Abstract
A critical event in the life cycle of a virus is its initial attachment to host cells. This involves recognition by the viruses of specific receptors on the cell surface, including glycans. Viruses typically exhibit strain-dependent variations in recognizing specific glycan receptors, a feature that contributes significantly to cell tropism, host specificity, host adaptation and interspecies transmission. Examples include influenza viruses, noroviruses, rotaviruses, and parvoviruses. Both rotaviruses and noroviruses are well known gastroenteric pathogens that are of significant global health concern. While rotaviruses, in the family Reoviridae, are the major causative agents of life-threatening diarrhea in children, noroviruses, which belong to Caliciviridae family, cause epidemic and sporadic cases of acute gastroenteritis across all age groups. Both exhibit enormous genotypic and serotypic diversity. Consistent with this diversity each exhibits strain-dependent variations in the types of glycans they recognize for cell attachment. This chapter reviews current status of the structural biology of such strain-dependent glycan specificities in these two families of viruses.
Introduction
Initial attachment of a virus particle to the host cell membrane represents a critical stage in the viral infectious cycle. Such an attachment is often mediated by the interactions of the virus surface protein with specific glycan components of cell-surface glycoproteins, glycolipids, or proteoglycans[1]. Viruses employ a wide variety of glycans for initial cell attachment ranging from charged glycan moieties such as sialic acid (Sia), recognized by influenza viruses [2], orthoreoviruses [3] and rotaviruses [4], heparan sulfate by parvoviruses [5–7] and herpes viruses [8], to neutral glycans such as histo-blood group antigens (HBGAs) by noroviruses [9–11] and rotaviruses [12, 13]. Within a particular virus species, significant variations in recognizing specific glycans resulting from genotypic alterations is a common feature that provides an underlying basis for cell tropism, host specificity, host adaptation, interspecies transmission and pathogenesis [14, 15]. Together with classical methods such as agglutination and cell-blockade assays, and volunteer studies, recent advances in glycan microarray screening [16–18] have been most useful in systematically profiling cell attachment glycans for viruses. Concurrently, crystallographic studies of viral cell attachment proteins in complex with glycans have provided valuable structural insight into how genotypic variations alter glycan specificity. Here, we review recent advances in our understanding of the structural basis of strain-dependent glycan specificity in rotaviruses and noroviruses, which represent two important gastrointestinal pathogens of global health concern.
Strain-dependent glycan specificity is rotaviruses
Rotavirus (RV), a multi-segmented dsRNA virus in the family Reoviridae, is the major cause of infantile gastroenteritis leading to ~450,000 deaths annually worldwide [19]. These viruses exhibit enormous genetic and strain diversity. In addition to point mutations and gene rearrangements, genetic reassortment between co-circulating strains, similar to influenza viruses, contributes to the expanding diversity of RVs [20, 21]. Current evidence indicates that many of the human RV (HRV) strains originated from animal reservoirs through reassortment and inter-species transmission [21, 22]. RVs have a complex icosahedral architecture with three concentric capsid layers encapsulating the eleven genomic dsRNA segments [23–25] (Fig. 1A). Based on neutralization specificity of the outer layer proteins (Glycoprotein VP7 and Protease-sensitive VP4), RVs are classified into G (VP7) and P (VP4) genotypes following a binary nomenclature system similar to influenza viruses [26]. Productive infection requires proteolytic-priming of the virus that results in the cleavage of VP4 into VP8* and VP5* [27, 28] (Fig. 1A inset). RV cell entry is a multistep process involving cellular glycans in the initial cell attachment step and multiple receptors during post-attachment steps [29, 30]. VP8* of the VP4 spike mediates interactions with cellular glycans, whereas VP5* is implicated in interactions with downstream receptors.
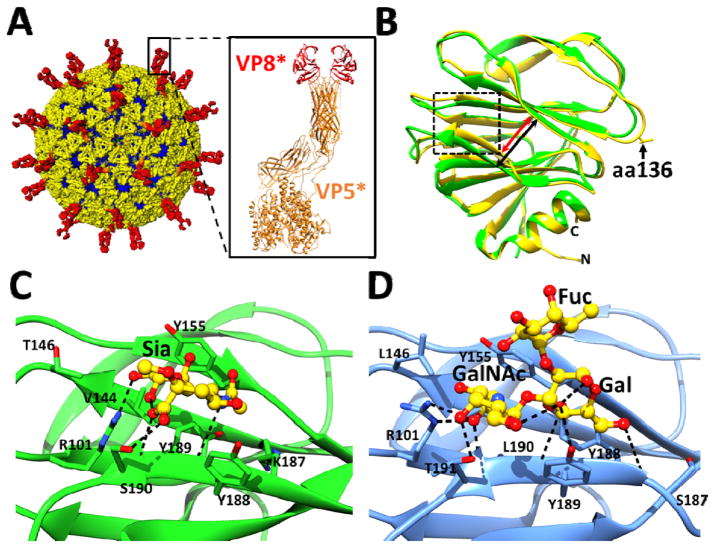
Figure 1.
