Abstract
An internal time-keeping mechanism has been observed in almost every organism studied from archaea to humans. This circadian clock provides a competitive advantage in fitness and survival (18, 30, 95, 129, 137). Researchers have uncovered the molecular composition of this internal clock by combining enzymology, molecular biology, genetics, and modeling approaches. However, understanding the mechanistic link between the clock and output responses has been elusive. In three model organisms, Arabidopsis thaliana, Drosophila melanogaster, and Mus musculus, whole-genome expression arrays have enabled researchers to investigate how maintaining a time-keeping mechanism connects to an adaptive advantage. Here, we review the impacts transcriptomics have had on our understanding of the clock and how this molecular clock connects with system-level circadian responses. We explore the discoveries made possible by high-throughput RNA assays, the network approaches used to investigate these large transcript datasets, and potential future directions.
Keywords: transcriptome, network analysis, transcriptional regulation, microarray analysis, systems biology
INTRODUCTION
As early as the eighteenth century, evidence began accumulating that some organisms possess an internal, persistent mechanism to measure the passing of time. In 1729, Jean Jacques d’Ortous de Mairan noticed that the leaves of a heliotrope plant moved rhythmically throughout the day (reviewed in 72). To test if this movement was independent of diurnal signals, he moved the plants to a dark cellar and observed that, even in the absence of light cues, the leaf movement persisted. Experiments demonstrating the presence of a circadian clock in animals followed in the early twentieth century (reviewed in 100). The existence of an endogenous timekeeping mechanism that was independent of any cues from the Earth’s rotation was confirmed when the fungus Neurospora crassa was grown in space. In the Spacelab, completely removed from earth-orbital cues, Neurospora maintained a rhythmic growth pattern of approximately 23 h in complete darkness (117). In addition to leaf movements in plants and fungal growth patterns, many other behaviors were identified as circadian regulated, from the pigmentation of arthropods to the sun-compass behavior of starlings. Similar to the observations in the heliotrope plant, Arabidopsis thaliana displays clock-regulated leaf movement and hypocotyl elongation (133). In mice, core body temperature, feeding, and wheel-running behavior are regulated by the clock. Lastly, in Drosophila melanogaster eclosion and egg-laying are two physiological outputs linked to the clock. By observing how environmental changes affect these physiological responses, a clear picture of the clock began to emerge. This internal clock maintains a rhythm of approximately 24 h even in constant conditions, thus the name circadian (Latin for “about a day”). These early physiological experiments also established that the clock regulates daily rhythms of physiology and behavior, is temperature compensated, and can be re-entrained to match altered environmental conditions.
The arrival of genetic techniques enabled the identification of the molecular components of the clock and provided insights into how environmental signals regulate the clock. The first clock gene, Period (PER), was discovered in Drosophila via forward genetics (reviewed in 5). The development of bioluminescent reporter gene constructs in the 1990s rapidly accelerated discoveries in Arabidopsis, flies, and rodents (reviewed in 126). Results from these genetic studies generated a detailed structure of the clock itself and several of the input mechanisms for each model organism (reviewed in 34, 53, 55, 138) (Figure 1). This review focuses on Arabidopsis, Drosophila, and Mus musculus, although many discoveries that contribute to our understanding of the clock were also initiated in other organisms. For analysis of these other organisms, readers are encouraged to visit the respective reviews (Neurospora crassa; 8, 25, 39) (Cyanobacteria; 20, 69) (Danio rerio; 10).
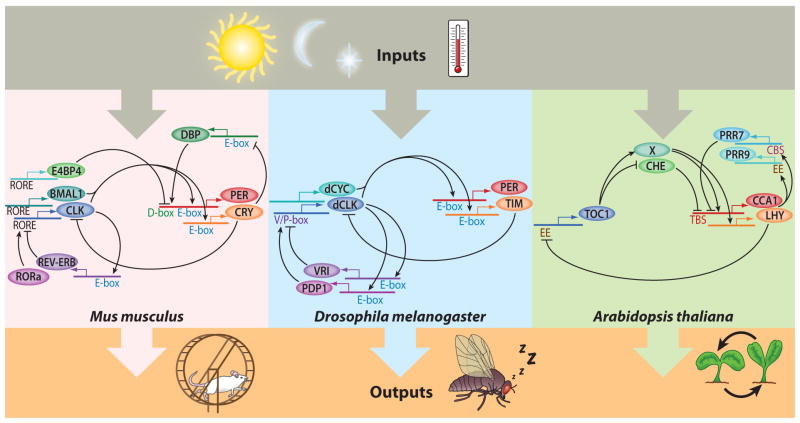
Figure 1.
A simplified schematic model diagram to highlight the similarities between the circadian-clock oscillators of Mus musculus, Drosophila melanogaster, and Arabidopsis thaliana. Abbreviations: BMAL1, brain and muscle ARNT-like 1; CBS, CCA1 binding site; CCA1, CIRCADIAN CLOCK ASSOCIATED 1; CHE, CCA1 hiking expedition; CLK, circadian locomotor output cycles protein kaput; CRY, cryptochrome; DBP, D-box binding protein; dCYC, Drosophila cycle; dCLK, Drosophila circadian locomotor output cycles protein kaput; E4BP4, E4 promoter-binding protein 4; EE, evening element; LHY, late elongated hypocotyl; PDP1, PAR-domain protein 1; PER, period; PRR7, pseudoresponse regulator 7; PRR9, pseudoresponse regulator 9; REV-ERB, reverse erb; RORa, RAR-related orphan receptor A; RORE, REV-ERB/ROR response element; TBS, TCP binding site; TIM, timeless; TOC1, timing of CAB expression 1; V/P-box, Vrille/PDP1 binding box; VRI, Vrille.
Homology exists between the molecular mechanisms of Drosophila and mouse clocks, although the genetic interactions of specific clock components vary (112, 138, 139). In Arabidopsis, while the components are not orthologous, the network architecture of positive and negative feedback loops is conserved (Figure 1) (34, 138). Although here we focus on transcriptional regulation, post-translational regulation plays an integral role and has been extensively reviewed (Arabidopsis; 34, 73) (Drosophila; 142) (mammalian; 27, 36).
Although the tools of genetics and molecular biology rapidly advanced the understanding of the core oscillator and input mechanisms, the mechanistic link between regulatory proteins and the observed circadian rhythmicity in behavior and environmental responses remained elusive. It was speculated that the link between the circadian clock and physiological outputs is composed of robust, complex networks and that straightforward genetic and enzymatic approaches may prove insufficient to unravel these connections. In such a robust network, multiple pathways may coordinately regulate a response. Thus, effects of a single gene mutation may be masked by compensation from other pathways. The observed role of the clock in regulating a wide range of physiological responses made it plausible that a link between the clock and these outputs was itself a complicated, robust network, requiring new tools to unravel these output pathways (103). At the time, array-based RNA quantification methods were being developed that enabled the measurement of global transcriptional changes in an organism. The observation that many of the core-clock components affect transcription suggests that transcriptional networks may be one facet of how the clock connects to output responses and that these new tools could potentially provide the much needed means to link the clock to the regulation of circadian output. Researchers in the circadian field took advantage of the opportunity to examine global transcriptional changes that occur throughout the day and determine which of these are under circadian control. This analysis of cycling transcripts has had broad-ranging impacts on the clock field that are still being explored.
The First Circadian Microarrays
Transcriptional microarrays were developed in the mid-1990s and were first applied to a circadian time course in 2000 by Harmer et al. (35) to profile Arabidopsis plants after release to constant light conditions. Plants were harvested every 4 h for 48 h, enabling the analysis of gene expression changes over a two-day period. Within two years, several additional whole-genome oligonucleotide array experiments were performed in Drosophila and mouse as well (reviewed in 23, 24, 26, 113). These initial arrays provided a first glimpse into the global circadian regulation of transcripts. However, beyond just a description of cycling genes, these experiments also provided insights into the role, extent, and mechanism of circadian clock. These first transcriptomic analyses produced many exciting discoveries: the identification of novel cis-elements overrepresented in transcripts peaking at specific times of day; the revelation that the expression of metabolic pathway components are temporally coordinated; and the discovery of circadian regulation of stress responses. Cumulatively, these results confirmed the pervasiveness of the clock in regulating physiology. The discoveries from these arrays have been extended even further in recent years by combining these arrays with each other, with additional arrays performed under different conditions, and with other high-throughput system-level experiments. These new approaches are giving researchers the opportunity to approach the study of the clock and circadian outputs at a system level, thus developing a whole-organism understanding of the roles and mechanisms of the circadian clock.
THE CIRCADIAN TRANSCRIPTOME
The extent of circadian cycling observed in the transcriptome was a surprise to many researchers. In Arabidopsis 6%, in Drosophila between 1% and 5%, and in mouse 5% to 10% of transcripts were identified as circadian (24, 35, 97; reviewed in 23). In addition, enhancer trapping in Arabidopsis found that one-third of the promoters examined showed circadian rhythmicity, confirming the impact of the clock on transcriptional regulation (82). This large number of cycling transcripts may be one explanation for why forward genetic screens, which were very successful at identifying core-clock components, were relatively poor at identifying components of the output pathways given that compensation by other output signals could mask the loss of any individual component. A second commonality between the Arabidopsis, Drosophila, and mouse [specifically, the suprachiasmatic nucleus (SCN)] arrays was the observation of a bimodal distribution of the cycling transcripts. That is, most circadian expressed genes showed peak expression either immediately before dawn or dusk, reflecting the importance of anticipating daily light transitions.
