INTRODUCTION
The biosynthesis of histidine in Salmonella typhimurium (official designation, Salmonella enterica serovar Typhimurium) and Escherichia coli has been an important system for the study of relationships between the flow of intermediates through a biosynthetic pathway and the control of the genes encoding the enzymes that catalyze the steps in the pathway. Earlier studies of histidine biosynthesis contributed to understanding mechanisms basic to the regulation of biosynthetic pathways, such as feedback inhibition, energy charge, and the setting of basal biosynthetic enzyme levels. The histidine biosynthetic pathway itself contains several interesting and unusual intermediates and enzymatic steps and forms a critical link between amino acid, purine, and thiamine biosynthesis. Fundamental concepts in gene regulation, such as attenuation, Rho factor-dependent polarity, the polycistronic organization of mRNA molecules, autogenous regulation, transcriptional pausing, and positive control of metabolic regulation, were formulated to explain aspects of histidine biosynthesis. In fact, the term “attenuation” was first coined to describe regulatory patterns of the histidine operon (132). In addition, the histidine biosynthetic pathway and the histidine operon have served as a powerful model system for studying fundamental evolutionary, metabolic, physiological, and genetic processes, such as gene duplications, transposition, mutagenesis, including the widely used Ames test (13, 101), and tRNA biosynthesis and function.
The first comprehensive review of histidine biosynthesis was written by Brenner and B. N. Ames in 1971 (42). This extraordinary article consolidated information about control of the histidine biosynthetic pathway and posed many of the questions about histidine biosynthesis and his operon control that were the subject of investigation in subsequent years. Later reviews, including the first two versions of this review in 1987 and 1996, and reviews by Blasi and Bruni (34) and Artz and Holzschu (25), covered the developments of the molecular structure of the his operon, the mechanisms of his operon metabolic regulation and attenuation control, and the function of the his regulatory loci, including identification of hisW mutations and one class of hisU mutations as alleles of gyrA and gyrB, respectively, which encode the subunits of DNA gyrase. The 1996 review by Alifano et al. (4) is noteworthy and describes histidine biosynthesis in organisms besides E. coli and S. typhimurium and provides one of the first summaries of the evolution of the histidine biosynthetic pathway.
Since the previous version of this review, exceptional progress has been made in a couple of research areas. Most of the enzymes that catalyze histidine biosynthesis have now been crystallized and had their structures determined. Many of the histidine biosynthetic intermediates are unstable, and the histidine biosynthetic enzymes catalyze some chemically unusual reactions. Therefore, these studies have led to considerable mechanistic insight into the pathway itself and have provided deep biochemical understanding of several fundamental processes, such as feedback control, allosteric interactions, and metabolite channeling. Highlights from these studies are summarized in this review. At the same time, some of the older physiological information from the previous versions of this review has been retained and updated because of its metabolic importance and use in understanding his operon control. One particularly satisfying generalization that can be drawn from the recent biochemical work is just how accurate the older results and models turned out to be, especially given the relative limitations of the methods then available. Considerable recent progress has also been made on aspects of his operon regulation, including the mechanism of pp(p)Gpp stimulation of his operon transcription, the molecular basis for transcriptional pausing and Rho factor-mediated polarity. These and other developments are summarized. The large number of genomic sequences that continue to be determined has sustained interest in the evolution of the histidine biosynthetic pathway and enzymes in eubacteria, archaea, fungi, and plants. Evolution is touched upon in this review as it relates to E. coli and S. typhimurium. Several additional reviews on the evolution of histidine biosynthesis in different organisms, including plants, have recently appeared (4, 85-87, 162, 179, 204).