Rotavirus cryo-EM structure and the crystal structures of cell attachment protein VP8*. (A) The triple layered particle (TLP) is colored with VP4 spikes in red, the VP7 layer in yellow, and the VP6 layer in blue. The cartoon representation of a VP4 spike (PDB ID: 3IYU) is shown with the VP8* domain colored in red and the VP5* domain in orange. (B) Structural overlay of sialidase insensitive P[14] VP8* structure (blue, PDB ID: 4DRV) with VP8* of sialidase-insensitive HR strain Wa (green, PDB ID: 2DWR). The width of the cleft between two twisted β-sheets in the P[14] VP8* is narrower (red arrow) than in the Wa VP8* structure (black arrow). Amino acid 136 is shown in stick and indicated by a black arrow. (C) Interaction between the P[3] VP8* of sialidase sensitive animal strain RRV (PDB ID: 1KQR) and Sia. The P[3] VP8* structure in presented in orange ribbon with the amino acid residues interacting with Sia shown as sticks, and bound Sia is shown as green sticks with oxygen and nitrogen atoms in red and blue, respectively. (D) Interactions between the P[14] VP8* of sialidase insensitive human strain Hal1166 with A-type HBGA. The P[14] VP8* structure in presented in blue ribbon with the amino acid residues interacting with A-HBGA shown as yellow sticks with oxygen and nitrogen atoms colored as in (C). Network of hydrogen bond interactions (dashed lines) are shown.
The cell attachment protein VP8* exhibits a galectin-like fold with two twisted β-sheets separated by a shallow cleft (Fig. 1B). Although structurally well conserved, sequence wise VP8* is the least conserved among RV structural proteins giving rise to a phylogeny consisting of at least 35 P genotypes. In animal RV (ARV) strains, for which infectivity is sensitive to sialidase treatment of cells, sialoglycans with terminal Sia are implicated in the initial attachment through its interactions with VP8*[4, 31, 32]. In contrast, the majority of HRV strains are insensitive to sialidase treatment[33]. Using cell-based assays and NMR, it was shown that one of the HRV strains (Wa) belonging to P[8] genotype, binds to gangliosides such as GM1 using internal Sia[34]. These studies led to the suggestion that while the sialidase-sensitive (s-s) strains recognize glycans with terminal Sia such as GD1a, the sialidase-insensitive (s-i) human strains bind to gangliosides such as GM1 with internal Sia, and that Sia is the key determinant for host cell recognition in all rotaviruses. Recent studies, however, called into question such a general paradigm for rotavirus cell attachment.
Crystal structures of VP8* of s-s ARV in complex with Sia (Fig. 1C) have shown that Sia binds near the cleft region. In the VP8* structures of some s-i HRV strains, particularly those that are globally dominant, such as DS1 and Wa belonging to P[4] and P[8] genotypes, respectively, this cleft is noticeably wider. In addition, there are significant differences in the amino acids that line the cleft (Fig. 1B) Although the crystal structure of a human VP8* with a wider cleft in complex with a glycan has not been yet reported, NMR and computer modeling studies propose that a wider cleft allows binding of gangliosides with internal Sia [34].
The recently determined structure of VP8* of an s-i HRV strain (HAL1166), belonging to G8P[14] genotype [33, 35], showed a narrow cleft similar to that observed in the VP8* of the s-s ARV strains. The P[14] strains, with their origins in even-toed ungulates [36], are being increasingly documented in human infections by global rotavirus surveillance [36–38]. Superimposition of ARV VP8* and P[14] HAL1166 VP8* structures showed that despite a narrow cleft, the amino acid composition in the cleft of P[14] VP8* is not compatible with Sia binding. To identify the type of glycans preferentially recognized by this VP8*, Hu et al., [12] carried out a glycan array screen comprised of 611 different glycans, including a variety of glycans with terminal or internal Sia and showed that P[14] VP8* specifically recognizes glycans with a terminal oligosaccharide sequence that is typical of A-type histo blood group antigens (HBGAs). HBGAs are genetically determined glycoconjugates present in mucosal secretions and epithelia and on red blood cells [39]. They are synthesized by sequential addition of a monosaccharide to a precursor disaccharide motif by glycosyl-transferases such as the fucosyl-transferases FUT2 and FUT3, and enzymes A and B to generate A-, B-, H- and Lewis (Le) HBGAs (see Fig 3A. in the review article by Le Pendu et al. in this volume). The relevance of A-type HBGA interaction with HAL1166 HRV VP8* in the context of virus infection was clearly established by infectivity assays to provide a novel paradigm for initial cell attachment of HRV strains [12].
