Abstract
Five major β-globin locus haplotypes have been established in individuals with sickle cell disease (SCD) from the Benin, Bantu, Senegal, Cameroon, and Arab-Indian populations. Historically, β-haplotypes were established using restriction fragment length polymorphism (RFLP) analysis across the β-locus, which consists of five functional β-like globin genes located on chromosome 11. Previous attempts to correlate these haplotypes as robust predictors of clinical phenotypes observed in SCD have not been successful. We speculate that the coverage and distribution of the RFLP sites located proximal to or within the globin genes are not sufficiently dense to accurately reflect the complexity of this region. To test our hypothesis, we performed RFLP analysis and high-density single nucleotide polymorphism (SNP) genotyping across the β-locus using DNA samples from either healthy African Americans with normal hemoglobin A (HbAA) or individuals with homozygous SS (HbSS) disease. Using the genotyping data from 88 SNPs and Haploview analysis, we generated a greater number of haplotypes than that observed with RFLP analysis alone. Furthermore, a unique pattern of long-range linkage disequilibrium between the locus control region and the β-like globin genes was observed in the HbSS group. Interestingly, we observed multiple SNPs within the HindIII restriction site located in the Gγ-globin intervening sequence II which produced the same RFLP pattern. These findings illustrated the inability of RFLP analysis to decipher the complexity of sequence variations that impacts genomic structure in this region. Our data suggest that high density SNP mapping may be required to accurately define β-haplotypes that correlate with the different clinical phenotypes observed in SCD.
Keywords: single nucleotide polymorphism, β-locus, haplotype, sickle cell disease, Haploview
Introduction
Sickle cell anemia is caused by a SNP in the 6th codon of the β-globin gene, however the disease has great phenotypic heterogeneity and many clinical sub-phenotypes have been described. This observation stimulated research to identify genetic determinants associated with disease severity such as β-haplotypes based on RFLP analysis [1-3]. Five major β-haplotypes designated as Benin, Bantu, Cameroon, Senegal, and India-Arabic based on populations with the designated patterns from different geographical areas, have been associated with SCD [4, 5]. These haplotypes were established with several RFLP sites in the genomic region stretching from the ε-globin to β-globin gene. Among them, the Senegal and India-Arabic haplotypes are generally associated with a benign clinical course in SCD, followed by the Benin haplotype. In contrast, individuals with the Bantu β-haplotype tend to have more severe SCD. These haplotype-phenotype associations are far from perfect, however. In African Americans, the Benin, Bantu and Senegal haplotypes are the major β-locus SNP patterns along with many rare haplotypes. This most likely is due to racial admixture in the African American population [6, 7].
Many attempts have been made to establish haplotype-phenotype relationships in SCD but a robust correlation has not emerged [8-12]. We postulate that the distribution and number of RFLP sites used historically to define β-haplotypes is not sufficient to define the full range of genetic variations in this region. To address this issue we performed high density SNP mapping since it is known that most SNPs do not produce changes in restriction digestion sites however, they could cause DNA structural changes. Recently researchers have begun to utilize new genomic techniques to identify SNPs in genes such as klotho, bone morphogenic protein 6, and annexin A2, whose protein products are related to protection of the cardiovascular system, bone formation, and osteonecrosis respectively in SCD patients [13]. The klotho gene encodes for a transmembrane protein which may protect the cardiovascular system through endothelium derived nitric oxide production. Bone morphogenic protein 6 belongs to transforming growth factor-β superfamily and is associated with growth of bone and cartilage. SNPs have also been identified in other genetic regions and the β-globin gene-like cluster which are associated with elevated fetal hemoglobin levels [14] in sickle cell patients.
A growing number of SNPs have been identified in the β-locus, however the data required to establish β-haplotypes in individuals with SCD is still lacking. Therefore, we believe that SNPs across the β-locus can be used to more accurately define haplotypes that may be associated with the various clinical phenotypes observed in SCD. A chip containing probes for 88 SNPs in the β-locus region was designed and used for genotyping DNA samples isolated from healthy and SCD African American subjects. Using Haploview analysis, we generated a greater number of haplotypes with the SNP-chip genotypes than with traditional RFLP analysis alone. We also demonstrated that RFLP analysis is not capable of capturing all genomic changes. Furthermore, a unique pattern of long-range linkage disequilibrium (LD) between the locus control region and the β-like globin genes was characterized in HbSS individuals using the SNP-chip approach. These results suggest that high-density SNP mapping is a better approach for defining β-haplotypes which can be used to correlate with the different clinical phenotypes observed in SCD and may have implications for basic research studies in globin gene regulation.
Materials and Methods
Subjects and DNA Isolation
Informed consent was obtained from subjects prior to drawing blood samples in accordance with guidelines of the Institutional Review Boards at the University of Texas at Dallas and collaborating institutions. All samples were obtained from African Americans as follows: eight HbAA and 10 HbSS subjects participated from the University of Texas Southwestern Medical Center (Dallas, TX); 32 samples were obtained from HbSS patients followed in the Sickle Cell Clinic at Howard University (Washington, DC). Twenty-one DNA samples from healthy HbAA individuals were purchased from Coriell Institute for Medical Research (Camden, NJ). All samples were used anonymously. Mononuclear cells were separated from whole blood cells using HISTOPAQUE-1077 separation (Sigma-Aldrich, St. Louis, MO). Genomic DNA was then extracted using Qiagen Flexigene DNA kit (Qiagen Inc., Valencia, CA). Whole-genome amplification was performed using the REPLI-g Kit (Qiagen Inc., Valencia, CA) following the manufacturer's instructions.
RFLP Analysis
Five well-established RFLP sites including the HincII site of 5′ ε-globin (5′ε-HincII), the HindIII sites at intervening sequence II of Gγ-globin (Gγ-IVSII-HindIII) and Aγ-globin (Aγ-IVSII-HindIII), HincII sites in ψβ-globin (ψβ-HincII) and the HincII site 3′ of ψβ-globin (3′ψβ-HincII) were used to define β-haplotypes by traditional methods. Genomic DNA was first amplified by polymerase chain reaction (PCR) using published primers [15, 16] followed by restriction digestion and agarose gel inspection. The presence of an A or T nucleotide at in the 6th codon of β-globin (β6) was confirmed for all DNA samples using the restriction enzyme DdeI and SNP chip analysis. The Gγ-IVSII-HindIII region was also sequenced for a subset of DNA samples (8 HbAA and 8 HbSS) to determine the correlation of RFLP digestion patterns with SNPs in this region.