Although all array experiments showed a similar fraction of the genome as cycling, there was little overlap in the specific transcripts identified in each study. In Drosophila, five papers profiling circadian expression of RNA levels in the fly head were published in 2001–2002. Cumulatively, 548 transcripts were identified as cycling, although only 1% (7 genes) were identified as circadian in all experiments, and approximately 25% between any two (5).
Multiple factors could contribute to this apparent lack of consensus. First, the transcripts not detected in multiple array sets may be highly susceptible to entrainment conditions. Therefore, these transcripts may be regulated by the clock only in specific conditions that are not consistent between laboratories. For example, in the five Drosophila array publications, two different strains of Drosophila were assayed, and slight variations in entrainment and sampling protocols existed between experiments (5). Another contribution to low overlap in genes identified as circadian could be the different algorithms used in each study to identify cycling transcripts (13, 64, 118). Further investigation and meta-analysis in Arabidopsis and murine systems confirm that environmental differences, sampling frequency, and algorithms used have a strong contribution to the low overlap of cycling transcripts in these experiments (5, 15, 26, 51, 71, 79).
Methods for Identifying Circadian-Regulated Transcripts
Different algorithms for identifying cycling transcripts can give conflicting results on the same dataset (64). Although advances are still being made in identifying periodically expressed transcripts, a clear consensus on the best method is lacking (see sidebar, Selecting the Optimal Cycling Algorithm). Therefore, this subject is worthy of its own review, and the brief overview of selected methods in Table 1 should serve as a starting point rather than an exhaustive list. Because these multiple time-point experiments are costly, an ideal method should be able to handle noise in the expression data that is inherent to biological samples, with limited, if any, replicates, and a short time series that covers only two periods. Further, since thousands of transcripts are examined, the probability exists that by chance some will be identified as cycling. Therefore, a multiple testing correction needs to be applied to determine significance. Once circadian transcripts are identified, it is also necessary to identify when during the day (phase) the transcripts are expressed. The algorithms currently available for performing these tasks are often adapted from signal-processing approaches and are applied to identify periodic expression in both cell-cycle and circadian experiments. All methods perform well in identifying transcripts with a strong difference in amplitude from peak to trough (64). However, the identification of cycling transcripts with weaker amplitude varies greatly from algorithm to algorithm. This variation in detecting weak signals suggests that the differences observed between algorithms and experiments could be due to noise that is typically prefiltered before processing in other systems. Such prefiltering is not possible in these biological experiments, although approaches have been developed to apply post-data-collection filtering (105). However, most algorithms currently in use attempt to identify periodic signals from unfiltered data. Conceptually, the methods can be divided generally into two categories: comparisons in the time domain through pattern matching and comparisons in the frequency domain through signal decomposition, although the principle of these two approaches is similar.
Table 1.
Sample of algorithms available for identifying transcripts with a circadian pattern of expression
| Algorithms for identifying cycling transcripts | ||||
|---|---|---|---|---|
| Algorithm name | Features | Accessibility | Phase call | References |
| Ahdesmaki 2005 | Robust periodicity test assuming non-Gaussian noise | MATLAB and R code available from authors | Requires a separate step | 1 |
| Ahdesmaki 2007 | Adapts robust regression methods for nonuniformly sampled data | MATLAB code http://www.cs.tut.fi/sgn/csb/robustregper/ |
Requires a separate step | 2 |
| Chudova 2009 | Applies a Bayesian mixture model to identify periodically expressed patterns | MATLAB code http://www.datalab.uci.edu/resources/periodicity |
Requires a separate step | 13 |
| COSOPT Straume 2004 | Matches transcript to cosine function | Available from authors | Optimal correspondence between cosine model and expression determines phase | 116 |
| GeneCycle 2009 | Combination of Wichert 2004, Ahdesmaki 2005, and Ahdesmaki 2007. Based on Fisher’s G test. | http://cran.r-project.org/web/packages/GeneCycle/ | Requires a separate step | 1, 2, 136 |
| Glynn 2005 | Lomb-Scargle periodogram improves detection with unevenly spaced time points | R code http://research.stowers-institute.org/efg/2005/LombScargle/ | Requires a separate step | 28 |
| HAYSTACK Michael 2008 | Matches transcript pattern to user-defined models of expression | http://haystack.cgrb.oregonstate.edu/haystack_help.html | Model with strongest correlation provides phase | 79 |
| Liang 2009 | Applies Laplace periodogram; improves performance when outliers exist | MATLAB code http://kuoching.googlepages.com |
Requires a separate step | 63 |
| de Lichtenberg 2005 | Develops a score based on permutations; scores transcripts based on periodicity and magnitude | Algorithm detailed in referenced manuscript | Algorithm provided | 64 |
| Luan and Li 2004 | Selects guide genes and performs a cubic B-spline–based periodic function | MATLAB program available from authors | Requires a separate step | 66 |
| Lu 2004 | Applies a Bayesian approach with periodic-normal mixture model | Algorithm detailed in referenced manuscript | Requires a separate step | 67 |
| Lu 2007 | Combines expression data from multiple species | Algorithm detailed in referenced manuscript | Requires a separate step | 68 |
| Ptitsyn 2007 | First assigns a phase to all transcripts and then filters the data before identifying cycling transcripts | Algorithm detailed in referenced manuscript | Selects based on highest correlation with a series of phase-shifted cosine curves | 105 |
| Spellman 1998 | Fourier transform | Algorithm detailed in referenced manuscript | Requires a separate step | 114 |
| Wichert 2004 | Fisher’s G test | Available from authors | Requires a separate step | 127 |
| Zhao W 2009 | Includes a step to incorporate prior knowledge in identification of cycling transcripts | Algorithm detailed in referenced manuscript | Requires a separate step | 140 |
The first methods that analyzed the transcripts in the time domain initially compared the expression pattern of each transcript to a cosine function (35). Most methods use a linear regression approach to determine the correlation between the transcript and the model, followed by a permutation test to determine the significance. The phase is determined by the phase of the model that is its closest match. By limiting the hypotheses being tested, these pattern-matching algorithms reduce the search space, thereby improving the sensitivity. Realizing that circadian expression did not always match a perfect cosine wave, investigators extended analysis from this basic cosine-matching approach to enable identification of nonsinusoidal shapes that repeat every 24 h (66, 79, 84). One limitation of these types of approaches is that the models must be predefined by the user. To address this limitation, one approach is to identify periodically expressed patterns of any shape using a Bayesian procedure to estimate the contribution of a periodic component in the observed expression (13).
Alternatively, approaches comparing transcripts in the frequency domain first perform transformation of the data. Originally, the Fourier transform was used to determine the spectral composition of each transcript. Transcripts with a strong spectrum signal that correlates to a period of around 24 h are selected as circadian. The phase of expression is determined in a separate step. This method performs well on evenly spaced data (116). Significance for these methods is usually determined by either permutation or a heuristic cutoff. Alternative methods, including the Lomb-Scargle periodogram and Laplace periodogram, have recently been applied to time-series data and may provide improved detection of cycling transcripts (28, 63).
One assumption in the significance determination in most methods is that the majority of transcripts are not circadian. However, Ptitsyn and colleagues challenge this assumption (105). They re-evaluate existing arrays in Arabidopsis and mouse and suggest that a majority of transcripts cycle (105–107). In support of their results, they point to the fact that components of the core transcriptional machinery themselves cycle. These results, the complications of identifying circadian transcripts, and the fact that transcripts of some circadian regulators such as CLOCK (circadian locomotor outputs kaput)/CYCLE do not cycle in some tissues indicate that perhaps less emphasis should be placed on determining if a transcript is cycling (111). Depending on the question under investigation, it may be worthwhile to focus less on which genes are cycling and more on the differences in expression between conditions or tissues or times of day. Differences in level and phase could be important regulatory mechanisms that would be missed if the attention is solely on a comparison of cycling transcripts.
Expanding the Discoveries from Transcriptional Arrays with Meta-Analysis and Network-Modeling Approaches
The lack of consensus on the specific genes that cycle dampened somewhat the initial excitement regarding the applications of global transcriptional profiling to the circadian field. However, several developments have bolstered the value of global transcriptional profiling. Additional experiments have been performed under varying entrainment and sampling conditions and in various genotypes and tissues. Meta-analysis, the combination of multiple large datasets, has allowed the circadian field to take advantage of these publicly available arrays, increasing the power of analysis and leading to new discoveries. Further, the integration of the transcript levels with other genome-wide datasets has reinvigorated the transcriptional approach and is not only improving the understanding of how the clock integrates output responses, but is also providing novel insights into the composition of the core clock itself (Figure 2).