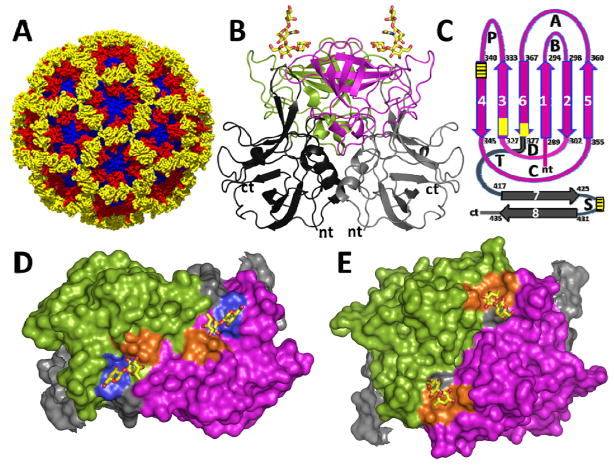
Figure 3.
(A) X-ray structure of Norwalk virus capsid (PDB ID: 1IHM). The shell domain (S) is shown in blue, the PI and P2 subdomains of the protruding P domain are shown in red and yellow, respectively. (B) Cartoon representation of the P-domain dimer (side view) from the GI.1 HNoV bound to H-type HBGA (PDB ID: 2ZL6). The HBGA binding site lies on the distal P2 subdomain. The P2 subdomain of the individual subunits of the dimer are colored in green and magenta respectively (here and subsequent Figs), and their P1 subdomains are colored in dark and lighter grey. (C) Topology diagram of the P domain highlighting the locations of HBGA binding sites in GI NoVs (yellow box) and GII NoVs (yellow box with lines). The antiparallel β strands (1–6) forming a barrel-like structure in the P2 subdomain are indicated by vertical arrows (magenta) and those in the P1 subdomain (7,8) that contribute to HBGA binding are indicated by grey. The variable loops connecting the β-strands are denoted A–D, P, T and S. (D) Surface representation (top view) of GI P-dimer (PDB ID: 2ZL5) showing distinct HBGA-binding sites (yellow circle) in each of the subunits. Residues that interact with Gal and Fuc moieties of the HBGA (shown in yellow stick representations) are shown in blue and gold colors, respectively. (E) Surface representation (top view) of the GII P domain dimer (PDB ID: 3SLN) showing the HBGA binding site shared between the opposing subunits in the dimer. Residues that interact with the Fuc moiety of the HBGA (shown in yellow stick representations) are colored in yellow.
The crystal structure of P[14] VP8* in complex with A-type HBGA [12] (Fig. 1D), which shows that the ligand binds in the same location in the cleft as Sia in the animal VP8* structure, demonstrates how subtle changes within the same structural framework of VP8* can lead to altered glycan specificity. Consistent with the observation that most of the residues in P[14] VP8* involved in binding to A-type HBGA are well conserved in the VP8*of feline origin P[9] HRV, cell-based assays showed that A-type HBGA is also the cell attachment glycan for P[9] genotype. Although crystal structures of other VP8* with glycans are yet to be determined, recent studies have clearly emphasized that HRVs show genotype-dependent glycan specificity and that binding to sialo-glycans is not obligatory [13]. These studies have shown that P[4] and P[8] HRVs recognize H-type HBGAs; whereas, a neonate-specific P[11] HRV, which is a bovine-human reassortant virus, specifically recognizes glycans, which are the precursor of H type 2 HBGA, also referred to as type II glycans [40].
Comparison of the available HRV VP8* structures suggest two distinct divergent patterns, one with a narrow cleft as found in the VP8* of P[14] and likely in that of P[9] HRV strains, and the other with a wider cleft as observed in the VP8* of Wa-like P[4] and DS1-like P[8] HRVs, which are suggested to have bovine and porcine origins, respectively (Fig. 1B). Although these two sets of RVs have found their way into human population, they have taken different evolutionary paths influenced by several factors such as interactions with co-circulating strains and types of animals they were able to infect. The wider cleft strongly correlates with a deletion at amino acid position 136, and a significant change at position 101 (Fig. 2A), which is a conserved residue contributing to glycan interaction in the VP8*s with a narrow cleft. From this correlation, together with known sensitivity to sialidase, RVs can be grouped into four classes (Fig. 2B). From the observation that A-HBGA binding site in s-i human P[14]-VP8* (class A) overlaps with Sia binding site in s-s ARVs (class C), we can predict that all VP8*s with a narrow cleft likely share the same site for glycan binding, although their glycan specificity may differ. A relevant question is what type of glycans are recognized by s-i ARVs (class D) which are predicted to have a narrow cleft. This is indeed a significant question considering that one of the genotypes, P[5], in this class is the predominant genotype in one of the RV vaccines, Rotateq™ currently in use [41]. Thus far, all the structures that have been determined with bound glycans have been only with VP8*s that have a narrow cleft. The glycan binding site for VP8*s in class B, which are predicted to have a wider cleft, and contain globally dominant strains as well as a geographically restricted neonate–specific P[11] and P[6] strains, remains to be characterized structurally.