β-Locus SNP-Chip
Eighty-eight SNPs from the β-locus (GenBank: U01317) were used to design a custom SNP-chip in collaboration with Asper Biotechnology (Tartu, Estonia) using arrayed primer extension (APEX) technology [17, 18]. The SNPs summarized in Table 1 were used to perform β-locus SNP genotyping. Briefly, APEX was performed on a two-dimensional array of 60-mer oligonucleotides immobilized on the chip via the 5′ end. DNA samples were PCR amplified and then hybridized to the chip. Four fluorescently labeled dideoxy nucleotide triphosphates were used for the APEX reaction. The fluorescent signal intensities of the chip was quantified by Genorama™ Genotyping Software (Asper Biotech) and genotypes established for the polymorphic site tested by each probe.
Table 1. Summary of Single Nucleotide Polymorphisms on the β-Locus SNP Chip.
| No. | ID | Pos. | SNP | No. | ID | Pos. | SNP | No. | ID | Pos. | SNP | No. | ID | Pos. | SNP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LCR | 23 | rs4910740 | 23370 | T/C | 45 | rs2855040 | 39045 | C/G | 66 | rs33944208 | 62049 | C/T | |||
| 1 | rs11826674 | 1270 | T/G | 24 | rs10160271 | 25990 | C/T | 46 | rs28440105 | 40688 | G/T | 67 | rs33941377 | 62050 | C/G |
| 2 | rs7119142 | 1580 | T/C | 25 | rs11036507 | 26351 | A/G | 47 | rs2402330 | 41022 | T/A | 68 | rs33980857 | 62107 | T/A |
| 3 | rs34272388 | 7573 | T/C | 26 | rs10160678 | 27102 | G/T | 48 | rs916111 | 41144 | A/T | 69 | rs34598529 | 62108 | A/G |
| 4 | 7579 | 7579 | C/G | 27 | rs11820733 | 29227 | C/T | 49 | rs916112 | 41209 | A/T | 70 | 62153 | 62153 | C/? |
| 5 | rs12292063 | 8254 | C/T | 28 | rs5010978 | 31343 | C/A | 50 | rs6578592 | 41347 | T/G | 71 | 62160 | 62160 | G/A |
| 6 | 8365 | 8365 | A/C | Gγ-globin | 51 | rs7924684 | 43756 | A/G | 72 | 62166 | 62166 | T/C | |||
| 7 | 8384 | 8384 | A/G | 29 | rs10128653 | 33028 | T/G | ψβ-globin | 73 | rs334 | 62206 | A/T | |||
| 8 | rs7119428 | 8580 | A/C | 30 | rs2855121 | 33198 | A/G | 52 | rs2071348 | 46334 | A/C | 74 | rs33926764 | 62211 | A/G |
| 9 | rs9736333 | 8598 | A/G | 31 | rs2855122 | 33253 | G/A | 53 | rs11036415 | 47670 | G/T | 75 | rs35799539 | 62219 | C/? |
| 10 | rs10837757 | 8616 | T/C | 32 | rs3020750 | 34154 | T/C | 54 | rs10488677 | 48982 | C/A | 76 | 62225 | 62225 | T/C |
| 11 | 9114 | 9114 | A/T | 33 | rs2860456 | 34161 | G/A | δ-globin | 77 | rs35890959 | 62247 | G/A | |||
| 12 | rs7936221 | 10463 | A/T | 34 | 34163 | 34163 | A/C | 55 | 54886 | 54886 | G/A | 78 | rs35724775 | 62284 | T/C |
| 13 | rs4638332 | 10494 | G/T | 35 | 34874 | 34874 | G/A | 56 | 55047 | 55047 | G/T | 79 | rs11549407 | 62434 | A/C |
| 14 | rs7946623 | 12346 | T/C | 36 | 35770 | 35770 | C/G | 57 | rs3752382 | 56514 | C/T | 80 | rs11549406 | 62560 | C/G |
| 15 | rs11036587 | 13372 | G/A | 37 | rs2236794 | 36224 | G/A | 58 | rs7112844 | 58260 | A/T | 81 | rs1803195 | 62567 | G/T |
| 16 | rs11036586 | 13393 | G/A | 38 | rs10768707 | 36570 | G/A | β-globin | 82 | rs10768683 | 62647 | C/G | |||
| 17 | rs11036571 | 15024 | C/A | 39 | rs2855032 | 36786 | C/T | 59 | 61144 | 61144 | G/A | 83 | rs7480526 | 62705 | G/T |
| ε-globin | 40 | rs2255519 | 36952 | C/T | 60 | rs12289247 | 61413 | G/T | 84 | rs7946748 | 62712 | C/T | |||
| 18 | rs7479652 | 17554 | G/A | 41 | rs2855126 | 37346 | G/C | 61 | rs11036364 | 61434 | T/C | 85 | rs1609812 | 63297 | C/T |
| 19 | rs3759068 | 18633 | A/G | 42 | rs2855034 | 37486 | C/T | 62 | rs10742584 | 61668 | T/C | 86 | rs12788013 | 63843 | C/G |
| 20 | rs3759069 | 18831 | C/T | Aγ-globin | 63 | rs10742583 | 61797 | T/C | 87 | rs10837631 | 64081 | A/T | |||
| 21 | rs3759071 | 19130 | T/C | 43 | rs11827654 | 37975 | C/T | 64 | 62027 | 62027 | G/A | 88 | rs10768682 | 64928 | A/G |
| 22 | rs2213165 | 20983 | C/G | 44 | rs2855039 | 38826 | A/G | 65 | 62036 | 62036 | C/T | ||||
The 88 SNPs included on the SNP-chip are listed. Nucleotide positions (Pos.) were established using the HBB record U01317 and distribution of SNPs across the β-globin locus are indicated at the beginning of each section. The βS SNP, rs334, is indicated in bold. The column labeled “SNP.” contains the wild-type/polymorphic nucleotide. dbSNP ID is used to identify a SNP in the database based on dbSNP build 127. Position numbers in the HBB record for SNPs not in the dbSNP database were used as unique identifiers. Abbreviations: LCR, locus control region.