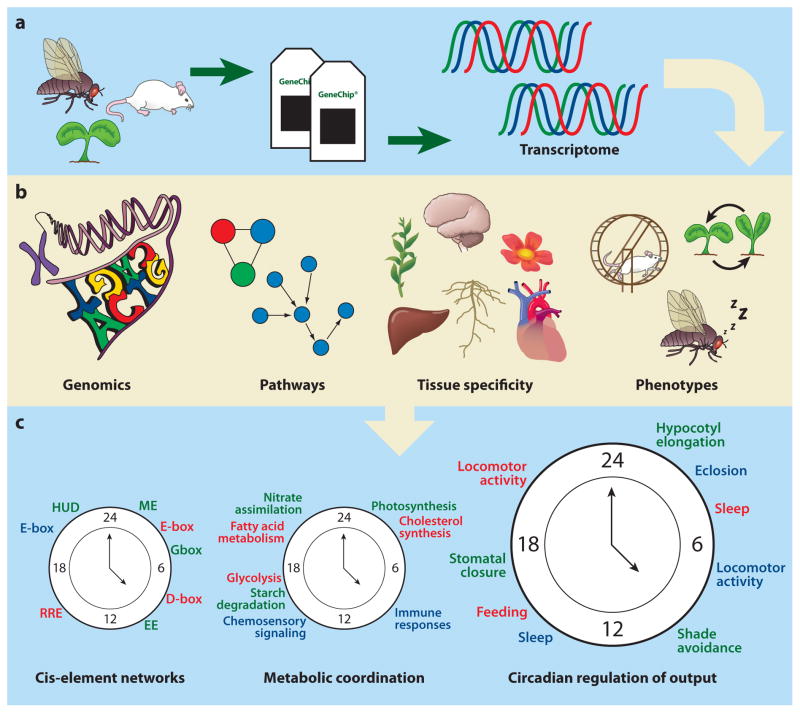
Figure 2.
Integration of the circadian transcriptome with other high-throughput levels of data. Circadian expression information can be integrated with multiple layers. (a) Expression levels of RNA transcripts are collected at multiple time points throughout a circadian cycle. (b) This circadian transcriptome can be integrated with genomics, protein pathway networks, information on tissue specificity, and phenotypic information. (c) By combining these multiple layers of global data, timing of molecular events, such as cis-element regulation and metabolic coordination, can be developed. The ultimate goal of these network-analysis approaches is to understand the regulatory mechanisms by which an organism’s internal circadian clock is able to temporally regulate biological processes.
Expression Analysis Offers Insights Into the Core-Clock Mechanisms
Although the primary objective for performing circadian transcriptional array experiments was to identify links to the output, these experiments also allowed closer examination of the cogs of the core clock itself. Reporter-gene constructs were instrumental in elucidating the components and interactions of the core clock. Expression arrays have revealed transcripts that cycle with peaks of expression at all times of day. Generating novel reporter constructs from the promoters of these genes, particularly those with peaks of expression at phases not thoroughly investigated, will provide useful tools to help unravel the cascade and interconnected network extending from the core clock.
One commonality between transcripts involved in the core-clock mechanism across all species is that they tend to have high-amplitude expression and robust cycling across multiple experiments and tissues. Comparison of amplitudes is often overlooked in cycling analysis. Perhaps identification of transcripts with robust cycling patterns could provide leads into new core-clock genes (15). Alternatively, identification of transcripts with high-amplitude circadian expression may identify novel cis-elements and robust candidates for additional reporter-gene constructs, allowing for improved forward genetic screens.
The input mechanisms that keep the clock in tune with the environment can also be evaluated from these transcriptional arrays. By comparing expression difference between varying entrainment conditions, the effects of these entrainment signals on the clock can be evaluated. For example, in Arabidopsis, a comparison between different entrainment conditions showed that the phase of some transcripts varied when entrained by photocycles or by thermocycles (79). However, when entrained by both temperature and light, the phase was closer to that observed in thermocycles, indicating that for these transcripts, temperature is a stronger regulator of their circadian phase of expression. This temperature-regulated shift in expression patterns supports earlier observations that Arabidopsis has two distinct mechanisms regulating output that can be distinguished by entrainment regimes (77). In Drosophila, although many genes are rhythmically regulated by temperature cycles, in the absence of a photocycle, shifting to constant temperatures abolishes the cycling of a majority of these transcripts (4). The transcripts that maintained cycling in constant conditions after thermocycle entrainment included the core-clock genes. Even though the temperature entrainment was gradual, with a low before dawn and a high at noon, these transcripts maintained phases similar to photoentrainment, implying integration of signals via a single molecular clock.
Effects of the Circadian Clock on Transcription are Pervasive
The pervasiveness of the circadian clock on transcription was observed in the first expression arrays and has been emphasized by the experiments of the last ten years. This level may still be an underestimation, as the analysis of cycling cytosolic proteins indicates that more proteins cycle than transcripts (108). Functional analysis and protein-pathway analysis show that many pathways are affected by the circadian clock from neuro-signaling to photosynthesis (Figure 3). Several tools are now available to help analyze a gene of interest for a potential circadian-regulated effect. In Arabidopsis, Diurnal (http://diurnal.cgrb.oregonstate.edu/) enables quick assessment of the circadian regulation of a gene in multiple entrainment conditions and mutants (84). In mammalian systems, through the BioGPS portal (http://biogps.gnf.org), a gene of interest can be queried for circadian expression patterns in different tissues and to see if small-interfering RNA (siRNA) constructs of the gene affect circadian period (44, 131, 141). Further, BioGPS allows customizable interrogation of multiple high-throughput datasets for each gene, allowing the integration of circadian analysis with other experimental conditions. Of course, not all regulation is at the transcriptional level. Therefore, as other layers of circadian regulation are investigated, these results can be integrated to available web tools for analysis.
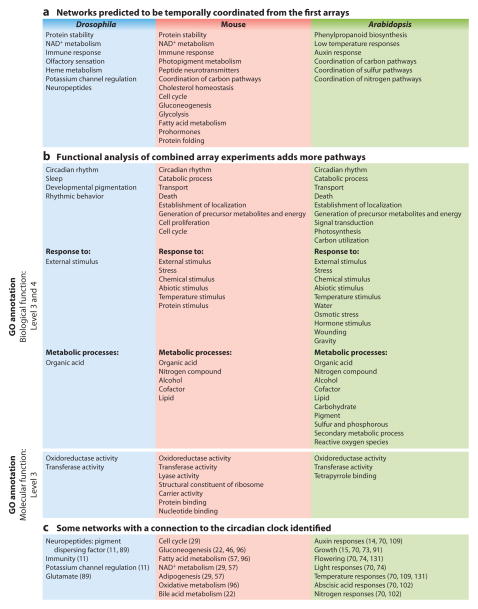
Figure 3.
Predicted pathway regulation by the circadian clock. (a) The first arrays identified many pathways as circadian regulated that have since been confirmed. Combining multiple arrays together allows for the identification of multiple pathways with transcripts enriched for circadian regulation. (b) Cycling transcripts were selected from meta-analysis studies of each organism (51, 79, 136). A selected subset of gene ontology (GO) terms that are enriched (adjusted p-value <0.05) for the GO category of biological function at levels 3 and 4 and the GO category of molecular function at level 3 are listed here. (c) Some examples of pathways with a verified molecular connection to the circadian clock.
NETWORK MODELS BUILT FROM WHOLE GENOME EXPRESSION ANALYSIS
Measurements of clock outputs are reminiscent of mathematically tractable sinusoidal functions. This observation and the mathematical interest in oscillators have made the circadian clock a favorite medium for the integration of mathematics and biology. Since the late 1960s, bottom-up models of clock function, primarily constructed through differential equations, have been developed, applied, tested, and refitted. The application of these models to the clock system has been fruitful, including the prediction of a two-component oscillator and, in Arabidopsis, the prediction that there were still unidentified components of the core clock. For an excellent review of these models, see reference 110.
The flood of transcript information from expression arrays also allows for a deductive type of modeling. This top-down or network-analysis approach combines large-scale data of multiple levels to reconstruct interactions between the layers. The goal of these network-analysis approaches is to combine these inter-actions to develop a model network, tracing information from external entrainment signals through the oscillator to signals resulting in output responses.
Generating these network models is possible through many means; here, we focus on results produced from integrating the transcriptional array results with other genome-wide layers of information. These combinations include integration of transcriptional data with genomics to develop cis-regulatory networks; with other RNA measurements, including non-coding RNA analysis; with protein level information such as enzymatic functional groups; and with organismal levels of information such as tissue specificity or entrainment conditions. Eventually, multiple layers of high-throughput information can be incorporated, with the ultimate goal of elucidating mechanisms by which genes, transcripts, proteins, and metabolites interact with the environment to establish a phenotype.
COMBINING THE TRANSCRIPTOME WITH THE GENOME TO DEVELOP CIS-REGULATORY NETWORKS
One of the most direct applications of expression data is the identification and integration of cis-regulatory elements to develop a cis-regulatory network. The first step in this analysis is to separate circadian transcripts into the phases of expression and identify the general structure of the cis-regulatory elements enriched in the promoters of transcripts expressed at each phase. In the case of complex or degenerate cis-elements, it is sometimes necessary to refine the exact identity of the cis-element. Finally, once the cis-elements involved are identified and characterized, a cis-regulatory network can be developed.
From the first circadian arrays in Arabidopsis, promoters of transcripts with similar expression patterns were analyzed for conserved cis-elements. A novel element was identified in the promoters of the genes expressed in the evening (35). Further investigation into this evening element (EE; AAATATCT) revealed that it is the direct target of circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY), which are components of the core-clock morning loop (33) (Figure 1). This link between the morning loop components CCA1/LHY and the promoters of genes containing evening elements could provide a mechanistic understanding of how the Arabidopsis clock may directly regulate many of the evening expressed output genes.
Additional clock enriched cis-regulatory elements were also identified by other groups: CCA1-binding site (CBS; AAAAAATCT), morning element (ME; CACTAACCAC), and hormone up at dawn (HUD; CACATG) (43, 78, 81) A study by Michael et al. (reviewed in 101) combined expression data from eleven different Arabidopsis time-series arrays. Based on this, they were able to refine the ME (CCACAC) and identified combinations of cis-elements that resulted in new cis-regulatory modules: The ME combined with the G-box (CACGTG) was enriched in morning expressed genes; the EE combined with the GATA element was enriched in evening expressed genes; and the promoters of midnight expressed genes were enriched for telobox (TBX; AAACCCT), starch synthesis-box (SBX; AAGCCC), and protein box (ATGCCC) (reviewed in 33). Similarly, Covington et al. (15) combined three circadian arrays and identified the EE, ME, GATA, and G-box as being overrepresented at specific times of day. The consistent identification of these elements by several studies attests to their proposed functional importance, which remains to be experimentally validated.