THE HISTIDINE BIOSYNTHETIC PATHWAY AND ENZYMES
Intermediates in the Pathway
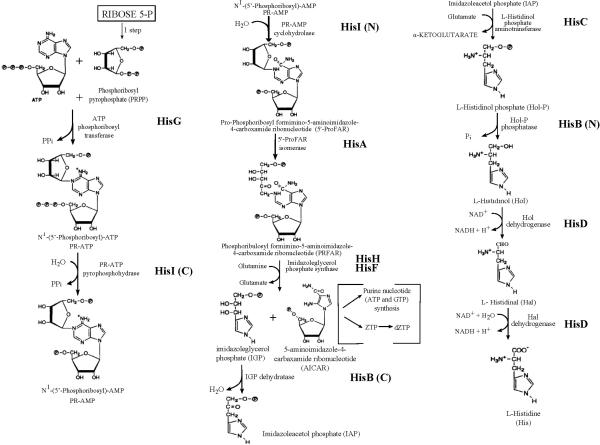
The pathway and details of histidine biosynthesis are the same in E. coli and S. typhimurium (34). In fact, the same pathway is used in all organisms that synthesize histidine, although there are variations in the genetic organization and grouping of the biosynthetic enzymes (4, 162, 166, 203, 204). Histidine has been postulated to have formed abiotically, and histidine-containing peptides may have been involved in the prebiotic synthesis of peptides and nucleic acid molecules (see (4, 85)). Analyses of the distribution of the histidine biosynthesis genes suggest that a series of duplication, elongation, and fusion events were involved in the evolution of this biologically ancient pathway (4, 179). The ten biochemical steps in this unbranched pathway include a number of complex and unusual reactions, and all nine chemical intermediates have been described chemically (Fig. 1). A new method to determine metabolome changes based on tandem capillary electrophoresis and mass spectrometry has shown that these intermediates accumulate rapidly in an E. coli hisD mutant abruptly starved for histidine (171). This starvation condition also induces the stringent response, which leads to increases in the levels of ppGpp, ATP, GTP, amino acids and changes in intermediates in pathways subjected to stringent control (171).
FIGURE 1.
Histidine biosynthetic pathway. The steps in the pathway are described in the text, and the enzymes that catalyze the reactions are listed in Table 1. When the enzymes are bifunctional (C) indicates that the carboxyl terminal is performing the reaction and (N) indicates that the amino terminal is performing the reaction.
The determination of the pathway was started by the work of B. N. Ames and coworkers (see references in (42)) and completed only recently by Klem and Davisson (137). For simplicity, the eight enzymes that catalyze the reactions (Table 1) are designated by the genes that encode them in the his operon. Three of the proteins, HisI, HisB, and HisD, are bifunctional in E. coli and S. typhimurium and carry out two separate reactions in the pathway (Table 1 and Fig. 1). Two other proteins, HisH and HisF, form an obligate heterodimer (Table 1). Phosphoribosylpyrophosphate (PRPP) and ATP are the initial substrates and connect histidine biosynthesis with the biosynthesis of pyrimidine nucleotides, purine nucleotides, pyridine nucleotides, thiamine, and tryptophan (see chapters XX, XX, XX, and XX, respectively, in this volume). The metabolic link between histidine and purine biosynthesis is indicated by numerous observations. For example, S. typhimurium mutants that have increased expression of the his operon and lack feedback control of the histidine biosynthetic pathway require adenine for growth at 42°C (128). Histidine starvation of hisF mutants, which cannot recycle 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) for purine biosynthesis (Fig. 1), results in a 15-fold drop in cellular ATP pools without severely lowering GTP, UTP, and CTP levels (125). This observation forms the basis of a reversible method to deplete cellular ATP pools to around 100 μM (125). Along the same line, E. coli strains carrying leaky frameshift mutations in the promoter-distal part of hisH grow when supplemented with purines, such as inosine, without histidine (178). These leaky hisH mutants also grow on medium lacking purines or histidine when the histidine biosynthetic pathway is partially feedback inhibited by a histidine analog (see below). All these observations can be explained by futile ATP consumption by the first part of the histidine biosynthetic pathway without recycling of the purine precursor, AICAR (Fig. 1). In addition, recent work has shown that metabolic flux in the purine and histidine biosynthetic pathways can influence the synthesis of thiamine, whose synthesis branches from the purine pathway (9). The requirement of S. typhimurium purH mutants for thiamine can be satisfied by histidine in a parent strain, but not in a mutant lacking feedback control. This result can be explained by a model in which AICAR accumulation by the purH mutant blocks thiamine biosynthesis, and histidine addition reduces the flux through the histidine pathway, reduces AICAR amount, and relieves the inhibition on thiamine biosynthesis (9).
TABLE 1.