β-Locus Haplotype Analysis
RFLP data were used to construct β-haplotypes based on the ability of the target restriction enzyme to digest (+) or not digest (-) the PCR products generated with gene-specific primers. Since RFLP genotypes are bi-allelic, these data could be used for Haploview 3.2 analysis (http://www.broad.mit.edu/haploview). This software utilizes the estimation-maximization algorithm to calculate D′, the coefficients for LD and to infer haplotypes. To perform Haploview analysis, nucleotide symbols were arbitrarily chosen to represent (+) or (-) pattern for all five RFLP sites. Once the haplotypes were inferred, nucleotide symbols were then replaced with corresponding (+) or (-) signs. By contrast, the SNP-chip genotypes were generated at the nucleotide level and the Haploview analysis was performed without this conversion.
Haploview probes genotypes to determine conformity with Hardy-Weinberg equilibrium. The SNPs with statistically significant departures from the Hardy-Weinberg equilibrium (p<0.001) or minor allele frequencies < 5% were excluded from data mining, whereas data for all five RFLP sites were included. SNPs with strong LD (D′ ≥ 0.8) were classified into haplotype block using the Four Gamete Rule, a variant of Wang's algorithm [19] and haplotype-tagging SNPs (htSNPs) were selected on a block-by-block basis to identify SNPs that carry non-redundant information about genomic structure.
Results
β-haplotypes were confirmed by RFLP Data
RFLP analysis was performed on genomic DNA samples isolated from 42 HbSS and 29 HbAA individuals for the presence (+) or absence (-) of the five restriction sites in the β-locus as shown in Fig. 1A. Four major β-haplotypes were constructed for HbSS subjects including 39.3% Benin (----+), 22.6% Bantu (-+---), and 3.6% Senegal (-+-++); the Atypical I (-----) haplotype occurred in 16.7% of subjects. The remaining 17.8% of subjects inherited rare or incomplete haplotypes. By contrast, 16 haplotypes were observed in HbAA subjects including 20.9% Atypical II (+----), 18.9% Bantu, 8.6% Benin, and 6.8% Senegal; the remaining twelve haplotypes contributed equally in 44.8% of HbAA subjects. Interestingly, the Atypical II haplotype was not observed in the HbSS group.
Fig. 1. β-haplotypes established using RFLP genotypes.
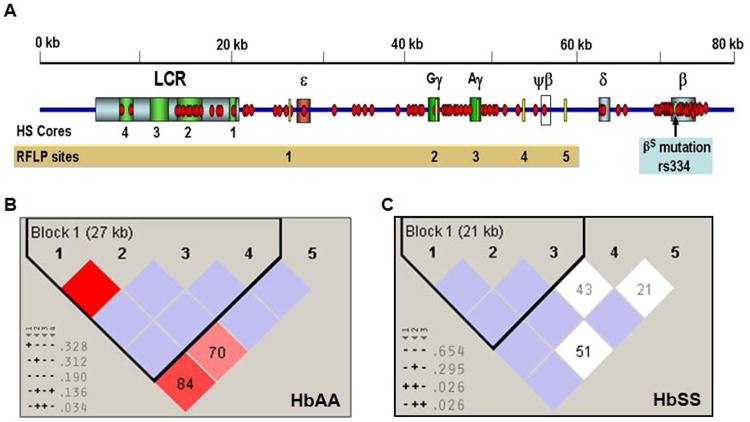
A) Schematic diagram showing the distributions of 88 SNPs (red dot) on the custom SNP-chip and five RFLP sites (yellow bar) that were analyzed. Each globin gene is indicated by a colored box and ψβ-globin is shown by a white box. The βS mutation (rs334) is shown as a green circle. Abbreviations: LCR, locus control region; HS, DNase I hypersensitive site. B) The linkage disequilibrium (LD) patterns and haplotypes established by Haploview analysis for the RFLP genotypes are shown for the HbAA subjects. Pair-wise computation was performed for the SNPs which are shown on each side of the boxes. The degree of LD is defined by value of D′ and LOD (the logarithm of the likelihood odds ratio), which is a measure of the confidence of D′ values. The red boxes indicate strong LD (LOD >2, D′ = 1), white boxes no LD (LOD < 2, D′ < 1), and pink (LOD = 2, D′ <1), and blue (LOD < 2, D′ =1) boxes indicate intermediated LD. The D′=1 unless indicated in the boxes where D′ is multiplied by 100. SNPs with strong LD were defined into haplotype blocks (black triangle) and the size of the region in LD is shown in parentheses. The inferred haplotypes and frequency observed are shown in the lower corner; htSNPs are indicated by a gray triangle beneath the RFLP number. Symbols: “+” = cut with the specific restriction enzyme used for analysis; “-“= no cut. C) The LD patterns and haplotypes for the RFLP genotypes established by Haploview analysis are shown for the HbSS group. The symbols are the same as defined in Panel B.
We next performed Haploview analysis of the RFLP data (see Materials and Methods for details). This software computes genotypic and allelic frequencies from the input data to infer haplotypes or the combination of alleles at multiple loci that are inherited together on the same chromosome. Haploview analysis identified a subset of the RFLP sites as haplotype-tagging-SNP (htSNPs) that carry non-redundant information and can define the diversity and total number of haplotypes in this region. Using RFLP data we identified 5′ε-HincII, Gγ-IVSII-HindIII, and Aγ-IVSII-HindIII as htSNPs in the HbAA and HbSS study groups (Fig. 1B and 1C) however the ψβ-HincII sites was a htSNP exclusively in the HbAA group. Four major haplotypes were inferred in the HbAA group whereas two major haplotypes were inferred in the HbSS group. By comparing haplotypes generated from the traditional and Haploview analysis, we concluded that less haplotypes were generated using Haploview however a greater number of haplotypes are consistently present in the HbAA subjects. There were also differences on the frequencies of haplotypes. When RFLP analysis was used, the five sites were treated equally, whereas Haploview analysis inferred haplotypes using non-redundant htSNPs indicating that the RFLP sites are not equally informative.
Haploview analysis also allowed us to divide the β-locus genomic region into haplotype blocks defined by SNPs in LD. Fig. 1B and 1C showed there was one haplotype block defined for both groups however for the HbAA group, the haplotype block stretched over a 27 kb region from 5′ ε-globin to ψβ-globin. By contrast, the haplotype block in the HbSS group included a 21-kb region from 5′ ε-globin to Aγ-globin. This data suggest decreased linkage between the RFLP sites in the HbSS group. Based on these observations, we postulated that the genomic structure in this region may have undergone molecular change under the pressure of natural selection.