In Drosophila, a circadian-regulated cis-element, the E-box, was identified prior to the array analysis. However, an exact consensus sequence was not clear (reviewed in 32). By combining genomic information from multiple species, Paquet et al. (98) extended cis-element analysis to identify an informative consensus sequence for the E-box in Drosophila. Potential direct output targets of the transcription factors clock (CLK) and cycle (CYC) were difficult to predict due to the degeneracy of its E-box (CANNTG). Therefore, to identify a more specific CLK/CYC target element, they started with the PER gene enhancer that contained the E-box. Using this enhancer as a seed, they compared the promoters of five other known CLK/CYC targets to identify a cis-element that was conserved among all 12 Drosophila species. From this analysis, they generated a position-specific weight matrix for nucleotides that comprised a two-part E-box in which the core of the first half, E1 (CACGTG), was followed by a 1- to 2-nucleotide spacer, then E2, a more degenerate E-box–like element. This longer E1-E2 model of the E-box consensus sequence was able to predict clock-regulated genes in the mouse, including PER1 and PER2, known targets of the mammalian E-box binding transcription factors CLOCK and brain and muscle aryl hydrocarbon nuclear translocator like 1 (BMAL1). A similar approach was also applied to identify the E-box consensus sequence based on conservation in mammalian genomes (86). This approach could be extended to identify or refine other circadian-regulated cis-elements.
Cis-regulatory elements involved in circadian regulation were experimentally defined in mammalian systems prior to the transcriptional arrays. However, expression analysis was used to refine the consensus sequence of previously identified cis-elements: E-box (CACGTG) targeted by CLOCK/BMAL1 transcription factors, D-box (TTATG[T/C] AA) targeted by D-Box binding protein (DBP) and E4BP4 transcription factors, and RRE ([A/T]A[A/T]NT[A/G]GGTCA) targeted by REVERB and RAR-related orphan receptors (ROR) (59, 97). These studies were the foundation for the development of a cis-element-based network (120). From this network, Ukai-Tadenuma et al. (122) predicted that the entire range of temporal expression observed in mice could be generated by varying the combinations of the morning responsive E-box, the evening responsive RRE, and the daytime active D-box in the promoters of activators and repressors. The availability of a transient expression system enabled the testing of this cis-regulatory logic in a beautiful example of synthetic biology. They controlled the temporal expression of an artificial activator, a GAL4-VP16 chimeric protein, and a repressor (GAL4 alone) with different combinations of these three clock-controlled elements. By altering the different phases of activator and repressor expression, they were able to generate expression of a target reporter gene driven by a GAL4 UAS promoter at specific times throughout the day, including morning, daytime, and evening.
Taken together these approaches reveal that although each experiment may not identify overlapping cycling transcripts, combining experiments can improve the discovery of regulatory elements. Such a meta-analysis was performed by Yan et al., who compared arrays in mice on 14 different tissues (136). In all tissues, they found that E-box, AP2, CRE, SP1, D-box, and EGR elements were enriched in the promoters of cycling genes. However, when comparing the phase of expression, they discovered differences between the tissues. For example, the D-box was enriched for a specific phase, circadian time (CT) 16, only in aorta and adrenal tissues. Other elements showed a tissue-dependent phase of expression; the E-box is primarily associated with CT12 in most tissues, but in skeletal muscle was overrepresented at CT0, even though most clock genes showed a similar phase of expression between the two tissues. Because overrepresentation analysis is sensitive to the number of genes, these differences could be an artifact of the total number of circadian genes identified in each tissue. However, if it is representative of a change, it could indicate that the upstream regulators of these components are either different in the various tissues or themselves have tissue-specific variation in expression.
The identification of cis-regulatory elements that coincide with specific time-of-day expression is a first step in understanding circadian regulation of transcripts. However, the presence of a circadian-regulated element in a promoter is not sufficient for implicating a specific transcription factor in the regulation of that transcript. As confirmation, follow-up experiments such as chromatin immunoprecipitation (ChIP) are necessary to validate the regulation implied by the presence of these elements. These experiments are still largely absent from the circadian literature, in part because of the additional challenge and cost associated with performing these assays at multiple time points throughout the day. Overcoming some of these challenges, Nakamichi et al. investigated the temporal binding of the pseudoresponse regulators PRR9, PRR7, and PRR5 to the LHY and CCA1 promoter over a 24 h period in Arabidopsis (88). These experiments revealed that the composition of these repressive factors at the CCA1 and LHY promoters changes in a time-of-day-dependent manner. PRR9 is present at the CCA1 and LHY promoters just after dawn while PRR7 and PRR5 occupy the promoters later in the day. Extending this analysis of the temporal binding of clock-regulated transcription factors to a whole-genome level through ChIP arrays (ChIP-chip) or ChIP high-throughput sequencing (ChIP-seq) time courses will be necessary for building the complete transcriptional output network from the clock.
COMBINING TRANSCRIPTION AND RNA REGULATION
Identifying Trans-Factors
After combining transcriptional and genomic information to identify cis-regulatory elements, investigators can address the mechanism by which mRNA accumulation is regulated. Expression levels of messenger RNA (mRNA) can be regulated both transcriptionally by trans-factors via the identified cis-elements or post-transcriptionally by RNA regulatory mechanisms. Incorporation of expression information can aid in the discovery of novel circadian trans-factors as demonstrated in a recent study by Pruneda-Paz and colleagues (102). In this study, they filtered the approximately 2,000 transcription factors in Arabidopsis and selected those that were identified as cycling in the Harmer et al. microarray (35). About 200 cycling transcription factors were selected, and a library with these proteins was generated for yeast-one-hybrid analysis. Using this library, they successfully identified CHE (CCA1 hiking expedition,), a transcription factor in the TCP (TB1, CYC, PCFs) family that is a repressor of CCA1. Because there are 23 TCP proteins in Arabidopsis, functional redundancy could explain why CHE was not identified in previous genetic screens. This approach of narrowing the search pool by 90% by selecting only cycling transcription factors could be adapted to other organisms and other molecular interactions.
Role of Noncoding RNA
In addition to transcriptional regulation, mRNA levels are also regulated at the post-transcriptional level partly by noncoding RNAs. Integrating the mRNA transcriptome with the expression pattern of nonprotein-coding RNA transcripts implicates new mechanisms of clock regulation. Although the initial expression arrays focused primarily on detecting mRNA levels from protein-coding transcripts, noncoding RNAs also play an important role in the regulation of the clock (58; reviewed in 12, 94). In Drosophila transcriptional profiling of 78 microRNAs (miRNA) identified several miRNAs that showed an altered expression pattern in clock-disrupted mutant flies, cyc°1, compared to wild type (WT) (135). In murine retinas, twelve miRNAs showed variation in expression between noon and midnight, making them interesting candidates for investigation as potential circadian-regulated miRNAs (132). Furthermore, a connection between miRNAs and clock components has been observed in murine SCN, where miRNA-132 is a positive regulator of PER1 expression (reviewed in 12).
In Arabidopsis, a more detailed analysis was performed using tiling arrays to map the cyclic expression of most of the genome (37). Surprisingly, they observed that seven percent of the natural antisense transcripts were expressed in a circadian pattern. Four circadian-regulated miRNAs were identified, as were a number of cycling introns. Although these phenomena are intriguing, further investigation into the physiological significance of these findings is required to establish a connection between posttranscriptional regulation and clock function. One promising link is the observation that CCA1 transcript stability is regulated by light quality (134). The destabilization of CCA1 by certain wavelengths of light provides a potential mechanism for synchronization of the clock components with the environment. Although preliminary, what is perhaps most exciting about these noncoding RNA experiments is that they confirm the repeated observations in circadian studies that evolutionary selection has produced a clock that takes advantage of regulation at almost all possible levels. This multiple-layer regulation may ultimately contribute to the robustness of the clock.
LOOKING BEYOND NUCLEIC ACIDS: INTEGRATION OF TRANSCRIPTOMICS WITH SYSTEM-LEVEL INFORMATION
Enrichment of Protein Networks
Concomitant with identification of cis-elements and analysis of DNA-RNA interactions, another means of leveraging high-throughput transcriptional data is to look for enrichment of known molecular pathways in coexpressed transcripts, known as functional enrichment. Circadian application of functional enrichment determines if, in a set of transcripts that peak at a similar time of day, there are more transcripts whose protein products function together in a known pathway than would be expected by chance. The first circadian array papers revealed that multiple transcripts for some pathways were expressed at the same time of day. Expression of phenylpropanoid biosynthetic genes, which play a protective role against UV radiation, peaked just before dawn in Arabidopsis (35). In mice, transcript expression was highest for metabolic genes involved in glycolysis, gluconeogenesis, and fatty acid metabolism at night when the animals are feeding (97). In Drosophila, genes involved in immune responses had peak transcript expression at the end of day and into the night, with the lowest expression at dawn (11). Since the publication of these first arrays, advances in the methods for identifying functional enrichment have made this a successful approach to interpreting transcriptional data in many systems, and over 60 tools are available to aid in such analysis (reviewed in 42). Detailed annotation of genes has enabled the development of pathway resources that generate ontologies identifying proteins that function together. In addition to manually generating lists of proteins involved in a common function, two commonly used categorization methods are the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/), which groups genes by participation in a common biological pathway, and gene ontology (GO), which groups genes by function in a common biological process, cellular compartment, or molecular function (3, 48–50). Studies in Arabidopsis have been particularly successful in exploiting these methods of determining functional enrichment to identify pathways regulated by the circadian clock.