Figure 2.
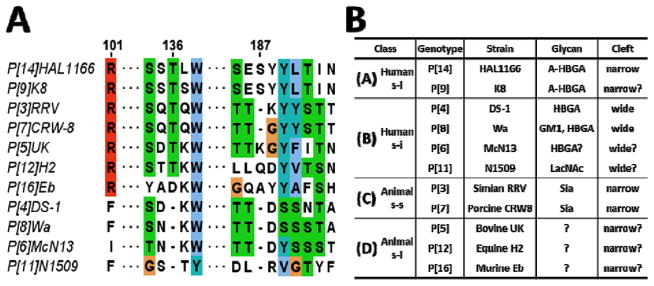
(A) Sequence alignment of representative VP8*s of different genotypes. The amino acids are colored using Clustal protein color scheme in Jalview [insert ref later]. (B) Classification of VP8* into A–D classes.
Strain-dependent glycan specificity in noroviruses
While the discovery that HBGAs function as cell attachment factors for some of the HRV strains is recent, involvement of these polymorphic glycoconjugates not only in cell attachment but also in conferring susceptibility has long been known in the case of human noroviruses (HNoVs) [9–11, 42]. These icosahedral viruses with a positive-sense RNA genome exhibit enormous genetic diversity and are classified into six genogroups (GI-GVI), and each genogroup is further subdivided into one or more genotypes [43]. Genogroups GI, GII and GIV contain human pathogens [44]. The HNoVs belonging to genogroup II and genotype 4 (GII.4) are the most prevalent, accounting for 70–80% of the noroviral outbreaks worldwide [45]. It is suggested that these GII.4 NoVs undergo epochal evolution, analogous to A/H3N2 influenza viruses, with the emergence of a new GII.4 variant every 2–4 years coinciding with a new epidemic peak [46, 47].
HNoVs are resistant to cell culture; however, co-expression of the major capsid protein VP1 and the minor protein VP2 results in the formation of virus-like particles (VLPs) that preserve the morphological and antigenic features of the authentic virions. The NoV capsid exhibits a T=3 icosahedral organization formed by 90 dimers of VP1 (Fig. 3A and 3B) and has two major domains, the S domain that forms the shell, and the P domain, with P1 and P2 subdomains, that protrudes from the S domain [48–51]. HNoVs bind to HBGAs through the distally-located hypervariable P2 subdomain (Fig. 3B). HNoVs provide an exquisite example of how genotypic variations allow for exploitation of the polymorphic nature of HBGAs in host population to counter herd immunity and cause epidemics. Bacterially-expressed P domain is used in all the crystallographic studies to characterize HNoV-HBGA interactions, as it duplicates the P domain dimeric structure observed in the capsid as well as the HBGA binding properties [52, 53]. Although the core structure of the P2 subdomain is composed of a six-stranded antiparallel β-barrel (Fig. 3C) that is invariant between the genogroups and within the genotypes, sequence variations allow for differences in strand lengths and loop structures to not only differentially alter the HBGA binding profiles between the strains but also alter the electrostatic surface topography contributing to antigenic variation [54, 55].
A fascinating observation from the crystallographic studies of various GI and GII genotypes with HBGAs is that the carbohydrate binding sites in GI and GII are distinctly different both in their locations and in their structural characteristics [49, 53–59] (Fig. 3C and 3D). This is consistent with observed differences in their HBGA binding profiles, and also is well borne-out by minimal sequence conservation in their P2 subdomains, including the amino acid residues that participate in the HBGA interactions. While the majority of interactions with HBGA in the GI P domain dimer are localized within each subunit of the dimer (Fig. 3C), they are shared between the opposing subunits in the GII P dimer (Fig. 3D). Another distinguishing feature is that the HBGA binding in GI is primarily centered around a Gal moiety (Fig. 4A), whereas in GII, it is centered around a Fuc (particularly the Fuc added by FUT2 during the synthesis of ABH HBGAs, referred to as SeFuc, which is differente from the Fuc, LeFuc, added by FUT3 during the synthesis of Lewis HBGAs) (Fig. 4B). In GI, in addition to conserved interactions with Gal, another exceptionally well conserved feature is the hydrophobic interaction between the Fuc moiety (as in the H-type) or the N-acetyl arm of N-acetylgalactosamine (in the A-type) with a conserved Trp residue in the P2 subdomain (Fig. 4A). This combinatorial requirement of Gal and hydrophobic interactions places restriction on the variety of HBGAs that GI HNoVs can bind. Many studies have failed to find binding of GI HNoVs to B-type HBGAs. Although B-type has a terminal Gal moiety, it lacks a group that could be involved in the hydrophobic interactions because of which the affinity for the B-type is significantly reduced. Such a combinatorial requirement does not appear to be the case for HBGA binding in GIIs allowing many of these HNoVs to bind all ABH HBGAs, which could be one of the factors in the greater prevalence of GII, particularly GII.4 HNoVs.