Unique haplotype patterns are identified in the β-locus by high-density SNP mapping
We next determined whether SNP-chip genotyping with more dense coverage across the β-locus would reveal greater differences in genomic structure within the two study groups than that produced by RFLP analysis. To achieve this end, we designed a custom SNP-chip based on APEX technology (see Materials and Methods). Because the globin genes are highly homologous and there exists many repetitive sequences throughout the locus, we were only able to identify 88 SNPs that could be reliably tested by APEX technology (Fig. 1A). The same DNA samples used in the RFLP analysis were genotyped using the SNP-chip. The Gγ-IVSII-HindIII site was included on the chip to retest the SNP at position 35770 (C/G) to confirm our RFLP data.
Genotypes for DNA samples isolated from 29 HbAA and 42 HbSS African Americans were generated using the SNP-chip. Haploview analysis identified 48 SNPs with minor allele frequency <5% in the HbAA group, while 61 SNPs had minor allele frequency <5% in the HbSS groups; three additional SNPs (rs9736333, rs3759071 and rs6578593) deviated from the Hardy-Weinberg Equilibrium in the HbSS group. After these exclusions, there were 40 and 24 SNPs included in the HbAA and HbSS groups respectively for further analysis to determine LD and major haplotype patterns in the β-locus.
The first procedure completed with the genotype data was the generation of “heat maps” as shown in Fig 2A and 2B. Each column in the heat map represents individual samples, while each row represents one SNP. The genotypes for the SNPs were color coded as follows: blue representing homozygous wild type; red for homozygous mutant alleles and yellow for heterozygous alleles. Note for some SNP positions there were samples for which genotyping data was not generated (white boxes). From these maps, we concluded that genotypes alone were not as meaningful. However, when individuals with similar genotype patterns were grouped together, then the heat maps revealed different genotype patterns for the two study groups as a whole. For the HbAA group, the appearance of heterozygous genotypes is very diverse and random compared to that observed for HbSS subjects. These findings suggest a dramatic difference in the β-locus genomic structures for the two groups. This difference was not observed with the RFLP genotypes (data not shown). We concluded that the β-locus in healthy subjects had a higher frequency of recombination events that that observed in the HbSS population.
Fig. 2. Haploview-inferred haplotypes using SNP-chip genotypes.
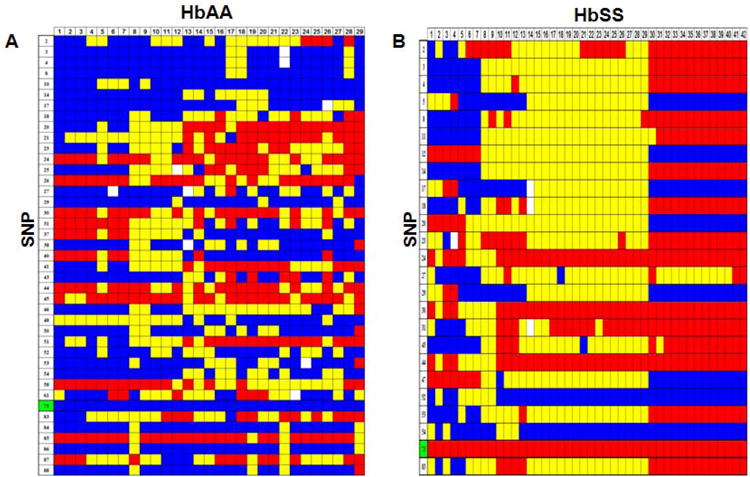
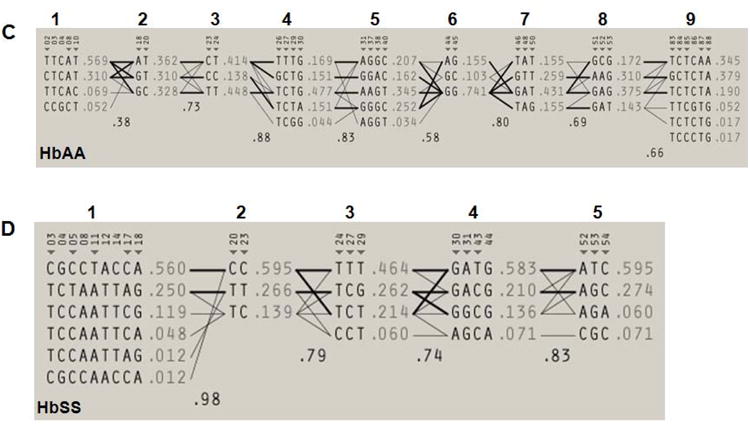
A) A genotype heat map was established for the HbAA subjects. Genotypes produced using the SNP-chip that satisfied the criteria detailed in Materials and Methods are shown. The rows represent the SNPs analyzed and the columns represent each DNA sample tested. SNPs are numbered according to that defined in Table 1; the βS SNP (rs334) is number 73 and is highlighted in green. At each SNP position, a blue box represents wild-type homozygous genotypes, a yellow box heterozygous and the red box homozygous mutant genotypes. White boxes indicate genotype data is missing for the SNP indicated. B) Genotype heat map for the HbSS subjects. The color code is the same as described in Panel A. C) Haploview software was used to infer haplotypes using SNP-chip genotype data for the HbAA subjects. The numbers in the gray shaded area represent the SNPs listed in Table 1. The inferred haplotypes in each haplotype block and linkage between blocks are shown. Nine haplotype blocks were defined (numbers above the gray shaded area). The lines between haplotype blocks indicate the linkage frequency with a thick line for > 10% and a thin line for a frequency from 1-10%. Haplotype-tagging SNPs (htSNPs) are indicated underneath the SNP numbers by a triangle (▼). Haplotype frequency within each haplotype block is shown next to each haplotype. The number between two blocks is the Hedrick's multi-allelic D′; D′ >= 0.8 is indicative of strong LD between the blocks. D) Inferred haplotypes and linkages between five haplotype blocks (numbers above the gray shaded area) for the HbSS subjects. The methods and symbols are the same as described in Panel C.
