tier of the SNc are particularly vulnerable (Damier et al., 1999). Thus the DA1A subtype could be of special clinical relevance, since it is located in the ventral tier of the SNc. To determine if molecular codes were linked to vulnerability, we evaluated the vulnerability of DA1A neurons to the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), commonly used to mimic a number of important pathological features of parkinsonism in mice. We observed a significant decrease in the number of midbrain Th+ cells as quantified by stereological analysis (4039 compared to 2352; p-value<0.01; Fig. 6a). As previously reported (German et al., 1996), cells located in the SNc were particularly vulnerable (58.0% cell lost, p-value < 0.0001), compared to VTA (39.9% cell lost, p-value < 0.005), IF (14.3% cell lost, p-value = 0.5), RR (23.4% cell lost, p-value = 0.25) and CLI (43.3% cell lost, p-value < 0.05). We tested the hypothesis that DA1A subtype could be more vulnerable to MPTP toxin and examined the vulnerability of Aldh1a1 labeled neurons of SNc and RR (defining DA1A). Indeed, Calb1 and Otx2 genes that have been associated to resistance to MPTP and PD are absent in this population (Di Salvio et al., 2010b; Liang et al., 1996). We found that DA1A neurons were significantly more vulnerable since we observed a 66.2% diminution of Aldh1a1+ cells in the SNc and RR of MPTP-treated mice compared to animals injected with saline (Fig. 6b). The remaining DA neurons were significantly less affected by MPTP, with only 39.1% DA cell loss compared to saline (p-value=0.001; Fig. 6c). In comparison, DA2B subtype that is defined by the coexpression Aldh1a1 and Otx2, and is observed in the ventromedial VTA, is not differently affected by MPTP compared to other DA populations (Fig. 6c). Additionally, when we counted Aldh1a1 positive and negative cells specifically in the SNc, we found that Aldh1a1+ cells (DA1A) are significantly more vulnerable than Aldh1a1- (DA1B) cells to MPTP (Aldha1+ showed 70% loss, Aldh1a1– showed 50% loss; p-value = 0.0337). MPTP vulnerability has been in part attributed to levels of Slc6a3 (Di Salvio et al., 2010b); this latter finding is interesting as both DA1A and DA1B display high Slc6a3 (Fig. S5e), yet the Aldh1a1+ DA1A cohort appears more vulnerable. Factors other than Slc6a3 levels, may therefore also impact DA vulnerability.
Figure 6. Dopamine neuron DA1A subtype is selectively vulnerable in an MPTP model.
(a) Example of Aldh1a1 (red) and Th (green) labeling in the midbrain of saline and MPTP treated animals. (b) MPTP induced a significant reduction in Th immunoreactivity of midbrain neurons. (c) DA1A subtype is more severely affected by the toxin as compared to other DA subtypes (labeled DA-1A); in contrast DA2B does not show differential vulnerability. Scale bar: low magnification = 200µm.
Discussion
This study demonstrates the existence of multiple molecularly distinct DA neuron subtypes using a novel single-cell qPCR approach. These subtypes all share a core genetic program to produce the neurotransmitter dopamine, upon which subtype-specific gene expression modules are superimposed. Our high-throughput approach identified several markers related to specific DA subpopulations, and more importantly their coordinated expression in single cells. We show that each DA subtype has signature expression profiles of transcription factors (Fig. S5b), channels (Fig. S5c), receptors (Fig. S5c), DA related genes (Fig. S5e), neuropeptides (Fig. S5f) and secretory factors (Fig. S5g). Furthermore, we show that some of these identified subtypes have distinct anatomical and functional properties. To our knowledge, our study is the first application of this technology to classify, in a polythetic manner, neuron subtypes within a neurotransmitter group. Although our study focused exclusively on the DA system, a similar approach could be used to dissect other neurotransmitter systems within a defined brain region.
Heterogeneous expression of markers in the DA system has been reported (Di Salvio et al., 2010b; Dougalis et al., 2012; Fu et al., 2011; Gerfen et al., 1987; Hnasko et al., 2012; McCaffery and Dräger, 1994; Smits et al., 2013). Previous studies have relied on in situ hybridization, or antibody co-labeling with Th, to localize marker expression. These studies provided an important starting point for the classification of DA neuron subtypes presented here. Since DA neurons are heterogeneous and intermingled, these approaches while informative, do not provide conclusive evidence of coordinated gene expression, an essential feature of a neuronal subtype. In contrast, the approach presented here reveals coordinated expression of cohorts of genes, thus allowing the classification of DA subtypes in a polythetic manner, an important goal in the field of neuronal classification (Bota and Swanson, 2007; Fishell and Heintz, 2013).