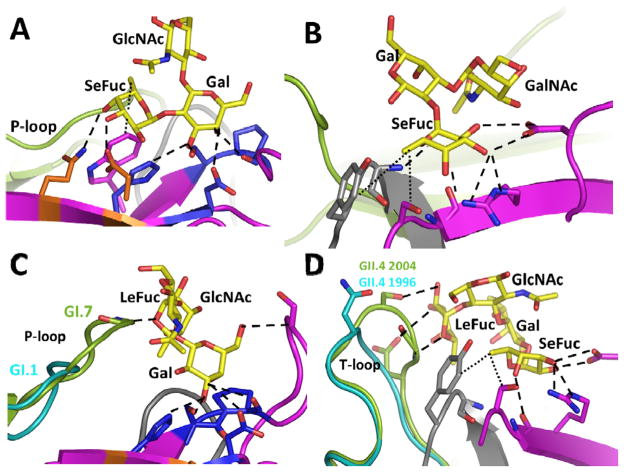
Figure 4.
(A) Gal-centric HBGA interactions in GI NoVs. Shown here as an example is interactions between GI.1 and H-type HBGA. A-type HBGA makes similar interaction with its Gal and N-acetamido groups of N-acetylgalactosamine similar to the Gal and Fuc moieties of the H-type. (B) Fuc-centric HBGA interactions in GII NoVs, shown here as an example is interactions between GII.4 P domain and A-type HBGA (PDB ID: 3SLD). (C) Alterations in length and structure of the P-loop that allows GI.7 bind non-secretor Lea (PDB I.D. 4P3I), GI.1 with a shorter P–loop cannot make similar interactions (D) Structural alterations in the T-loop that allows 2004 GII.4 variant to interact additionally with di-fucosyl secretor Lewis HBGA (Leb) (PDB ID: 3SLD), similar interactions with Leb are not possible in the 1996 GII.4 variant (cyan). The interacting P domain residues are shown as sticks with oxygen and nitrogen atoms in red and blue, respectively.
Recent crystallographic studies of different genotypes in GI and GII have also highlighted how sequence alterations in the P domain within the genogroup members also contribute to varied HBGA binding profiles [49, 53–60]. A striking observation from these studies leading to a generalizable concept is that HBGA binding in HNoVs involves two sites; one that is highly conserved, allowing the preservation of Gal and Fuc dominant nature of interactions in GI and GII, respectively, and the other that is highly susceptible to structural alterations because of sequence variations, allowing for differential binding to Lewis HBGAs. In GI HNoVs, sequence changes differentially alter their ability to bind non-secretor Lewis HBGA, Lea. Although in general secretor-positive status is strongly correlated with HNoV infection, recent epidemiological studies show an increase in the prevalence of GI outbreaks worldwide, with different genotypes such as GI.4, GI.6, GI.3, and GI.7 predominating in different geographical regions (9–13). By comparative analysis of the crystal structures of the GI.1 [53], which does not bind Lea, with that of GI.7 [55] and GI.2 [58], which show binding, Shanker et al, [55] have proposed that the threshold length and structure of the P loop are the critical determinants for Lea binding (Fig. 4C). Similarly, comparative analysis of the P domains structures of 1996 and 2004 GII.4 temporal variants show that structural changes in the T loop modulate the binding strength of difucosyl Lewis HBGAs between the variants (Fig. 4D), and thus contribute to epochal evolution, perhaps by differential targeting of the GII.4 variants to Lewis-positive secretor-positive individuals [54].
Another prominent feature highlighted by these comparative analyses is that A and B loops are susceptible to significant changes [54]. Interestingly, in GI.1 NV, the B loop contains a residue critical for binding of HBGA blocking antibodies [61], and the corresponding loop in the P domains of murine NoV (GV) [62], and rabbit hemorrhagic disease virus (animal calicivirus) [50] contains the neutralization antigenic sites. Thus, this region is potentially a major site for differential antigenic presentations contributing to serotypic differences among not only in NoVs but caliciviruses in general.