We next performed a detailed analysis of the SNP-chip data using Haploview software. A pair-wise computation between SNPs alleles and genotype frequencies was completed to determine the degree of LD across the β-locus to identify htSNPs, and to define the haplotype blocks and inferred β-haplotypes. We first constructed the major haplotypes across the entire β-locus for comparison with RFLP analysis by calculating the linkage between haplotype blocks or multi-allelic D′ (Fig. 2C and 2D). Nine haplotype blocks were defined for the HbAA subjects that produced 42 haplotypes including six at a frequency from 3.4-5.1% and 36 minor haplotypes accounting for 76.4% of the genetic variation in this group. A similar analysis of the HbSS subjects showed five haplotype blocks as shown in Fig. 2D. By contrast, 21 haplotypes were defined in the β-locus including four major haplotypes at a frequency of 40.4%, 13.1%, 10.7% and 5.9% account for 70.1% of genomic variation. Finally, we identify 28 htSNPs for the HbAA and 16 htSNPs for the HbSS subjects (Fig. 2C and 2D, indicated by inverted triangles) that are sufficient to define the major haplotypes in each group. When these data were compared to the RFLP analysis, there was no correlation between haplotypes generated by the two methods. The SNP-chip results clearly demonstrated that a greater number of haplotypes and differences in the genomic structure of the β-locus exist between the HbAA and HbSS study groups which were not identified by RFLP analysis.
Genomic complexity in the Gγ-IVSII-HindIII site demonstrated by SNP-chip analysis
For SNP-chip analysis, we targeted the C to G (C/G) SNP at nucleotide 35,770 which destroys the HindIII site (AAGCTT) in the Gγ-globin intervening sequence II region. However, the SNP-chip genotypes generated were not in agreement with the RFLP results for the Gγ-IVSII-HindIII digestion pattern. We observed a discrepancy in that the RFLP result was (+- or --) in the presence of a CC genotype obtained from SNP-chip results. Since the HindIII site has been commonly used in RFLP studies to define β-haplotypes, we believed that direct sequencing was necessary to determine the basis of the inconsistent results. Sixteen samples from 8 HbAA and 8 HbSS subjects were sequenced for this region. The results are summarized in Fig. 3. At position 35,770 all samples had a homozygous CC genotype. Interestingly, two additional SNPs at positions 35,769 and 35,772 were identified that correlated with the RFLP digestion patterns. The SNP at 35,769 has not been reported, but the SNP at 35772 was registered in the dbSNP database (rs2070972; G/T).
Fig. 3. Correlation of HindIII digestion with direct sequencing of the Gγ-IVSII region.
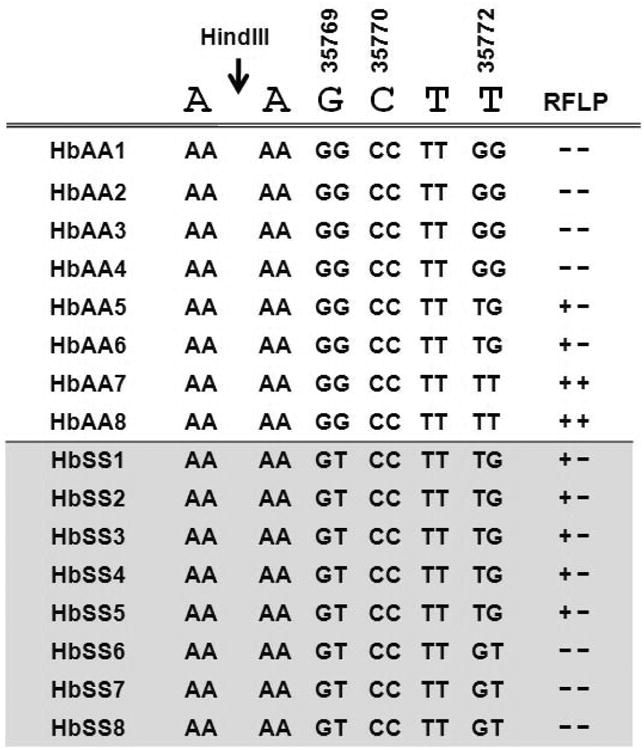
The PCR products generated for RFLP analysis was used for direct sequencing analysis. The HindIII restriction site AAGCTT and position of three nucleotides according to the HBB record (U01317) are shown at the top of the figure. Each row represents data for individual DNA samples; each nucleotide position and genotype is shown for both alleles based on the sequencing results. For RFLP patterns the positive symbol (+) refers to HindIII digestion, while the negative symbol (-) no cut.
The sequencing data showed that HbAA subjects had GG genotypes at 35,769 whereas HbSS subjects were heterozygous GT. For the SNP at 35,772 there was greater variability in the HbAA group however the HbSS subjects were all heterozygous GT. By comparison with the RFLP digestion patterns and the predicted effect on HindIII digestion, for HbAA subjects the 35,772 SNP was predictive of the RFLP result (-- or +-) produced by a variable sequence of AAGCT[T/G]. For the HbSS samples, two RFLP patterns were observed which correlated with heterozygosity at position 35,769 or 35,772 (AA[G/T]CT[T/G]). The sequencing and digestion data combined showed the association of AATCTG, and AATCTT DNA sequence with the HbSS subjects. It remains to be addressed whether these variations can be correlated with the various clinical phenotypes observed in SCD. Mutations in the intervening sequences in other β-like globin genes have been associated with various hemoglobinopathies [20, 21] and γ-gene expression in K562 cells [22]. Our results demonstrated that HindIII digestion alone could not accurately distinguish the underlying genomic structural differences at the nucleotide level in the Gγ-globin gene.
High-density β-locus genotyping reveals unique htSNPs in HbSS subjects
In our next analysis, we compared the frequencies of the associated alleles for 22 SNPs from both groups. As shown in Table 2, there were only two SNPs, rs6578593 and rs7480526, which occurred at similar frequencies in both groups. Thirteen SNPs including six in the hypersensitive sites, four in ε-globin, two in γ-globin (rs10128653 in Gγ- and rs11827654 in Aγ-globin) and one in ψβ-globin showed higher allele frequency in the HbSS group. Seven SNPs displayed the opposite patterns with higher frequency in HbAA subjects. With the vast number of SNPs identified, it is beneficial to identify htSNPs that can be used to capture the major haplotype variants without analyzing all SNPs in a given region. Interestingly we discovered three SNPs that were unique to either the HbSS or HbAA group indicating different patterns of inheritance for the two groups. Identifying specific htSNPs will be useful for population studies or characterization of clinical sub-groups in SCD patients.