The downstream targets of many plant hormones are enriched for regulation at specific times of day, implicating a role for the clock in coordinating hormone responses (14, 41, 83; reviewed in 109). One example is the phytohormone auxin, an important regulator of plant growth (14). It was observed over 60 years ago that the effects of exogenous auxin on a plant depend on the time of day that it is applied (reviewed in 109). Functional analysis of Arabidopsis circadian arrays identified enrichment for both auxin regulatory factors and transcripts involved in inactivation of auxin via conjunction (14, 41). Targets of auxin signaling are also enriched for circadian regulation with a peak of expression in the middle of the day, indicating a link between the clock and output responses (14, 41). This link to the clock via the circadian regulation of auxin signaling and auxin targets may explain the diurnal changes in auxin sensitivity observed in earlier studies.
Functional enrichment of a biological pathway can be combined with cis-element analysis to identify potential mechanisms for co-ordinated pathway regulation. In Arabidopsis, a literature-generated list of 182 phytohormone genes was examined for circadian phase enrichment (78). This functional enrichment analysis identified coordinated expression of most of these transcripts peaking at dawn in short day photoperiods and at dusk in continuous light. This timing coincides with the peak of hypocotyl growth, which is regulated by light, phytohormones, and the circadian clock (21). In short day photoperiods, maximum hypocotyl growth occurs at dawn, while in constant-light conditions growth peaks at dusk (92). To identify regulatory mechanisms for this enrichment, the promoters of these co-regulated phytohormone genes were analyzed, and the HUD cis-element was found to be enriched. Expression of a HUD element reporter construct mimicked the expression pattern observed in the phytohormone genes, indicating that HUD may be the cis-regulatory element through which the clock regulates phytohormone genes and therefore hypocotyl growth. The HUD element now offers a tractable opportunity to identify the upstream regulators and mechanisms by which the clock controls growth through hormones, the first steps to identifying a functional link between the clock and hormone-regulated growth.
Tissue Specificity in Circadian Expression: The Third Dimension of Transcription
In addition to integrating transcripts with other molecular levels of regulation, DNA, RNA, and protein, circadian research is also incorporating systems-level analysis. Comparisons of how circadian regulation of transcripts differs between tissues are providing insights into the regulation, function, and role of tissue-specific clocks.
As discussed above, combining the transcripts identified as cycling in different mammalian tissues aided in the identification of clock-related cis-elements. However, when comparing different tissues, there was relatively little overlap between transcripts identified as cycling, even though environmental conditions were identical and the same algorithm was applied in each experiment (11, 40, 97, 115). This raised questions about the differences between tissues in the clock and its output. Investigators interested in pursuing tissue-specific differences in mammalian clock outputs initially looked at cycling transcripts in individual cell lines. However, one study observed a hundredfold more cycling transcripts in livers dissected from mouse than in NIH3T3 cells and U2OS cells (fibroblast and osteosarcoma culture cells, respectively) (44). This striking reduction of cycling transcripts in the cell-culture lines indicated that noncell-autonomous signals are likely required to maintain robust circadian transcription. However, the severely low numbers of cycling transcripts in the cell-culture lines in this study were not observed in other studies, suggesting that further investigation is necessary (24, 31, 76).
To determine the contribution of a nontissue-autonomous signal to circadian expression in the liver, mice with a liver-specific disruption in the circadian clock were generated (56, 60). Circadian expression of liver transcripts was compared between WT mice and transgenic mice with a disrupted liver clock due to a liver-specific inducible Rev-erbα construct. This comparison revealed that even without a functional liver clock, two classes of genes were still circadian regulated: cholesterol synthesis and chaperone proteins. However, since these mice still had a functional SCN clock, it is possible that they maintained rhythmic feeding, a potential driver of circadian expression in the liver. The role of the SCN in regulating circadian transcripts in the liver by maintaining rhythmic feeding was investigated by Vollmers and colleagues (123). In this study, they controlled the access to food in WT and Cry1−/−:Cry2−/− double knockout mice, which are arrhythmic in behavior. Comparison of circadian liver transcripts under controlled feeding regimens in the different genetic backgrounds revealed few differences, suggesting that feeding is an important entrainment cue of circadian expression in the liver. Furthermore, the majority of hepatic cycling genes remained rhythmic based on feeding, a behavior that is regulated by the clock in the SCN. However, one surprising result of the Vollmers et al. study was the extent to which feeding impacted the number of cycling transcripts. Almost 90% of the nearly 3000 transcripts that were circadian in controlled feeding conditions lost rhythmic expression during fasting. Both the dramatic reduction in the number of cycling transcripts during fasting and the reduction in cycling transcripts in constant dark-grown plants implicates energy signaling as an important link between the clock and rhythmic transcription for a large portion of the transcriptome (79).
Although ideal, the expense associated with repeating time-course transcriptional arrays in multiple tissue samples is often a limitation. In Drosophila, tissue-specific clock expression offers an alternative approach. One of the most exciting discoveries in the Drosophila circadian clock has been the neurological anatomy of the clock. Drosophila clock genes appear to be expressed primarily in the peripheral sensory neurons and central brain neurons, and a cell-specific clock architecture has been described for neurons in the fly brain (reviewed in 90). Nagoshi et al. specifically profiled clock neurons and identified transcripts that were enriched in these neurons (85). In this analysis, they identified Nocturnin, a deadenylase with no previously known function in the Drosophila clock. When expression of Nocturnin is reduced by RNA interference (RNAi), flies maintained rhythmic expression of PER under constant light conditions in contrast to WT. Thus, comparison of tissue-specific expression was able to identify a novel component of the Drosophila clock. This approach could also be advantageous to organisms without such specific clock architecture. For example, by sampling different tissues at the same circadian time and comparing expression of known clock genes, one might be able to identify tissue-specific differences in clock regulation for that specific time.
In a recent study in Arabidopsis, transcriptional arrays comparing the circadian expression in root and shoot tissues indicated that expression of clock genes in the root is regulated by the shoot, possibly through signals derived from photosynthesis (47). This observation of noncell-autonomous regulation of the clock is in contrast to previous experiments with aerial tissues where isolated tissues can maintain circadian rhythms of gene expression (87; reviewed in 45, 74). In a single tobacco plant, tissue-specific entrainment, established by applying foil covers at different phases, showed that the aerial tissues maintained their phase of entrainment, irrespective of the entrainment signals neighboring tissue received (87, 119). These results indicate that noncell-autonomous signals between aerial tissues were not strong enough to overcome the cell-autonomous signals (119). An important question in the plant circadian field will be to investigate the difference between the apparent nonautonomous regulation observed in the root clocks and the evidence for cell-autonomous regulation in the aerial tissues (reviewed in 34, 45). It remains unknown whether the differences between root and shoot are due to a tissue-specific lack of autonomy or differences in sensitivity to entrainment conditions. Further investigation into the distinct regulation of output genes in the root will provide insights into tissue-specific functions of the clock.
Together, these investigations into cell autonomy are beginning to answer questions about what tissue-specific outputs are circadian regulated and how the effects are coordinated within the whole system. Cell-type specific analysis of circadian expression may be enabled by sophisticated tissue separation methods including laser assisted microscopic dissection and fluorescent-labeled cell sorting (reviewed in 16, 61, 89). Studies in developmental regulation have made excellent use of combining temporal and spatial data to generate descriptive networks in Drosophila and Arabidopsis (9; reviewed in 99). Extension of these approaches to the clock will enable the development of a cell-specific regulatory map for specific outputs and help discern cell-autonomous and nonautonomous control under various conditions.
Comparison of the Cycling Transcriptome Between Genotypes
Comparisons of transcriptional differences between genotypes are also shedding light on regulation and output from the circadian clock. Genetic differences generated by natural selection between ecotypes, haplotypes, or species can be linked to changes in transcription to identify different regulatory mechanisms. Conserved components between genotypes are potentially critical clock components, while mechanisms that differ between genotypes may have a function in adaptation to specific environmental settings.
Although these comparisons are in the early stages in the circadian field, initial experiments are revealing the potential of such analysis. For example, Yan et al. (136) compared mouse, rat, rhesus macaque, and human transcript arrays, and observed that the antiphasic relationship between E-box and RRE cis-elements was preserved across diverse species. This observation implies that the regulatory mechanisms of these cis-elements maintain alternate regulation of their targets and that the antiphasic expression of transcripts under control of these elements is under positive selection. Because the phase of many cycling genes differed between the species, further investigation into these changes may reveal how the clock adapts in diurnally versus nocturnally active organisms.
Even within a species, the comparison of different mutant genotypes, such as lhy-1 and lux (LUX Arrhythmo) in Arabidopsis, can provide insights into clock function (38, 79). These single clock mutants show drastic effects on the circadian expression of many transcripts, although it is difficult to discern direct and indirect effects. More subtle differences, available from natural variation, can provide a means for linking genetic differences to clock-based adaptation. In Arabidopsis, the 1001 genome project will provide the genetic differences in DNA sequences between various ecotypes (125). Clock-specific phenotypic measurements have already been made for many of these ecotypes in the form of leaf movement assays (80). The polymorphisms in the genome can be connected to the phenotypic variation by examining the expression differences between these ecotypes. Differences in the circadian transcriptome between ecotypes can potentially be linked to causative genetic changes. Furthermore, since the natural environment that these ecotypes have adapted to is known, the differences in the circadian transcriptome may provide a link to ecology and natural selection.