Concluding remarks
Strain-dependent recognition of polymorphic HBGAs by gastroenteric viral pathogens HRVs and HNoVs is rather unique among human viruses. Of note here is that the microbial pathogen Helicobacter pylori, which causes gastric cancer, also exhibits strain-dependent binding to HBGAs for colonization. An interesting question is whether this is a mere coincidence or that there is a larger evolutionary significance. For both HNoVs and H. pylori it is well recognized that these glycans are susceptibility determinants. In the case of HRVs, it is yet unclear if HBGAs are susceptibility factors; although some recent data [63, 64] indicate the possibility, more studies are needed. Recent advances in our understanding of glycan specificity for these viruses raise several questions. Current available data indicate that these viruses use HBGAs for initial attachment to host cells; however, whether interactions with HBGA also affect downstream signaling pathways as a part of the virus entry process requires further investigation. A fascinating discovery is that a neonatal HRV specifically recognizes type II glycans, which is abundantly present in human breast milk. What is the significance of this specific binding in the context of pathogenesis, will be subject of further studies. The structural basis of how VP8* of HRVs with a wider cleft, observed in the globally dominant strains and also in the neonate-specific strains, bind to glycans needs to be elucidated. Although there has been extensive structural studies on how HBGAs are recognized by HNoVs, structural understanding of how antibodies, particularly those that block HBGA binding [65], interact with NoVs require future studies. Importance of such ‘neutralizing’ antibodies is underscored by recent studies showing circulating antibodies that block HBGA binding correlate with protection from NoV-associated illness [66].
Highlights.
Paradigm-shifting discovery that binding to sialoglycans is not obligatory for rotavirus cell attachment.
Polymorphic histo blood group antigens are cell attachment factors for sporadic and globally dominant human rotavirus strains.
Neonate-specific rotavirus strain binds specifically to type 2 glycans
Glycan specificity in GI noroviruses is not restricted to secretor-HBGAs and some strains bind to non-secretor Lewis HBGAs
Temporal sequence variations in GII.4 norovirus variants results in differential binding specificity for di-fucosyl secretor Lewis HBGAs.
Acknowledgments
We acknowledge support from NIH grants RO1 AI36040 (BVVP), RO1 AI080656, R01 AI105101 (MKE), PO1 AI057788 (MKE, RA, BVVP), and a grant (Q1279) from Robert Welch foundation (BVVP).
References
- 1.Olofsson S, Bergstrom T. Glycoconjugate glycans as viral receptors. Ann Med. 2005;37:154–172. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- 2.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 3.Reiter DM, Frierson JM, Halvorson EE, Kobayashi T, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides. PLoS pathogens. 2011;7:e1002166. doi: 10.1371/journal.ppat.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21:885–897. doi: 10.1093/emboj/21.5.885. Provided the first structural details of sialic acid binding in the VP8* of a sialidase-sensitive animal rotavirus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. Journal of virology. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hueffer K, Parrish CR. Parvovirus host range, cell tropism and evolution. Curr Opin Microbiol. 2003;6:392–398. doi: 10.1016/s1369-5274(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 7.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. Journal of virology. 2010;84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shieh MT, WuDunn D, Montgomery RI, Esko JD, Spear PG. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology. 2002;122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutson AM, Atmar RL, Marcus DM, Estes MK. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. Journal of virology. 2003;77:405–415. doi: 10.1128/JVI.77.1.405-415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, et al. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. The Journal of infectious diseases. 2003;188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- 12**.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. This is the first paper demonstrating A-type HBGA is the cell atatchment factor for a human rotavirus strain and structurally characterizing the HBGA binding to VP8* [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. Journal of virology. 2012;86:4833–4843. doi: 10.1128/JVI.05507-11. These studies show other human rotavirus strains recognize HBGA in a type-specific manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends in microbiology. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore SF, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv Virus Res. 2007;70:183–232. doi: 10.1016/S0065-3527(07)70005-2. [DOI] [PubMed] [Google Scholar]
- 16.Rillahan CD, Paulson JC. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X, Yu H, Chen X, Lasanajak Y, Tappert MM, Air GM, Tiwari VK, Cao H, Chokhawala HA, Zheng H, et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF. Shotgun glycomics: a microarray strategy for functional glycomics. Nat Methods. 2011;8:85–90. doi: 10.1038/nmeth.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tate JE, Chitambar S, Esposito DH, Sarkar R, Gladstone B, Ramani S, Raghava MV, Sowmyanarayanan TV, Gandhe S, Arora R, et al. Disease and economic burden of rotavirus diarrhoea in India. Vaccine. 2009;27 (Suppl 5):F18–24. doi: 10.1016/j.vaccine.2009.08.098. [DOI] [PubMed] [Google Scholar]