Our study has limitations. First our classification was achieved in early postnatal brain, at a stage when DA neurons are still immature in terms of pacemaking activity (Chan et al., 2007) and glutamate receptor expression (Bellone et al., 2011). It is therefore possible that some markers on which the classification is based, are specific to this developmental timepoint. However, the fact that we could observe all markers that we tested in the adult brain would argue that our single cell array analysis is highly informative, despite the neonatal time point. Second, it is possible that DA neurons with low/absent Slc6a3 (Lammel et al., 2008) were excluded from these analyses, due to inefficient recombination of the reporter by Slc6a3::Cre. Third, it is possible that there are biases in the dissociation and FACS sorting method that could disproportionately skew the numbers of each DA subtype. This could explain the discrepancy between the proportion of each subtype observed in the array, compared to what was observed in the adult by immunofluorescence and in situ hybridization, although these latter methods may also have inherent bias. Fourth, this method requires some knowledge of gene expression, in this case provided by important earlier studies (Chung et al., 2005; Greene et al., 2005; Grimm et al., 2004). Additionally, expanding the number of genes and cells sampled could increase the power of this method, and could yield additional DA subtypes, or further subdivide currently identified DA subtypes. Ultimately, unbiased single cell mRNA sequencing could be used to provide further resolution for DA cell classification. Despite these limitations, this study represents an important advance; the method used here allows simultaneous, efficient, and quantitative assessment of key DA markers in single cells. Based on these coordinated gene expression profiles, we were able to distinguish multiple molecularly distinct DA subtypes with unique properties, one with a specific projection, and one selectively vulnerable. Further work will be required to illustrate the projections, physiological characteristics, and functions of the DA subtypes defined here.
Defining DA neurons on a molecular basis will allow a better understanding of the vulnerability of selective DA neuron subsets. In PD, ventral tier SNc neurons are more vulnerable than either dorsal tier or VTA DA neurons (Damier et al., 1999). Our data suggest that the Aldh1a1 population in the ventral tier appears to be particularly vulnerable in an MPTP mouse model. It is of interest that Aldh1a1, an aldehyde dehydrogenase, is inhibited by the fungicide benomyl, for which high exposure was associated with an increased PD risk (Fitzmaurice et al., 2012). Additionally, the DA1A subtype is characterized by significantly lower levels of Foxa2 and En1, as well as higher levels of Snca, which could in part underpin selective vulnerability in dosage-sensitive backgrounds (Kittappa et al., 2007; Sgado et al., 2006; Singleton et al., 2003). Our data therefore could offer additional insights into the selective vulnerability of DA neurons.
Understanding the molecular complexity of midbrain DA subtypes might help refine and develop new therapeutic approaches for DA related diseases. Indeed, DA signaling and receptors are currently the main and potential target in the treatment of PD, schizophrenia, ADHD, addiction, and depression, among others. An important limitation of current DA therapies is side effects, largely due to the indiscriminate targeting of the DA system. For instance, systemic levodopa, the main treatment to PD, induces psychotic symptoms including hallucinations and impulsive behaviors in up to 40% of patients (Chou et al., 2007; Diederich et al., 2009). Conversely, typical and atypical antipsychotics display adverse effects including motor symptoms such as Parkinsonism or tardive dyskinesia. Elucidating DA subtypes may help uncovering novel candidates for a new generation of targeted therapeutics.
The stem cell field has highlighted the urgent need to better understand the molecular basis of DA neuron diversity, towards programming human embryonic or induced pluripotent stem cells (iPS) towards specific DA fates (Studer, 2012). DA subtypes derived from stem cells would serve as more accurate models for DA related disease, and would facilitate regenerative medicine. Our study provides a molecular barcode for several DA subtypes including subtype DA1A, of great intense interest due to its location at the ventral aspect of the SNc, and its selective vulnerability. In this regard, revisiting previous lineage analyses aimed at uncovering distinct DA lineages (Joksimovic et al., 2009), will benefit from this array of molecular markers. Additionally, our study reveals transcription factor codes that may underpin the development of DA heterogeneity (Fig. S5b), which could be important for embryonic stem cell programming of DA subtypes.