Table 2. SNP Frequency and htSNPs in the HbAA and HbSS groups.
| Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | ID | Position | SNP | Associated Allele | HbAA (%) | HbSS (%) | HbAA htSNP | HbSS htSNP | β-Locus Region |
| 2 | rs7119142 | 1580 | T/C | C | 36.2 | 76.2 | √ | HS4 | |
| 3 | rs34272388 | 7573 | T/C | C | 5.4 | 57.1 | √ | HS2 | |
| 4 | 7579 | 7579 | C/G | G | 5.4 | 57.1 | |||
| 8 | rs7119428 | 8580 | A/C | C | 5.2 | 57.1 | √ | ||
| 14 | rs7946623 | 12346 | T/C | C | 12.1 | 57.1 | |||
| 17 | rs11036571 | 15024 | C/A | A | 8.9 | 25.6 | √ | HS1 | |
| 18 | rs7479652 | 17554 | G/A | A | 36.2 | 62.2 | √ | √ | ε-globin |
| 20 | rs3759069 | 18831 | C/T | C | 32.8 | 59.5 | √ | √ | |
| 21 | rs3759071 | 19130 | T/C | T | 31 | 46.4 | |||
| 23 | rs4910740 | 23370 | T/C | T | 44.8 | 25.6 | √ | √ | |
| 24 | rs10160271 | 25990 | C/T | C | 13.8 | 6 | √ | √ | |
| 27 | rs11820733 | 29227 | C/T | T | 16.7 | 46.4 | √ | √ | |
| 29 | rs10128653 | 33028 | T/G | G | 5.2 | 26.2 | √ | √ | Gγ-globin |
| 30 | rs2855121 | 33198 | A/G | A | 15.5 | 7.1 | √ | √ | |
| 31 | rs2855122 | 33253 | G/A | G | 41.4 | 20.7 | √ | √ | |
| 43 | rs11827654 | 37975 | C/T | T | 24.1 | 58.3 | √ | Aγ-globin | |
| 44 | rs2855039 | 38826 | A/G | A | 15.5 | 7.1 | √ | ||
| 46 | *rs6578593 | 40688 | G/T | T | 31 | 36.9 | √ | ||
| 52 | rs2071348 | 46334 | A/C | C | 17.2 | 7.1 | √ | √ | ψβ-globin |
| 53 | rs11036415 | 47670 | G/T | T | 14.3 | 59.5 | √ | √ | |
| 54 | rs10488677 | 48982 | C/A | A | 15.5 | 6 | √ | ||
| 73 | rs334 | 62206 | A/T | A | 100 | 0 | β-globin | ||
| 83 | *rs7480526 | 62705 | G/T | G | 37.9 | 33.3 | √ | ||
The allele frequency for 23SNPs including some htSNPs (√) for the HbAA and HbSS groups are summarized. SNP selection criteria are described in the text. The βS SNP (rs334) is indicated in bold letters. The column labeled “Variation” refers to wild-type/polymorphic nucleotide. The frequency was calculated for the associated allele. The “*“ indicates SNPs showing similar allele frequencies for both group. The nucleotide positions were located using the HBB record U01317; SNP distribution across the β-globin locus is indicated. Abbreviations: HS, hypersensitive site.
In agreement with the diverse genotypes and high number of (β-haplotypes in the HbAA group, nine haplotype blocks were generated and weak LD was observed in this group demonstrated by white boxes across the (β-locus (Fig. 4A). However, we observed five haplotype blocks in the HbSS group as shown in Fig. 4B. In contrast to the weak LD in the HbAA group, very strong LD was exhibited in the HbSS group between SNPs in the locus control region and several downstream SNPs located in ε-globin (rs7479652, rs3759069, rs4910740 and rs11820733), Gγ-globin (rs10128653 and rs2855122), Aγ-globin (rs11827654, rs2402330), ψβ-globin (rs2071348), and the β-globin gene (rs7480526) which is illustrated by the stretch of red boxes across the (β-locus region. The locus control region is required for developmental stage-specific globin gene expression through interaction with downstream promoter regions [4, 23-25]. Of note, rs2855122 was recently shown to be involved in drug-mediated Gγ-globin fetal hemoglobin induction by butyrate [26]. Thus, strong LD between these SNPs may affect the genomic structure of this region, which may contribute to the regulation of γ-globin gene transcription.
Fig. 4. Linkage Disequilibrium and haplotype block patterns across the β-locus.
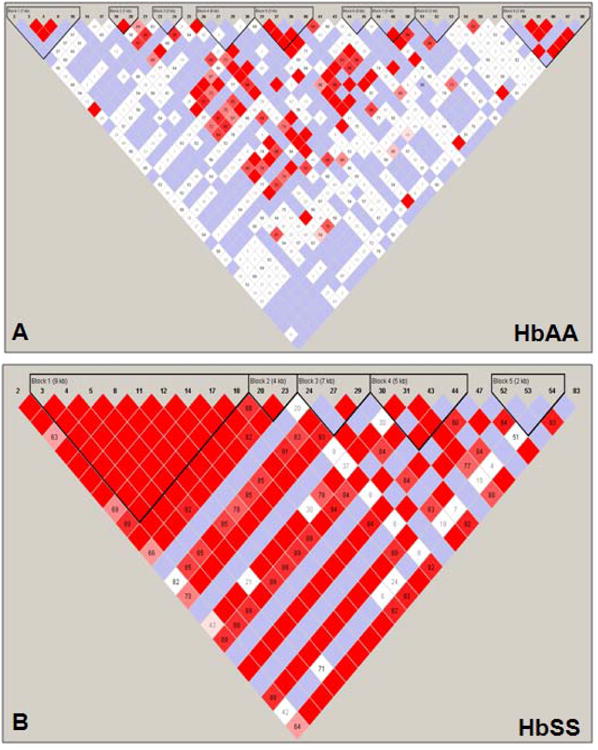
A) Shown are the LD patterns and haplotype blocks established by Haploview analysis with the SNP-chip data for the HbAA group. The numbers on top represent the SNPs used for the analysis. Haplotype blocks are shown by inverted triangles. Within each haplotype block, the distance of the region covered is indicated in kilo-bases (kb). The color code is the same as defined in the legend for Fig. 1B. B) The LD patterns and haplotype blocks established by the Haploview analysis with the SNP-chip genotype data for the HbSS group are shown. The symbols are the same as defined in Panel A
Haploview analysis of SNP-chip data also allowed us to establish the LD patterns across the entire (β-locus for both study groups. In Fig. 2C and 2D, the values for multi-allelic D′ are shown between haplotype blocks; LD between blocks with a D′≥0.8 is indicative of strong linkage. We observed weak linkage across the β-locus of HbAA subjects (mean multi-allelic D′=0.69) in contrast to strong linkage in the HbSS subjects (D′=0.84). This suggests that the presence of βS mutation and natural selection pressure had a long-range effect on the locus control region and downstream β-like globin genes.