- 20.Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Griffin D, Lamb R, Martin M, Roizman B, Strauss S, editors. Fields Virology. Vol. 2. Philadelphia: Wolters Kluwer Health; Lippincott, Williams and Wilkins; 2007. pp. 1917–1975. [Google Scholar]
- 21.Gray J, Iturriza-Gomara M. Rotaviruses. Methods Mol Biol. 2011;665:325–355. doi: 10.1007/978-1-60761-817-1_18. [DOI] [PubMed] [Google Scholar]
- 22.Matthijnssens J, De Grazia S, Piessens J, Heylen E, Zeller M, Giammanco GM, Banyai K, Buonavoglia C, Ciarlet M, Martella V, et al. Multiple reassortment and interspecies transmission events contribute to the diversity of feline, canine and feline/canine-like human group A rotavirus strains. Infect Genet Evol. 2011;11:1396–1406. doi: 10.1016/j.meegid.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. Journal of virology. 2009;83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad BV, Wang GJ, Clerx JP, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 25*.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. First near atomic resolution dedscriptio of mature rotavirus particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arias CF, Romero P, Alvarez V, Lopez S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estes MK, Graham DY, Mason BB. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J Virol. 1981;39:879–888. doi: 10.1128/jvi.39.3.879-888.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker M, Prasad BV. Rotavirus cell entry. Curr Top Microbiol Immunol. 2010;343:121–148. doi: 10.1007/82_2010_34. [DOI] [PubMed] [Google Scholar]
- 30.Lopez S, Arias CF. Rotavirus-host cell interactions: an arms race. Curr Opin Virol. 2012 doi: 10.1016/j.coviro.2012.05.001. 2012/06/05. [DOI] [PubMed] [Google Scholar]
- 31.Blanchard H, Yu X, Coulson BS, von Itzstein M. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*) J Mol Biol. 2007;367:1215–1226. doi: 10.1016/j.jmb.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Lopez S, Arias CF. Early steps in rotavirus cell entry. Curr Top Microbiol Immunol. 2006;309:39–66. doi: 10.1007/3-540-30773-7_2. [DOI] [PubMed] [Google Scholar]
- 33.Ciarlet M, Estes MK. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80 (Pt 4):943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 34*.Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, et al. Sialic acid dependence in rotavirus host cell invasion. Nat Chem Biol. 2009;5:91–93. doi: 10.1038/nchembio.134. These studies show that silaidase-insensitive rotavirus strain recognizes glycans with internal sialic acid for cell attachment leading to te initial suggestion that that the binding to sialic acid is obligatory for rotaviruses irrespective of their sensitivity to sialidase treatment. [DOI] [PubMed] [Google Scholar]
- 35.Gerna G, Sears J, Hoshino Y, Steele AD, Nakagomi O, Sarasini A, Flores J. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- 36.Matthijnssens J, Potgieter CA, Ciarlet M, Parreno V, Martella V, Banyai K, Garaicoechea L, Palombo EA, Novo L, Zeller M, et al. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chitambar SD, Arora R, Kolpe AB, Yadav MM, Raut CG. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 38.Fukai K, Saito T, Inoue K, Sato M. Molecular characterization of novel P[14], G8 bovine group A rotavirus, Sun9, isolated in Japan. Virus Res. 2004;105:101–106. doi: 10.1016/j.virusres.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Marionneau S, Cailleau-Thomas A, Rocher J, Le Moullac-Vaidye B, Ruvoen N, Clement M, Le Pendu J. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie. 2001;83:565–573. doi: 10.1016/s0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- 40**.Ramani S, Cortes-Penfield NW, Hu L, Crawford SE, Czako R, Smith DF, Kang G, Ramig RF, Le Pendu J, Prasad BV, et al. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. Journal of virology. 2013;87:7255–7264. doi: 10.1128/JVI.03518-12. First studies to show that a neonate-specific strain of rotavirus binds to type II glycans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthijnssens J, Joelsson DB, Warakomski DJ, Zhou T, Mathis PK, van Maanen MH, Ranheim TS, Ciarlet M. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology. 2010;403:111–127. doi: 10.1016/j.virol.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 42.Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. The Journal of infectious diseases. 2002;185:1335–1337. doi: 10.1086/339883. [DOI] [PubMed] [Google Scholar]
- 43.Ramani S, Atmar RL, Estes MK, Eohnauovd COGJ. Epidemiology of human noroviruses and updates on vaccine development. Curr Opin Gastroenterol. 2014;30:25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Kroneman A, Verhoef L, Harris J, Vennema H, Duizer E, van Duynhoven Y, Gray J, Iturriza M, Bottiger B, Falkenhorst G, et al. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the Foodborne Viruses in Europe network from 1 July 2001 to 30 June 2006. J Clin Microbiol. 2008;46:2959–2965. doi: 10.1128/JCM.00499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. Epochal evolution of GGII. 4 norovirus capsid proteins from 1995 to 2006. Journal of virology. 2007;81:9932–9941. doi: 10.1128/JVI.00674-07. These epidimiological studies show evidence for epochal evolution in GII.4 noroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol Rev. 2008;225:190–211. doi: 10.1111/j.1600-065X.2008.00680.