Finally, the present study provides entry points that will allow genetic access to subtypes of DA neurons. However, since none of the markers used here are expressed exclusively in a DA subtype, intersectional platforms using multiple recombinase systems would be required to manipulate these subtypes in an otherwise intact brain (Awatramani et al., 2003). Provided with an appropriate Flp line, these genetic entry points could be used with mouse genetic based platforms (Awatramani et al., 2003; Jensen et al., 2008; Kim et al., 2009; Ray et al., 2011; Robertson et al., 2013; Yamamoto et al., 2009), or more recently developed viral platforms, which allow dual recombinase mediated intersectional gene activation (Fenno et al., 2014). Projection and functional analyses following intersectional genetic labeling have been successful in dissecting the connections of serotonin and noradrenergic populations in the hindbrain (Jensen et al., 2008; Robertson et al., 2013). In the hindbrain, these neuronal populations span multiple developmentally defined rhombomeres, and therefore could be dissected using developmentally defined rhombomere-specific Cre lines (Awatramani et al., 2003; Farago et al., 2006; Jensen et al., 2008; Robertson et al., 2013). In the midbrain DA system, there are no such convenient segmental divisions. Thus our data, providing genetic entry points to defined DA subtypes, will further facilitate functional investigations using chemogenetic or optogenetic technology (Ray et al., 2011; Rogan and Roth, 2011; Tye and Deisseroth, 2012), as well as transcriptomic and connectomic analyses, with the ultimate goal of disentangling this pleiotropic neurotransmitter system.
Material and methods
Mice and flow cytometry
Mice were maintained and euthanized according to the protocols approved by the Northwestern University Animal Care and Use Committee. The Slc6a3::Cre and Ai9 mice used in this study were both maintained on C57Bl/6 background and were described previously (Bäckman et al., 2006; Madisen et al., 2010). In this mouse, the vast majority of DA neurons are recombined. For the single-cell study, the midbrain of three P4 mice were carefully dissected to grossly the size of 1mm × 2mm × 2mm. Midbrain neurons were dissociated using Papain Dissociation System (Worthington Biochemical Corporation) according to the manufacturer’s instructions. Cells were re-suspended in Neurobasal medium (Life technologies) and single cell were immediately sorted in a 96-well plate with the help of Northwestern University Flow Cytometry Facility and a Cancer Center Support Grant (NCI CA060553). FACS sorting was performed on an Aria II SORP sorter using a fine tune sort precision protocol. This approach provides a reliable single cell sorting by adequately calculating the drop delay, but also increase the proportion of empty well. Three 96-well plates for gene expression analysis were filled with single fluorescently-labeled neurons, for the exception of 6 wells used for calibration curves, 3 wells used for positive control (10, 20, and 50 cells), and 3 wells used for negative control (e.g. Fig. S1). For immunofluorescence and fluorescence in situ hybridization, perfused adult mice (older than P56) were used unless stated otherwise.
Primers design and validation
Each primer pairs were designed, generated, and experimentally validated by Fluidigm. Primers were designed to span exon junctions, when possible, with an average amplicon size of 82bp (Table S1). Assay performance was evaluated via serial dilution of a commercial reference sample. The ΔCT (or Cq) was calculated for all pairwise combinations of assays in this request and the slope of ΔCT versus log2 template dilution calculated and averaged. The mean slope of ΔCT versus template concentration for cDNA templates indicates relative error in measuring 2-fold changes for the given assay compared to the other assays in this request (Schmittgen and Livak, 2008). If two assays have equal efficiencies and there is no measurement error, the slope will be zero. 98.9% of assays fell in the range −0.1 to 0.1.