Discussion
Historically, RFLP analysis has been used as the preferred approach to establish β-haplotypes. Our results showed that three of the most commonly used RFLP sites were identified as htSNPs in the HbSS group suggesting that the five RFLP sites are not equally informative. Furthermore, these sites are located in region of moderate LD in the HbSS group. We also demonstrated that three SNPs in the Gγ-IVSII-HindIII site produced variable RFLP readouts based on genotypes which could affect the function of this intronic region and Gγ-globin transcription rates. Furthermore, a greater number of haplotypes were inferred from the SNP-chip data than with RFLP analysis, which could be used to define various sub-phenotypes. It has been suggested that genotyping a few widely spaced sites may result in missing important genomic variations [27]. Thus, our data demonstrate that the RFLP approach is not sufficient to reveal the genomic complexity of the β-locus and that high-density SNP mapping using new technology is preferred to discover a wide range of genetic determinants.
Despite the fact that HbSS is caused by a single A/T mutation in the sixth codon of the β-globin gene there are many different clinical phenotypes associated with this monogenic disorder. Patients with elevated fetal hemoglobin levels generally have less severe disease. Research efforts to identify epigenetic factors that alter γ-gene transcription synthesis have shown that SNPs in the β-locus or other distant genomic regions contribute to γ-globin regulation. Mutations including the -158 XmnI (C/T) SNP in the Gγ-globin gene promoter [28, 29] and distant SNPs in the 6q22.3-q23.2, 8q11-q12 and Xp22.2-p22.3 regions have been associated with elevated fetal hemoglobin levels [30-32]. It is believed that other genetic markers in the β-locus also exerted a significant effect on fetal hemoglobin synthesis [33].
To determine the correlation of SNPs in the β-locus with phenotypic outcomes in SCD, we constructed a SNP chip containing 88 SNPs across this region to study the LD of SNPs in the two study groups. SNP genotyping revealed differences between the healthy HbAA and HbSS subjects which were not observed by RFLP analysis. The healthy group showed diverse haplotype patterns with moderate LD over a 35 kb region but weak LD over the entire β-locus. By contrast, the HbSS group showed strong LD between SNPs stretching over 60 kb. A recent publication also described strong long-range linkage across the βS mutation in African and Afro-Caribbean HbSS populations [34].
Thus, our observations demonstrate that the traditional RFLP approach does not accurately reflect the complexity of the β-locus. Our study is the first detailed genomic analysis of this region in African Americans using modern genomics techniques. SCD is increasingly viewed as a disease whose phenotype is affected by an array of modifier genes. The clinical manifestations of SCD are most likely a result of variations on the β-locus in addition to other genomic regions. Therefore, high-density β-locus and genome-wide SNP mapping with a greater sample size should impact efforts to ascertain genotype-phenotype relationship in SCD.
Acknowledgments
This work was supported by training fund from the National Heart, Lung, and Blood Institute to Li Liu, PhD through the Southwestern Comprehensive Sickle Cell Center (U54 HL 70588).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wainscoat JS, Hill AV, Boyce AL, et al. Evolutionary relationships of human populations from an analysis of nuclear DNA polymorphisms. Nature. 1986;319:491–3. doi: 10.1038/319491a0. [DOI] [PubMed] [Google Scholar]
- 2.Kan YW, Lee KY, Furbetta M, et al. Polymorphism of DNA sequence in the beta-globin gene region. Application to prenatal diagnosis of beta 0 thalassemia in Sardinia. N Engl J Med. 1980;302:185–8. doi: 10.1056/NEJM198001243020401. [DOI] [PubMed] [Google Scholar]
- 3.Feldenzer J, Mears JG, Burns AL, et al. Heterogeneity of DNA fragments associated with the sickle-globin gene. J Clin Invest. 1979;64:751–5. doi: 10.1172/JCI109519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oner C, Dimovski AJ, Altay C, et al. Sequence variations in the 5′ hypersensitive site-2 of the locus control region of beta S chromosomes are associated with different levels of fetal globin in hemoglobin S homozygotes. Blood. 1992;79:813–9. [PubMed] [Google Scholar]
- 5.Nagel RL, Fleming AF. Genetic epidemiology of the beta s gene. Baillieres Clin Haematol. 1992;5:331–65. doi: 10.1016/s0950-3536(11)80023-5. [DOI] [PubMed] [Google Scholar]
- 6.Antonarakis SE, Boehm CD, Serjeant GR, et al. Origin of the beta S-globin gene in blacks: the contribution of recurrent mutation or gene conversion or both. Proc Natl Acad Sci U S A. 1984;81:853–6. doi: 10.1073/pnas.81.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powars D, Hiti A. Sickle cell anemia. Beta s gene cluster haplotypes as genetic markers for severe disease expression. Am J Dis Child. 1993;147:1197–202. doi: 10.1001/archpedi.1993.02160350071011. [DOI] [PubMed] [Google Scholar]
- 8.Kutlar A. Sickle cell disease: a multigenic perspective of a single gene disorder. Hematology. 2005;10(Suppl 1):92–9. doi: 10.1080/10245330512331390069. [DOI] [PubMed] [Google Scholar]
- 9.Steinberg MH. Predicting clinical severity in sickle cell anaemia. Br J Haematol. 2005;129:465–81. doi: 10.1111/j.1365-2141.2005.05411.x. [DOI] [PubMed] [Google Scholar]