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen R, Neill JD, Estes MK, Prasad BV. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8048–8053. doi: 10.1073/pnas.0600421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49*.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. Provided the first atomic level details of a norovirus capsid. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Xu F, Liu J, Gao B, Liu Y, Zhai Y, Ma J, Zhang K, Baker TS, Schulten K, et al. Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography. PLoS pathogens. 2013;9:e1003132. doi: 10.1371/journal.ppat.1003132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katpally U, Voss NR, Cavazza T, Taube S, Rubin JR, Young VL, Stuckey J, Ward VK, Virgin HWt, Wobus CE, et al. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. Journal of virology. 2010;84:5836–5841. doi: 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan M, Meller J, Jiang X. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. Journal of virology. 2006;80:7322–7331. doi: 10.1128/JVI.00233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53*.Choi JM, Hutson AM, Estes MK, Prasad BV. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9175–9180. doi: 10.1073/pnas.0803275105. These studies provided a possible structural explanation for restricted HBGA binding specificty in GI noroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Shanker S, Choi JM, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII. 4 epidemic variant: implications for epochal evolution. Journal of virology. 2011;85:8635–8645. doi: 10.1128/JVI.00848-11. These studies provided a structural description of how temporally changing residues in the GII.4 epidemic varaints alter HBGA binding specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Shanker S, Czako R, Sankaran B, Atmar RL, Estes MK, Prasad BV. Structural analysis of determinants to HBGA binding specificity in GI noroviruses. J Virol. 2014;88:6168–6180. doi: 10.1128/JVI.00201-14. These studies provide a structural explanation for differential biniding of non secretor HBGA in GI noroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Cao S, Lou Z, Tan M, Chen Y, Liu Y, Zhang Z, Zhang XC, Jiang X, Li X, Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. Journal of virology. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. Provided the first structural characterization of HBGA binding in norovirus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bu W, Mamedova A, Tan M, Xia M, Jiang X, Hegde RS. Structural basis for the receptor binding specificity of Norwalk virus. Journal of virology. 2008;82:5340–5347. doi: 10.1128/JVI.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Kubota T, Kumagai A, Ito H, Furukawa S, Someya Y, Takeda N, Ishii K, Wakita T, Narimatsu H, Shirato H. Structural basis for the recognition of Lewis antigens by genogroup I norovirus. Journal of virology. 2012;86:11138–11150. doi: 10.1128/JVI.00278-12. First structural analysis of non-secretor HBGA binding in GI noroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansman GS, Biertumpfel C, Georgiev I, McLellan JS, Chen L, Zhou T, Katayama K, Kwong PD. Crystal structures of GII.10 and GII. 12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. Journal of virology. 2011;85:6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Tan M, Xia M, Hao N, Zhang XC, Huang P, Jiang X, Li X, Rao Z. Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS pathogens. 2011;7:e1002152. doi: 10.1371/journal.ppat.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z, Sosnovtsev SV, Bok K, Parra GI, Makiya M, Agulto L, Green KY, Purcell RH. Development of Norwalk virus-specific monoclonal antibodies with therapeutic potential for the treatment of Norwalk virus gastroenteritis. Journal of virology. 2013;87:9547–9557. doi: 10.1128/JVI.01376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62*.Katpally U, Wobus CE, Dryden K, Virgin HWt, Smith TJ. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. Journal of virology. 2008;82:2079–2088. doi: 10.1128/JVI.02200-07. First structural description of a norovirus binding to neutralzing antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63**.Trang NV, Vu HT, Le NT, Huang P, Jiang X, Anh DD. Association between Norovirus and Rotavirus Infection and Histo-Blood Group Antigen Types in Vietnamese Children. Journal of clinical microbiology. 2014;52:1366–1374. doi: 10.1128/JCM.02927-13. Initial studies implicating HBGAs as sussceptibity factors for rotaviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Imbert-Marcille BM, Barbe L, Dupe M, Le Moullac-Vaidye B, Besse B, Peltier C, Ruvoen-Clouet N, Le Pendu J. A FUT2 Gene Common Polymorphism Determines Resistance to Rotavirus A of the P[8] Genotype. The Journal of infectious diseases. 2014;209:1227–1230. doi: 10.1093/infdis/jit655. Initial studies implicating HBGAs as sussceptibity factors for rotaviruses. [DOI] [PubMed] [Google Scholar]
- 65.LoBue AD, Lindesmith L, Yount B, Harrington PR, Thompson JM, Johnston RE, Moe CL, Baric RS. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine. 2006;24:5220–5234. doi: 10.1016/j.vaccine.2006.03.080. [DOI] [PubMed] [Google Scholar]
- 66.Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, Graham DY, Atmar RL. Serological correlate of protection against norovirus-induced gastroenteritis. The Journal of infectious diseases. 2010;202:1212–1218. doi: 10.1086/656364. [DOI] [PMC free article] [PubMed] [Google Scholar]