Single-cell gene expression qPCR
Cells were sorted directly into 5µL of the Reverse Transcription (RT) mix solution 1, composed of 1.2µL VILO Reaction Mix (SuperScript VILO cDNA Synthesis Kit, Life technologies), 0.3µL 20U/µL SUPERase-In, 0.25µL NP-40 Detergent Surfact-Amps Solution (Fisher Scientific), 3.25µL of Nuclease-free H2O (Teknova). RNA was denatured 90s at 65°C. The plate was quickly chilled on ice for 5min and centrifuged briefly at 4°C. We added 1uL to each well of RT mix Solution 2 composed of 0.15µL of 10X SuperScript Enzyme Mix, 0.12µL of T4 Gene 32 Protein (New England Biolabs) and 0.73µL of Nuclease-free H2O. Reverse transcription was carried in thermal cycler under the following condition: 5min at 25°C, 30min at 50°C, 25min at 55°C, 5min at 60°C, and 10min at 70°C. Subsequently, in the same tube, cDNA went through sequence-specific amplification by denaturing at 95°C for 15s, and annealing and amplification at 60°C for 4 min for 18 cycles. These preamplified products were diluted 5-fold prior to the analysis with Universal PCR Master Mix and inventoried TaqMan gene expression assays (ABI) in 96.96 Dynamic Arrays on a BioMark System (Fluidigm). Ct values were calculated from the system’s software (BioMark Real-time PCR Analysis; Fluidigm). For quality control, the melt curve was established and all peaks falling outside user-defined threshold was removed from analysis (Fig. S2).
Single-cell data processing and analysis
Overall, 274 single-cell were sorted into 96 well plates. Out of these cells, we removed cells with weak or no expression of Gapdh, defined by greater than two standard deviations from the median, were removed from the analysis. In addition, we removed cells with weak expression of genes defining DA neurons (Ddc, Th, Slc6a3, and Slc18a2) using the same criteria. These cells were assumed to be non-DA cells, and were removed from the analysis, leaving a final 159 DA neurons to be further analyzed. In addition, despite depicting a normal standard curve, two genes Pvalb and Aldh1a2 were undetectable in all cells analyzed, and these genes were thus excluded from further analysis and replaced by the housekeeping genes Actb and Hprt. All Ct values (e.g. Fig. S1) obtained from the BioMark System were converted into relative expression levels by subtracting the values from the threshold value of 24 (Fig. 1b and Fig. S3). All Ct values above 24 were considered below detectability level. As the Ct scale is logarithmic (a difference of one Ct corresponds to a doubling of measured transcript), the values obtained were normalized to the endogenous control by subtracting, for each cell, its Gapdh Ct value (delta Ct). The fold difference was obtained by using the highest gene expression cell sample for each gene, and used as a reference to calculate the delta delta Ct (comparative Ct method). In order to group cells with similar expression profile, we used cluster analysis by correlation coefficient similarity on logarithmic expression data. Cells were assigned to a specific DA subtype based on this cluster analysis. To compare gene expression level between identified populations, since the homogeneity of variance assumption was violated, normalized gene levels between subtypes were compared between DA subtypes using a Kruskal-Wallis and Dunn’s post-hoc analysis with Bonferroni correction of alpha value.
Immunofluorescence
Staining were performed on adult (more than two months of age) wildtype and Slc6a3::Cre; Ai9 mice for minimum of N=3 for all antibodies. To map the DA2A subtype, we crossed a mouse in which Cre was knocked-in Slc32a1 locus (Vong et al., 2011), with a Rosa locus reporter in which a floxed stop cassette, was followed by nuclear LacZ RosaNZG; (Yamamoto et al., 2009). Mice were perfused with cold 4% PFA in PBS, the brains were dissected and fixed overnight in the same solution, cryoprotected in 30% sucrose PBS solution, and sectioned at 25µm using a freezing microtome. Tissue sections were stained with the following primary antibodies: mouse anti-Calbindin (Sigma-Aldrich; 1:1000), rabbit anti-Calbindin (Chemicon; 1:500), rat anti-Dat (Santa Cruz; 1:50), goat anti-FoxA2 (Santa Cruz; 1:50), goat anti-LacZ (Biogenesis; 1:1500), rabbit anti-LacZ (Cappel; 1:1500), rabbit anti-Nr4a2 (Santa Cruz; 1:500), goat anti-Otx2 (Neuromics; 1:200), rabbit anti-Aldh1a1 (Abcam; 1:200), rabbit anti-Sncg (Abcam; 1:1000), rabbit anti-Sox6 (Abcam; 1:500), rabbit anti-Vip (Abcam; 1:500), rabbit anti-SatB1 (Abcam; 1:1,000), and sheep anti-TH (Pel-Freez; 1:300). Sections were imaged by confocal microscopy (Leica DM6000 CFS). For each DA subtype mapped in Fig. 4–5, a combination of markers specific to this subtype was analyzed. Cells were counted on representative sections, superimposed to the reference section from the Allen Brain Atlas, and averaged across animals. Each colored square in Fig. 4–5 represents five neurons of the DA subtype at the approximate location, whereas each black square represents five DA neurons of another subtype.