In plants, the cycling transcriptome of Arabidopsis was compared to another dicotyledonous species, Populus trichocarpa (poplar), and a more distantly related monocotyledonous species, Oryza sativa (rice) (S. Filichkin & T. Mockler, personal communication). The expression patterns of most clock-associated genes displayed similar cycling profiles among the three species, peaking within three hours of their respective orthologs. The preservation of cycling between rice and Arabidopsis, species separated by approximately 200 million years, suggests a strong conservation of the circadian output networks in plants (130). In multiple instances, predicted poplar, rice, and Arabidopsis orthologs involved in analogous metabolic processes showed similar expression profiles with peak expression at the same time of day. This comparison of circadian regulation offers the opportunity to identify conserved mechanisms of circadian regulation. For example, cycling genes in all three species showed conservation of circadian- and diurnal-associated promoter elements including the EE, CBS, GATA, G-box, ME, TBX, and SBX. Conversely, in other examples, the orthologous genes were phased to different times of day in the different plants, suggesting species-specific diversification for some metabolic pathways.
These experiments comparing cycling genes in plants and mammals are potentially just the beginning of much larger intra- and inter-species comparisons. The availability of high-throughput RNA sequencing (RNA-seq) will enable the sequencing of individual transcriptomes. RNA-seq opens up the potential to investigate the circadian transcriptional regulation of any organism, removing the limits imposed by focusing only on sequenced organisms. With this opportunity the circadian transcriptome of species or individuals with unique circadian phenotypes, unusual adaptation mechanisms, or the ability to survive in extreme conditions can be compared to the transcriptome of the well-studied model organisms. By expanding the range of species that can be examined, the output pathways studied will also no longer be limited to those quantifiable in the model organisms. These comparisons will potentially expand our understanding of the role the circadian clock plays in providing a competitive advantage and how the clock impacts adaptation. However, these investigations will require standardized methods to identify similar transcripts between species, an analysis that is advancing, but is not trivial (65). Therefore, methods of identifying orthologous transcripts need to be established to maximize the benefits of such comparative transcriptomic approaches.
Future Directions for Systems-Level Integration
While transcription plays an important role in clock-controlled output, it is only one of many layers of regulation. Integrating transcriptional studies with other levels of high-throughput data will undoubtedly be a rich resource for discovery. Such layers include protein-protein interaction networks, epigenetic analysis, proteomic analyses, metabolism studies, and phenotypic studies. Phosphorylation and protein stability are critical components of the core-clock mechanism, and expression of many posttranslational regulatory proteins themselves are circadian regulated (11, 27, 34, 36, 73, 75, 97, 142). Global analysis of diurnal changes in posttranslational modifications, such as examination of the circadian phosphoproteome, can be combined with diurnal expression patterns to evaluate the regulatory connections in both directions. A challenge for the near future will be to connect the daily cycling of the transcriptome with circadian-regulated metabolic changes (reviewed in 29, 57). Metabolomic analysis remains a challenging task, even in model organisms, owing to the diversity and vastness of the metabolome (91). However, the direct connection that metabolites offer to phenotypes make this a challenge worth pursuing. Advances in metabolic reconstruction networks will provide the opportunity to resolve some of these challenges and develop a link between transcriptional regulation and metabolic changes (reviewed in 93, 96). Finally, maintaining a circadian perspective while comparing phenotypic and transcriptional responses will enable researchers to incorporate regulation by an organism’s internal clock with observed behaviors and environmental responses. For example, Wilkins et al. (128) observed that time of day had a significant effect on the transcriptional drought response in Populus clones.
A hallmark of the circadian clock appears to be multiple interconnected loops, therefore, when combining these multiple levels, distinguishing between cause and effect in regulatory mechanisms will present a challenge. An example of such a feedback loop involving metabolites is observed in Arabidopsis. Levels of the signaling molecule cyclic adenosine diphosphate ribose (cADPR) are circadian regulated, yet treatment with nicotinamide, an antagonist of cADPR signaling, alters expression of core-clock genes (19). The application of inducible constructs provides an opportunity to resolve these regulatory feedback loops by delivering genetic perturbations at specific phases in the circadian cycle (54, 56, 60). Global analysis of changes that occur at the transcriptome, proteome, and metabolome levels after such phase-specific perturbations will enable the distinction between direct and indirect effects, providing directionality and structure to circadian networks.
The eventual goal will be to integrate many of these levels simultaneously. In a small-scale example of the concept, Wang et al. incorporated transcription, known regulation from literature, protein-protein interaction, phosporylation, cis-element, and protein-drug interaction (as external perturbations) on 80 cycling genes to develop a network of the circadian clock in mammals (124). These network-developing efforts are only worth-while if they are accessible to biologists. So far this work has advanced primarily due to the heroic efforts of multiple investigators who have successfully combined diverse and sometimes incomplete datasets. In order to advance this approach to the next level, experiments designed from conception to facilitate multiple layers of analysis will be required. Researchers in other systems, particularly in yeast, are laying the groundwork for such experiments, where several layers of data were collected for over 100 segregants of a cross between a laboratory and wild-type yeast strain (6, 7, 17). The genomic information from the segregants was combined with measures of expression changes, protein levels, and growth phenotypes in response to various environmental perturbations. In addition, techniques for integrating these layers are also being improved (52). The added challenge for chronobiologists is that these approaches will also require the integration of time.
CONCLUSION
One of the goals of the first arrays ten years ago was to understand the link between the molecular mechanism and the many physiological outputs regulated by the circadian clock. The whole-genome expression information obtained by these arrays provided a first step in that direction, but the follow-up steps to connect the dots have perhaps been more challenging than anyone anticipated. Most surprising was the large number of transcripts that display circadian regulation, and this has some-what complicated the understanding of the relationship from clock to output. With more transcripts cycling than the number of conditions and time points sampled, making a causal link between regulator and output was difficult. However, network analysis, by integrating expression with other system-wide levels of information, is beginning to overcome this limitation.
Now that we realize the extent and significance of circadian transcriptional effects, experiments can be designed to address the challenges presented by large temporal changes in the transcriptome. Lessons learned from bioinformatic studies in generating networks can be applied to the circadian field. Given that the ultimate goal is to understand how the clock links to phenotype, transcriptional arrays will need to be accompanied by detailed phenotypic measurements. Interesting physiological measurements that could be investigated in this manner include sleep in flies, feeding activity in mice, and flowering time and stomatal opening in Arabidopsis. The next step will be to combine phenotypic and transcriptomic observations across various genotypes or haplotypes to identify causative mutations. Eventually, combination of these network-modeling approaches with traditional thermodynamic models will provide a detailed model that links the molecular regulation of the clock to circadian phenotypes.
SUMMARY POINTS.
The circadian clock has been shown to affect many pathways in mammals, plants, and insects. This pervasiveness is reflected in the large number of transcripts that show circadian regulation throughout the day.
The algorithms used for identifying cycling transcripts are still in development, and an unbiased comparison of methods is needed.
The combination of circadian-transcriptomic data with genomic information has enabled the elucidation of both cis- and trans-elements involved in circadian regulation, leading to the development of cis-regulatory networks.
Noncoding RNA elements, some of which are rhythmically expressed, provide an additional mechanism of circadian regulation.
Pathway and ontology analysis combined with transcriptomics has led to the identification of circadian-regulated functional pathways.
Tissue specificity offers a third dimension of regulation in the link between the core clock and circadian outputs.
Comparisons of the circadian regulation of transcripts between species can provide insights into the contribution of the circadian clock to evolution and adaptation.
FUTURE ISSUES.
The comparison of genotypic variation through genome-wide association studies or through comparative sequencing of species variants will offer a link between genetic changes and variation in circadian-regulated outputs.
Integrating circadian-regulated transcriptional changes with temporal changes in posttranslational protein modifications will improve our understanding of the pathways of clock-regulated output.
The development of metabolic networks for model organisms will enable the connection of circadian expression changes to metabolic changes that occur throughout the day.
The use of inducible-promoter constructs will facilitate the distinction between direct and indirect effects, providing opportunity to determine the regulatory direction of interactions.
Combining multiple levels of information simultaneously, including genomic variation, transcriptional changes, proteomics, metabolomics, and phenotypic variation, will be the basis for the construction of an inclusive clock-regulatory network.
The first points of convergence between bottom-up and top-down models will provide a tractable understanding of the link between the molecular mechanisms of the clock and the phenotypic responses that are circadian regulated.
SELECTING THE OPTIMAL CYCLING ALGORITHM.