- 10.Inati A, Taher A, Bou Alawi W, et al. Beta-globin gene cluster haplotypes and HbF levels are not the only modulators of sickle cell disease in Lebanon. Eur J Haematol. 2003;70:79–83. doi: 10.1034/j.1600-0609.2003.00016.x. [DOI] [PubMed] [Google Scholar]
- 11.el-Hazmi MA, Warsy AS, Bashir N, et al. Haplotypes of the beta-globin gene as prognostic factors in sickle-cell disease. East Mediterr Health J. 1999;5:1154–8. [PubMed] [Google Scholar]
- 12.Rieder RF, Safaya S, Gillette P, et al. Effect of beta-globin gene cluster haplotype on the hematological and clinical features of sickle cell anemia. Am J Hematol. 1991;36:184–9. doi: 10.1002/ajh.2830360305. [DOI] [PubMed] [Google Scholar]
- 13.Baldwin C, Nolan VG, Wyszynski DF, et al. Association of klotho, bone morphogenic protein 6, and annexin A2 polymorphisms with sickle cell osteonecrosis. Blood. 2005;106:372–5. doi: 10.1182/blood-2005-02-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastiani P, Wang L, Nolan VG, et al. Fetal hemoglobin in sickle cell anemia: Bayesian modeling of genetic associations. Am J Hematol. 2008;83:189–95. doi: 10.1002/ajh.21048. [DOI] [PubMed] [Google Scholar]
- 15.Papachatzopoulou A, Menounos PG, Kolonelou C, et al. Mutation screening in the human epsilon-globin gene using single-strand conformation polymorphism analysis. Am J Hematol. 2006;81:136–8. doi: 10.1002/ajh.20580. [DOI] [PubMed] [Google Scholar]
- 16.Sutton M, Bouhassira EE, Nagel RL. Polymerase chain reaction amplification applied to the determination of beta-like globin gene cluster haplotypes. Am J Hematol. 1989;32:66–9. doi: 10.1002/ajh.2830320113. [DOI] [PubMed] [Google Scholar]
- 17.Schrijver I, Kulm M, Gardner PI, et al. Comprehensive arrayed primer extension array for the detection of 59 sequence variants in 15 conditions prevalent among the (Ashkenazi) Jewish population. J Mol Diagn. 2007;9:228–36. doi: 10.2353/jmoldx.2007.060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kringen P, Bergamaschi A, Due EU, et al. Evaluation of arrayed primer extension for TP53 mutation detection in breast and ovarian carcinomas. Biotechniques. 2005;39:755–61. doi: 10.2144/000112000. [DOI] [PubMed] [Google Scholar]
- 19.Wang N, Akey JM, Zhang K, et al. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71:1227–34. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agouti I, Bennani M, Ahmed A, et al. Thalassemia intermedia due to a novel mutation in the second intervening sequence of the beta-globin gene. Hemoglobin. 2007;31:433–8. doi: 10.1080/03630260701613210. [DOI] [PubMed] [Google Scholar]
- 21.Eram SM, Azimifar B, Abolghasemi H, et al. The IVS-II-1 (G→a) beta0-thalassemia mutation in cis with HbA2-Troodos [delta116(G18) Arg→Cys (CGC→TGC)] causes a complex prenatal diagnosis in an Iranian family. Hemoglobin. 2005;29:289–92. doi: 10.1080/03630260500310828. [DOI] [PubMed] [Google Scholar]
- 22.Donovan-Peluso M, Acuto S, Swanson M, et al. Expression of human gamma-globin genes in human erythroleukemia (K562) cells. J Biol Chem. 1987;262:17051–7. [PubMed] [Google Scholar]
- 23.Lu ZH, Steinberg MH. Fetal hemoglobin in sickle cell anemia: relation to regulatory sequences cis to the beta-globin gene. Multicenter Study of Hydroxyurea. Blood. 1996;87:1604–11. [PubMed] [Google Scholar]
- 24.Ofori-Acquah SF, Lalloz MR, Layton DM. Localisation of cis regulatory elements at the beta-globin locus: analysis of hybrid haplotype chromosomes. Biochem Biophys Res Commun. 1999;254:181–7. doi: 10.1006/bbrc.1998.9901. [DOI] [PubMed] [Google Scholar]
- 25.King DC, Taylor J, Zhang Y, et al. Finding cis-regulatory elements using comparative genomics: some lessons from ENCODE data. Genome Res. 2007;17:775–86. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sangerman J, Lee MS, Yao X, et al. Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2. Blood. 2006;108:3590–9. doi: 10.1182/blood-2006-01-023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss KM, Clark AG. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 2002;18:19–24. doi: 10.1016/s0168-9525(01)02550-1. [DOI] [PubMed] [Google Scholar]
- 28.Miller BA, Salameh M, Ahmed M, et al. Analysis of hemoglobin F production in Saudi Arabian families with sickle cell anemia. Blood. 1987;70:716–20. [PubMed] [Google Scholar]
- 29.Miller BA, Olivieri N, Salameh M, et al. Molecular analysis of the high-hemoglobin-F phenotype in Saudi Arabian sickle cell anemia. N Engl J Med. 1987;316:244–50. doi: 10.1056/NEJM198701293160504. [DOI] [PubMed] [Google Scholar]
- 30.Ma Q, Wyszynski DF, Farrell JJ, et al. Fetal hemoglobin in sickle cell anemia: genetic determinants of response to hydroxyurea. Pharmacogenomics J. 2007;7:386–94. doi: 10.1038/sj.tpj.6500433. [DOI] [PubMed] [Google Scholar]
- 31.Garner C, Silver N, Best S, et al. Quantitative trait locus on chromosome 8q influences the switch from fetal to adult hemoglobin. Blood. 2004;104:2184–6. doi: 10.1182/blood-2004-02-0527. [DOI] [PubMed] [Google Scholar]
- 32.Dover GJ, Smith KD, Chang YC, et al. Fetal hemoglobin levels in sickle cell disease and normal individuals are partially controlled by an X-linked gene located at Xp22.2. Blood. 1992;80:816–24. [PubMed] [Google Scholar]
- 33.Zertal-Zidani S, Ducrocq R, Sahbatou M, et al. Foetal haemoglobin in normal healthy adults: relationship with polymorphic sequences cis to the beta globin gene. Eur J Hum Genet. 2002;10:320–6. doi: 10.1038/sj.ejhg.5200809. [DOI] [PubMed] [Google Scholar]
- 34.Hanchard N, Elzein A, Trafford C, et al. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52. doi: 10.1186/1471-2156-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]