Fluorescent In situ hybridization
For fluorescent in situ hybridization (ISH) was performed with single-stranded digoxigenin (Roche) and fluorescein labeled riboprobes directed against the following mRNAs: Th (Genback accession#: NM_009377), Aldh1a1 (NM_013467), Ndnf (A930038C07_Rik), Slc32a1 (Genback accession#: NM_009508), Sncg (Genback accession#: NM_011430). The ISH protocol was adapted from Joksimovic et al. (2009).
MPTP protocol
Adult C57BL/6 mice (n=16) received four intraperitoneal (i.p.) injections of freshly diluted MPTP-HCl (20 mg/kg; Sigma, St. Louis, MO, USA) dissolved in saline 0.9%. MPTP was administered in an acute manner at 2-hour intervals as previously described (Jackson-Lewis and Przedborski, 2007). Remaining animals (n=8) received i.p. vehicle injections at the time of MPTP administration. Mice were sacrificed 7 days following the last MPTP injection through intracardiac perfusion with either PBS 0.1M only or PBS followed by PFA 4%. Brains were collected, further fixed in PFA and cryoprotected in 20% sucrose.
Supplementary Material
Acknowledgements
The authors would like to thank Jennifer Darnell and Jason McKinney (Fluidigm) for technical assistance. The authors thank Dr. Richard J Miller and Dr. D. James Surmeier for useful comments on the manuscript. This research was supported by R21 (1R21NS072703-01) and Northwestern Memorial Foundation (Paul Ruby Foundation for Parkinson’s Research) grants to R.B.A. This research was also supported by FRSQ, MJFF, and CIHR grants to J-F.P, a Paul Ruby Foundation grant to J.Z., as well as FRSQ grant to J.D-O. K-Y.A.K. would like to acknowledge the following grants: NIH CTSA funding to Northwestern University Feinberg School of Medicine (UL1 TR000150, UL1 RR025741). F.C. is recipient of a National Research Career award from the Fonds de recherche du Québec en santé providing salary support and funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
JF Poulin designed and performed the experiments and wrote the manuscript; J Zou performed several co-labeling experiments, J Drouin-Ouellet and F Cicchetti provided MPTP treated brain sections and initial analysis, KYA Kim helped with statistical analysis, RB Awatramani supervised the study and helped write the manuscript.
References
- Ang S-L. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal Subtype-Specific Genes that Control Corticospinal Motor Neuron Development In Vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, Tomac AC. Characterization of a mouse strain expressing Cre recombinase from the 3’ untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- Bellone C, Mameli M, Lüscher C. In utero exposure to cocaine delays postnatal synaptic maturation of glutamatergic transmission in the VTA. Nat Neurosci. 2011;14:1439–1446. doi: 10.1038/nn.2930. [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota M, Swanson LW. The neuron classification problem. Brain Res Rev. 2007;56:79–88. doi: 10.1016/j.brainresrev.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson’s disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Chou KL, Borek LL, Friedman JH. The management of psychosis in movement disorder patients. Expert Opin Pharmacother. 2007;8:935–943. doi: 10.1517/14656566.8.7.935. [DOI] [PubMed] [Google Scholar]
- Chung CY, Licznerski P, Alavian KN, Simeone A, Lin Z, Martin E, Vance J, Isacson O. The transcription factor orthodenticle homeobox 2 influences axonal projections and vulnerability of midbrain dopaminergic neurons. Brain. 2010;133:2022–2031. doi: 10.1093/brain/awq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Seo H, Sonntag KC, Brooks A, Lin L, Isacson O. Cell type-specific gene expression of midbrain dopaminergic neurons reveals molecules involved in their vulnerability and protection. Hum Mol Genet. 2005;14:1709–1725. doi: 10.1093/hmg/ddi178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain II. Patterns of loss of dopamine-containing neurons in Parkinson's disease. Brain. 1999;122:1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- Darvas M, Palmiter RD. Restriction of dopamine signaling to the dorsolateral striatum is sufficient for many cognitive behaviors. Proc Natl Acad Sci USA. 2009;106:14664–14669. doi: 10.1073/pnas.0907299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Omodei D, Acampora D, Simeone A. Otx2 expression is restricted to dopaminergic neurons of the ventral tegmental area in the adult brain. Int J Dev Biol. 2010a;54:939–945. doi: 10.1387/ijdb.092974ms. [DOI] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci. 2010b;13:1481–1488. doi: 10.1038/nn.2661. [DOI] [PubMed] [Google Scholar]