Although many methods have been proposed, a clear consensus on the optimal method for identifying circadian-regulated transcripts has not been reached. The main reason is that the quality of the dataset complicates the comparisons between algorithms. Therefore, to properly evaluate the performance of each algorithm, a synthetic dataset supplemented with preselected cycling and noncycling expression patterns is needed. Each approach could then be evaluated against this generated dataset for false-positive and false-negative calls to determine the accuracy and sensitivity of each method. Until such a benchmark is established, the ideal approach will depend on the question being investigated. For example, if the goal is to identify only strongly circadian-regulated transcripts for detailed follow-up experiments, a highly-selective approach would be preferred to reduce the number of false-positive calls. In this case, selecting transcripts consistently identified as cycling by multiple algorithms will provide a list of transcripts with robust cycling expression patterns for further investigation. Alternatively, if the goal of the experiment is to identify functional enrichment or cis-element enrichment, increasing the sensitivity by using a less-stringent approach may outweigh the costs of increasing the number of false-positive calls. Therefore, for an analysis in which an inclusive list is desired, pooling the results from multiple algorithms by including transcripts identified as cycling in at least one may be the optimal approach. Finally, if the experimental goal is a comparison of expression, e.g., between tissues, conditions, or species, simply eschewing the identification of cycling transcripts and directly comparing the time courses may be the most beneficial.
Glossary
- Transcriptome
global, transcribed RNA population of an organism
- SCN
superchiasmatic nucleus
- Entrainment
process by which the circadian clock of an organism is adapted to the time cues from the environment
- Meta-analysis
combination of multiple, often large, datasets originating from different laboratories and composed of a variety of measurement types
- CLOCK
circadian locomotor outputs kaput
- Functional analysis
evaluation of a set of genes, usually grouped by expression profile, for an enriched common annotation such as a biological process or cellular location
- Organic acid Nitrogen compound Alcohol
- EE
evening element (AAATATCT)
- CCA1
circadian clock associated 1
- LHY
late elongated hypocotyl
- CLK
clock
- CYC
cycle
- E1
E-box 1 (CACGTG)
- CT
circadian time
- Core-clock mechanism
specific molecular mechanisms that maintain an endogenous 24 h rhythm in an organism
- Network analysis
interactions derived from combining data from global-scale assays such as transcriptomics or proteomics
Contributor Information
Colleen J. Doherty, Email: cdoherty@ucsd.edu.
Steve A. Kay, Email: skay@ucsd.edu.
LITERATURE CITED
- 1.Ahdesmaki M, Lahdesmaki H, Gracey A, Shmulevich L, Yli-Harja O. Robust regression for periodicity detection in nonuniformly sampled time-course gene expression data. BMC Bioinf. 2007;8(1):233. doi: 10.1186/1471-2105-8-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahdesmaki M, Lahdesmaki H, Pearson R, Huttunen H, Yli-Harja O. Robust detection of periodic time series measured from biological systems. BMC Bioinf. 2005;6(1):117. doi: 10.1186/1471-2105-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3(4):e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boothroyd CE, Young MW. The in(put)s and out(put)s of the Drosophila circadian clock. Ann N Y Acad Sci. 2008;1129:350–57. doi: 10.1196/annals.1417.006. [DOI] [PubMed] [Google Scholar]
- 6.Brem RB, Kruglyak L. The landscape of genetic complexity across 5700 gene expression traits in yeast. Proc Natl Acad Sci USA. 2005;102(5):1572–77. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296(5568):752–55. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 8.Brunner M, Kaldi K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol Microbiol. 2008;68(2):255–62. doi: 10.1111/j.1365-2958.2008.06148.x. [DOI] [PubMed] [Google Scholar]
- 9.Buffin E, Gho M. Laser microdissection of sensory organ precursor cells of Drosophila microchaetes. PLoS One. 2010;5(2):9285. doi: 10.1371/journal.pone.0009285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahill G. Clock mechanisms in zebrafish. Cell Tissue Res. 2002;309(1):27–34. doi: 10.1007/s00441-002-0570-7. [DOI] [PubMed] [Google Scholar]
- 11.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22(21):9305–19. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng HM, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007;6(24):3034–38. doi: 10.4161/cc.6.24.5106. [DOI] [PubMed] [Google Scholar]
- 13.Chudova D, Ihler A, Lin KK, Andersen B, Smyth P. Bayesian detection of nonsinusoidal periodic patterns in circadian expression data. Bioinformatics. 2009;25(23):3114–20. doi: 10.1093/bioinformatics/btp547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5(8):e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covington M, Maloof J, Straume M, Kay S, Harmer S. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Day RC, Grossniklaus U, Macknight RC. Be more specific! Laser-assisted microdissection of plant cells. Trends Plant Sci. 2005;10(8):397–406. doi: 10.1016/j.tplants.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Demogines A, Smith E, Kruglyak L, Alani E. Identification and dissection of a complex DNA repair sensitivity phenotype in baker’s yeast. PLoS Genet. 2008;4(7):e1000123. doi: 10.1371/journal.pgen.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd AN, Salathia N, Hall A, Kevei E, Toth R, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–33. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 19.Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, et al. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science. 2007;318(5857):1789–92. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 20.Dong G, Golden SS. How a cyanobacterium tells time. Curr Opin Microbiol. 2008;11(6):541–46. doi: 10.1016/j.mib.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17(1):63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 22.Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107(6):1972–80. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffield GE. DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol. 2003;15(10):991–1002. doi: 10.1046/j.1365-2826.2003.01082.x. [DOI] [PubMed] [Google Scholar]
- 24.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12(7):551–57. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 25.Dunlap JC, Loros JJ. The Neurospora circadian system. J Biol Rhythms. 2004;19:414–24. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- 26.Etter PD, Ramaswami M. The ups and downs of daily life: profiling circadian gene expression in Drosophila. BioEssays. 2002;24(6):494–98. doi: 10.1002/bies.10109. [DOI] [PubMed] [Google Scholar]
- 27.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–56. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 28.Glynn EF, Chen J, Mushegian AR. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. Bioinformatics. 2006;22(3):310–16. doi: 10.1093/bioinformatics/bti789. [DOI] [PubMed] [Google Scholar]
- 29.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green RM, Tingay S, Wang Z, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129(2):576–84. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundschober C, Delaunay F, Pühlhofer A, Triqueneaux G, Laudet V, et al. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J Biol Chem. 2001;276(50):46751–58. doi: 10.1074/jbc.M107499200. [DOI] [PubMed] [Google Scholar]
- 32.Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms. 2004;19(5):348–60. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- 33.Harmer SL, Kay S. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–40. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60(1):357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 35.Harmer SL, Hogenesch JB, Straume M, Chang H, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290(5499):2110–13. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 36.Harms E, Kivimae S, Young MW, Saez L. Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms. 2004;19(5):361–73. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- 37.Hazen S, Naef F, Quisel T, Gendron J, Chen H, et al. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol. 2009;10(2):R17. doi: 10.1186/gb-2009-10-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102:10387–92. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heintzen C, Liu Y. The Neurospora crassa circadian clock. Adv Genet. 2007;58:25–66. doi: 10.1016/S0065-2660(06)58002-2. [DOI] [PubMed] [Google Scholar]
- 40.Hogenesch JB, Panda S, Kay S, Takahashi JS. Circadian transcriptional output in the SCN and liver of the mouse. In: Derek JAG, Chadwick J, editors. Molecular Clocks and Light Signaling. 2004. pp. 171–83. [PubMed] [Google Scholar]
- 41.Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, et al. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30(3):333–49. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A–regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003;133:1605–16. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5(4):e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13(1):83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imaizumi T, Kay SA, Schroeder JI. Circadian rhythms: daily watch on metabolism. Science. 2007;318(5857):1730–31. doi: 10.1126/science.1151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322(5909):1832–35. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–84. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Suppl 1):D354–57. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keegan KP, Pradhan S, Wang J, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3(11):e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kersey P, Apweiler R. Linking publication, gene and protein data. Nat Cell Biol. 2006;8(11):1183–89. doi: 10.1038/ncb1495. [DOI] [PubMed] [Google Scholar]
- 53.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23(1):713–42. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 54.Knowles SM, Lu SX, Tobin EM. Testing time: Can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? J Biol Rhythms. 2008;23(6):463–71. doi: 10.1177/0748730408326749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Suppl 2):R271–77. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 56.Kornmann B, Schaad O, Reinke H, Saini C, Schibler U. Regulation of circadian gene expression in liver by systemic signals and hepatocyte oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:319–30. doi: 10.1101/sqb.2007.72.041. [DOI] [PubMed] [Google Scholar]
- 57.Kovac J, Husse J, Oster H. A time to fast, a time to feast: the crosstalk between metabolism and the circadian clock. Mol Cells. 2009;28(2):75–80. doi: 10.1007/s10059-009-0113-0. [DOI] [PubMed] [Google Scholar]
- 58.Kramer C, Loros JJ, Dunlap JC, Crosthwaite SK. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature. 2003;421:948–52. doi: 10.1038/nature01427. [DOI] [PubMed] [Google Scholar]
- 59.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto K, et al. Analysis and synthesis of high-amplitude cis-elements in the mammalian circadian clock. Proc Natl Acad Sci USA. 2008;105(39):14946–51. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamia KA, Storch K, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105(39):15172–77. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J, Levesque M, Benfey PN. High-throughput RNA isolation technologies. New tools for high-resolution gene expression profiling in plant systems. Plant Physiol. 2005;138(2):585–90. doi: 10.1104/pp.105.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deleted in proof