- Diederich NJ, Fénelon G, Stebbins G, Goetz CG. Hallucinations in Parkinson disease. Nat Rev Neurol. 2009;5:331–342. doi: 10.1038/nrneurol.2009.62. [DOI] [PubMed] [Google Scholar]
- Dougalis AG, Matthews GAC, Bishop MW, Brischoux F, Kobayashi K, Ungless MA. Functional properties of dopamine neurons and co-expression of vasoactive intestinal polypeptide in the dorsal raphe nucleus and ventro-lateral periaqueductal grey. Eur J Neurosci. 2012;36:3322–3332. doi: 10.1111/j.1460-9568.2012.08255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM. Molecular profiling of neurons based on connectivity. Cell. 2014;157:1230–1242. doi: 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Heintz N. The Neuron Identity Problem: Form Meets Function. Neuron. 2013;80:602–612. doi: 10.1016/j.neuron.2013.10.035. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice AG, Rhodes SL, Lulla A, Murphy NP, Lam HA, O’Donnell KC, Barnhill L, Casida JE, Cockburn M, Sagasti A, et al. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in Parkinson disease. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1220399110. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yuan Y, Halliday G, Rusznák Z, Watson C, Paxinos G. A cytoarchitectonic and chemoarchitectonic analysis of the dopamine cell groups in the substantia nigra, ventral tegmental area, and retrorubral field in the mouse. Brain Structure & Function. 2011 doi: 10.1007/s00429-011-0349-2. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Baimbridge KG, Thibault J. The neostriatal mosaic: III. Biochemical and developmental dissociation of patch-matrix mesostriatal systems. J Neurosci. 1987;7:3935–3944. doi: 10.1523/JNEUROSCI.07-12-03935.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German DC, Nelson EL, Liang CL, Speciale SG, Sinton CM, Sonsalla PK. The neurotoxin MPTP causes degeneration of specific nucleus A8, A9 and A10 dopaminergic neurons in the mouse. Neurodegeneration. 1996;5:299–312. doi: 10.1006/neur.1996.0041. [DOI] [PubMed] [Google Scholar]
- Greene JG, Dingledine R, Greenamyre JT. Gene expression profiling of rat midbrain dopamine neurons: implications for selective vulnerability in parkinsonism. Neurobiol Dis. 2005;18:19–31. doi: 10.1016/j.nbd.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Grimm J, Mueller A, Hefti F, Rosenthal A. Molecular basis for catecholaminergic neuron diversity. Proc Natl Acad Sci USA. 2004;101:13891–13896. doi: 10.1073/pnas.0405340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Sun LL, Clarke ND, Robson P. Resolution of Cell Fate Decisions Revealed by Single-Cell Gene Expression Analysis from Zygote to Blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasue RH, Shammah-Lagnado SJ. Origin of the dopaminergic innervation of the central extended amygdala and accumbens shell: a combined retrograde tracing and immunohistochemical study in the rat. J Comp Neurol. 2002;454:15–33. doi: 10.1002/cne.10420. [DOI] [PubMed] [Google Scholar]
- Hayes L, Zhang Z, Albert P, Zervas M, Ahn S. Timing of Sonic hedgehog and Gli1 expression segregates midbrain dopamine neurons. J Comp Neurol. 2011;519:3001–3018. doi: 10.1002/cne.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Hjelmstad GO, Fields HL, Edwards RH. Ventral tegmental area glutamate neurons: electrophysiological properties and projections. The Journal of Neuroscience. 2012;32:15076–15085. doi: 10.1523/JNEUROSCI.3128-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra EJ, Oerthel von L, van der Linden AJA, Schellevis RD, Scheppink G, Holstege FCP, Groot-Koerkamp MJ, van der Heide LP, Smidt MP. Lmx1a is an activator of Rgs4 and Grb10 and is responsible for the correct specification of rostral and medial mdDA neurons. Eur J Neurosci. 2013;37:23–32. doi: 10.1111/ejn.12022. [DOI] [PubMed] [Google Scholar]