- 63.Liang K, Wang X, Li T. Robust discovery of periodically expressed genes using the laplace periodogram. BMC Bioinforma. 2009;10(1):15. doi: 10.1186/1471-2105-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Lichtenberg U, Jensen LJ, Fausboll A, Jensen TS, Bork P, Brunak S. Comparison of computational methods for the identification of cell cycle–regulated genes. Bioinformatics. 2005;21(7):1164–71. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 65.Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu S, et al. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol. 2010;10(1):41. doi: 10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luan Y, Li H. Model-based methods for identifying periodically expressed genes based on time course microarray gene expression data. Bioinformatics. 2004;20(3):332–39. doi: 10.1093/bioinformatics/btg413. [DOI] [PubMed] [Google Scholar]
- 67.Lu X, Zhang W, Qin ZS, Kwast KE, Liu JS. Statistical resynchronization and Bayesian detection of periodically expressed genes. Nucleic Acids Res. 2004;32(2):447–55. doi: 10.1093/nar/gkh205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y, Mahony S, Benos P, Rosenfeld R, Simon I, Breeden L, Bar-Joseph Z. Combined analysis reveals a core set of cycling genes. Genome Biol. 2007;8(7):R146. doi: 10.1186/gb-2007-8-7-r146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends Microbiol. 2007;15(9):381–88. doi: 10.1016/j.tim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 70.Mas P, Yanovsky MJ. Time for circadian rhythms: plants get synchronized. Curr Opin Plant Biol. 2009;12(5):574–79. doi: 10.1016/j.pbi.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 71.Matsumoto A. Genome-wide screenings for circadian clock genes in Drosophila. Sleep Biol Rhythms. 2006;4(3):248–54. [Google Scholar]
- 72.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McClung CR. Comes a time. Curr Opin Plant Biol. 2008;11(5):514–20. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 74.McClung CR. Circadian rhythms in plants: a millennial view. Physiol Plant. 2000;109(4):359–71. [Google Scholar]
- 75.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34(10):483–90. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menger GJ, Allen GC, Neuendorff N, Nahm S, Thomas TL, et al. Circadian profiling of the transcriptome in NIH/3T3 fibroblasts: comparison with rhythmic gene expression in SCN2.2 cells and the rat SCN. Physiol Genomics. 2007;29(3):280–89. doi: 10.1152/physiolgenomics.00199.2006. [DOI] [PubMed] [Google Scholar]
- 77.Michael TP, Salomé PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA. 2003;100(11):6878–83. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6(9):e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, et al. Network discovery pipeline elucidates conserved time-of-day–specific cis-regulatory modules. PLoS Genet. 2008;4(2):e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302(5647):1049–53. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 81.Michael TP, McClung CR. Phase-specific circadian clock regulatory elements in Arabidopsis. Plant Physiol. 2002;130:627–38. doi: 10.1104/pp.004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 2003;132:629–39. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49(3):481–87. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 84.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, et al. The diurnal project: diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–63. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 85.Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, et al. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13(1):60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahata Y, Yoshida M, Takano A, Soma H, Yamamoto T, et al. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol Biol. 2008;9(1):1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakamichi N, Matsushika A, Yamashino T, Mizuno T. Cell autonomous circadian waves of the APRR1/TOC1 quintet in an established cell line of Arabidopsis thaliana. Plant Cell Physiol. 2003;44(3):360–65. doi: 10.1093/pcp/pcg039. [DOI] [PubMed] [Google Scholar]
- 88.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N, Sakakibara H. Pseudo-response regulators 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nelson T, Tausta SL, Gandotra N, Liu T. LASER microdissection of plant tissue: What you see is what you get. Annu Rev Plant Biol. 2006;57(1):181–201. doi: 10.1146/annurev.arplant.56.032604.144138. [DOI] [PubMed] [Google Scholar]
- 90.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. 2008;18(2):R84–R93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 91.Nobeli I, Thornton JM. A bioinformatician’s view of the metabolome. BioEssays. 2006;28(5):534–45. doi: 10.1002/bies.20414. [DOI] [PubMed] [Google Scholar]
- 92.Nozue K, Covington M, Duek P, Lorrain S, Fankhauser C, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–61. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 93.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Neill JS, Hastings M. Circadian clocks: timely interference by microRNAs. 2007;17(17):R760–R62. doi: 10.1016/j.cub.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 95.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–64. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palsson B. Metabolic systems biology. FEBS Lett. 2009;583(24):3900–4. doi: 10.1016/j.febslet.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. 2002;109(3):307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 98.Paquet ER, Rey G, Naef F. Modeling an evolutionary conserved circadian Cis-element. PLoS Comput Biol. 2008;4(2):e38. doi: 10.1371/journal.pcbi.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petricka JJ, Benfey PN. Root layers: complex regulation of developmental patterning. Curr Opin Genet Dev. 2008;18(4):354–61. doi: 10.1016/j.gde.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Ann Rev Physiol. 1993;55(1):17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 101.Priest HD, Filichkin SA, Mockler TC. Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol. 2009;12:643–49. doi: 10.1016/j.pbi.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 102.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323(5920):1481–85. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pruneda-Paz JL, Kay SA. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010;15(5):259–65. doi: 10.1016/j.tplants.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ptitsyn AA, Gimble JM. Analysis of circadian pattern reveals tissue-specific alternative transcription in leptin signaling pathway. BMC Bioinforma. 2007;8(Suppl 7):S15. doi: 10.1186/1471-2105-8-S7-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ptitsyn AA, Zvonic S, Gimble JM. Digital signal processing reveals circadian baseline oscillation in majority of mammalian genes. PLoS Comput Biol. 2007;3(6):e120. doi: 10.1371/journal.pcbi.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ptitsyn A. Comprehensive analysis of circadian periodic pattern in plant transcriptome. BMC Bioinforma. 2008;9(Suppl 9):S18. doi: 10.1186/1471-2105-9-S9-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ptitsyn A, Zvonic S, Gimble J. Permutation test for periodicity in short time series data. BMC Bioinforma. 2006;7(Suppl 2):S10. doi: 10.1186/1471-2105-7-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 109.Robertson FC, Skeffington AW, Gardner MJ, Webb AAR. Interactions between circadian and hormonal signaling in plants. Plant Mol Biol. 2008;69(4):419–27. doi: 10.1007/s11103-008-9407-4. [DOI] [PubMed] [Google Scholar]
- 110.Roenneberg T, Chua EJ, Bernardo R, Mendoza E. Modeling biological rhythms. 2008;18(17):R826–35. doi: 10.1016/j.cub.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 111.Rutila JE, Suri V, Le M, So W, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93(5):805–14. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 112.Sandrelli F, Costa R, Kyriacou CP, Rosato E. Comparative analysis of circadian clock genes in insects. Insect Mol Biol. 2008;17(5):447–63. doi: 10.1111/j.1365-2583.2008.00832.x. [DOI] [PubMed] [Google Scholar]
- 113.Sato TK, Panda S, Kay SA, Hogenesch JB. DNA arrays: applications and implications for circadian biology. J Biol Rhythms. 2003;18(2):96–105. doi: 10.1177/0748730403252245. [DOI] [PubMed] [Google Scholar]
- 114.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, et al. Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 116.Straume M. DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. In: Brand L, Johnson ML, editors. Methods in Enzymology. Vol. 383: Numerical Computer Methods. Pt D. New York: Academic; 2004. pp. 149–66. [DOI] [PubMed] [Google Scholar]
- 117.Sulzman F, Ellman D, Fuller C, Moore-Ede M, Wassmer G. Neurospora circadian rhythms in space: a reexamination of the endogenous-exogenous question. Science. 1984;225(4658):232–34. doi: 10.1126/science.11540800. [DOI] [PubMed] [Google Scholar]
- 118.Tai YC, Speed TP. On gene ranking using replicated microarray time course data. Biometrics. 2009;65(1):40–51. doi: 10.1111/j.1541-0420.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- 119.Thain SC, Hall A, Millar AJ. Functional independence of circadian clocks that regulate plant gene expression. 2000;10(16):951–56. doi: 10.1016/s0960-9822(00)00630-8. [DOI] [PubMed] [Google Scholar]
- 120.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 121.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277(16):14048–52. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 122.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10(10):1154–63. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 123.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106(50):21453–58. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Y, Zhang X, Chen L. A network biology study on circadian rhythm by integrating various ’omics data. OMICS: A J Integr Biol. 2009;13(4):313–24. doi: 10.1089/omi.2009.0040. [DOI] [PubMed] [Google Scholar]
- 125.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10(5):107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Welsh DK, Kay SA. Bioluminescence imaging in living organisms. Curr Opin Biotechnol. 2005;16(1):73–78. doi: 10.1016/j.copbio.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 127.Wichert S, Fokianos K, Strimmer K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics. 2004;20(1):5–20. doi: 10.1093/bioinformatics/btg364. [DOI] [PubMed] [Google Scholar]
- 128.Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. Genotype and time of day shape the Populus drought response. Plant J. 2009;60(4):703–15. doi: 10.1111/j.1365-313X.2009.03993.x. [DOI] [PubMed] [Google Scholar]
- 129.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–86. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 130.Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA. 1989;86(16):6201–5. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282(34):25053–66. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 133.Yakir E, Hilman D, Harir Y, Green RM. Regulation of output from the plant circadian clock. FEBS J. 2007;274(2):335–45. doi: 10.1111/j.1742-4658.2006.05616.x. [DOI] [PubMed] [Google Scholar]
- 134.Yakir E, Hilman D, Hassidim M, Green RM. CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol. 2007;145(3):925–32. doi: 10.1104/pp.107.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang M, Lee J, Padgett R, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9(1):83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4(10):e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12(9):970–81. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 138.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2(9):702–15. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 139.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119(23):4793–95. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 140.Zhao W, Serpedin E, Dougherty ER. Identifying genes involved in cyclic processes by combining gene expression analysis and prior knowledge. EURASIP J Bioinforma Syst Biol. 2009;2009:683463. doi: 10.1155/2009/683463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139(1):199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178(3):1147–55. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]