- Jackson-Lewis V, Przedborski S. Protocol for the MPTP mouse model of Parkinson's disease. Nat Protoc. 2007;2:141–151. doi: 10.1038/nprot.2006.342. [DOI] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Anderegg A, Roy A, Campochiaro L, Yun B, Kittappa R, McKay R, Awatramani R. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proceedings of the National Academy of Sciences. 2009;106:19185–19190. doi: 10.1073/pnas.0904285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Cook MN, Carey MR, Shen C, Regehr WG, Dymecki SM. Linking genetically defined neurons to behavior through a broadly applicable silencing allele. Neuron. 2009;63(493):305–315. doi: 10.1016/j.neuron.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RDG. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Häckel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008;57:760–773. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisseroth K, Malenka RC. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012 doi: 10.1038/nature11527. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CL, Sinton CM, Sonsalla PK, German DC. Midbrain dopaminergic neurons in the mouse that contain calbindin-D28k exhibit reduced vulnerability to MPTP-induced neurodegeneration. Neurodegeneration. 1996;5:313–318. doi: 10.1006/neur.1996.0042. [DOI] [PubMed] [Google Scholar]
- Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126–e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Lock H, Chefer VI, Shippenberg TS, Hjelmstad GO, Fields HL. Kappa opioids selectively control dopaminergic neurons projecting to the prefrontal cortex. Proc Natl Acad Sci USA. 2006a;103:2938–2942. doi: 10.1073/pnas.0511159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Lock H, Hjelmstad GO, Fields HL. The ventral tegmental area revisited: is there an electrophysiological marker for dopaminergic neurons? J Physiol (Lond) 2006b;577:907–924. doi: 10.1113/jphysiol.2006.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffery P, Dräger UC. High levels of a retinoic acid-generating dehydrogenase in the meso-telencephalic dopamine system. Proc Natl Acad Sci USA. 1994;91:7772–7776. doi: 10.1073/pnas.91.16.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni EG, Gerety LP, Knoll AT, Cohen BM, Carlezon WA. Behavioral and anatomical interactions between dopamine and corticotropin-releasing factor in the rat. The Journal of Neuroscience. 2006;26:3855–3863. doi: 10.1523/JNEUROSCI.4957-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong H-W, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, et al. An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Ray RS, Corcoran AE, Brust RD, Kim JC, Richerson GB, Nattie E, Dymecki SM. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333:637–642. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SD, Plummer NW, de Marchena J, Jensen P. Developmental origins of central norepinephrine neuron diversity. Nat Neurosci. 2013;16:1016–1023. doi: 10.1038/nn.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sgado P, Alberi L, Gherbassi D, Galasso SL, Ramakers GM, Alavian KN, Smidt MP, Dyck RH, Simon HH. Slow progressive degeneration of nigral dopaminergic neurons in postnatal Engrailed mutant mice. Proc Natl Acad Sci USA. 2006;103:15242–15247. doi: 10.1073/pnas.0602116103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, et al. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JPH. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Smits SM, von L, Oerthel, Hoekstra EJ, Burbach JPH, Smidt MP. Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons. PLoS ONE. 2013;8:e76037–e76037. doi: 10.1371/journal.pone.0076037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AM, Jennings JH, Ung RL, Blair GA, Weinberg RJ, Neve RL, Boyce F, Mattis J, Ramakrishnan C, Deisseroth K, et al. A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron. 2013;80:1039–1053. doi: 10.1016/j.neuron.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L. Derivation of dopaminergic neurons from pluripotent stem cells. Prog Brain Res. 2012;200:243–263. doi: 10.1016/B978-0-444-59575-1.00011-9. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat Protoc. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KMK, Deisseroth KK. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci. 2012;13:251–266. doi: 10.1038/nrn3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye KM, Prakash R, Kim S-Y, Fenno LE, Grosenick L, Zarabi H, Thompson KR, Gradinaru V, Ramakrishnan C, Deisseroth K. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenvliet JV, Santos Dos MTMA, Kouwenhoven WM, Oerthel von L, Lim JL, van der Linden AJA, Koerkamp MJAG, Holstege FCP, Smidt MP. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development. 2013;140:3373–3384. doi: 10.1242/dev.094565. [DOI] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S, Lowell BB. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallén A, Zetterström RH, Solomin L, Arvidsson M, Olson L, Perlmann T. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999;253:737–746. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- Xue Z, Huang K, Cai C, Cai L, Jiang C-Y, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.