Properties of enzymes encoded by the his operon and major his regulatory loci
| Enzyme | Gene | Mol wt of monomera (# amino acids) | Association state of enzymeb and references for crystal structuresc | Kinetic parameters of enzymes from E. coli and S. typhimuriumd | Inhibitors | References for kinetic parameters and, inhibitors |
|---|---|---|---|---|---|---|
| ATP phosphoribosyltransferase (EC 2.4.2.17) | hisG | 33,271 (299) | Dimer when PRPP and ATP bound (active state); hexamer; when histidine and AMP bound (inhibited state) | Km (apparent) (ATP) = 70-110μM; Km(apparent) (PRPP) = 11-30 μM; Km (conc. independent) (ATP) =800 μM; Km (conc. independent) (PRPP) =110 μM; ping-pong bi-bi mechanism | Histidine (Ki = 60-380 μM); 2-thiazole-alanine; AMP (Ki [apparent] = 860 μM); ADP; AMP + histidine > AMP; ppGpp + histidine >histidine; PR-ATP (product) | (42, 71, 159-161, 206, 208) |
| Histidinol dehydrogenase (EC 1.1.1.23) | hisD (bi-functional) | 46,113 (432) | Dimer with 2 Zn2+ | Km (NAD+) = 600-1,300 μM; Km (Hol) =11-18 μM; Kd (Zn2+) = 50 μM; acid-base catalysis | Histamine (Ki = 40 μM); imidazole (Ki = 1.5 mM) | (109, 151, 210, 211) |
| Histidinol phosphate aminotransferase (EC 2.6.1.9) | hisC | 39,319 (356) | Dimer | Km (IAP) = 80-120 μM; Km (glutamate) = 7 mM; ping-pong bi-bi mechanism | Hol-P (product; Ki = 52-160 μM); IAP (substrate; Ki = 0.9 mM); various substrate analogs | (1, 120) |
| IGP dehydratase (C-terminal domain) (EC 4.2.1.19): histidinol phosphatase (N-terminal domain) (EC 3.1.3.15) | hisB (bi-functional) | 40,283 (352) | 24-mer in the presence of Mn+2; Hol Pase binds Zn+2 | Km (IPG) = 700 μM; Km (Hol-P) = 300 μM; HisB-N (Hol-P) = 54 μM and kcat = 2 × 103 s−1 | 3-Amino-1,2,4-triazole and herbicides inhibit dehydratase; Ca+2 inhibits Hol Pase | (41, 103, 181, 191) |
| IGP synthase subunit (glutaminase) | hisH | 21,655 (196) | Heterodimer with HisF (see below) | IGP synthase: Kmapp (glutamine) = 240 μM; Kmapp (PRFAR) = 1.5 μM; kcat ≈ 9 s−1 | Acivicin; Kiapp = 1.5 μM | (64, 137, 202) |
| Phosphoribosylformimino-5-amino-1-phosphoribosyl -4-imidazole carboxamide isomerase (EC 5.3.1.16) | hisA | 26,035 (245) | Monomer | At 25° C, Km (ProFAR) = 1.6 μM and kcat = 4.9 s−1; at 37° C, Km (ProFAR) = 1.8 μM and kcat = 14.3 s−1; | Unknown | (113) |
| IGP synthase subunit (cyclo-ligase) | hisF | 28, 457 (258) | Heterodimer with HisH (see above) | Cyclo-ligase in absence of HisH subunit; Km (PRFAR) = 21 μM; Km (NH4Cl) = 266 mM | Unknown | (137) |
| Phosphoribosyl-ATP pyrophosphohydrolase (“HisE” activity in C-terminal domain); phosphoribosyl-AMP cyclohydrolase (“HisI” activity in N-terminal domain) (EC 3.5.4.19) | hisI (bifunctional; formerly hisIE) | 22,764 (203) | Dimer; HisI binds Zn+2 and requires Mg+2 | Pyrophosphohydrolase (HisE) remains unknown; cyclohydrolase (HisI) of M. vannielii Km (PR-AMP) = 9.9; kcat = 4.1 s−1 | EDTA | (69) |
| Histidyl-tRNA synthetase (EC 6.1.1.21) | hisS | 46,898 (424) | Dimer | Aminoacylation: Km (His) = 3.2-25 μM; Km(ATP) = 140 μM; Km (tRNAHis) = 0.04 – 1.5 μM; Kcatapp = 3 s=1; pyrophosphate exchange: Kmapp(His) = 87 μM; kcatapp (His) = 91 s−1; Kmapp(ATP) = 641 μM; kcatapp (ATP) = 170 s−1 | α-Methylhistidine; histidinol; adenosine; AMP; ADP; AMP + PPi > AMP; His-tRNAHis (product) | (42-44, 46, 96, 97, 130) |
| Pseudouridine synthase I | truA (hisT) | 30,399 (270) | Dimer | Km (tRNA) 1.8 μM and Kcat = 0.2 s−1 at 15°C | (68, 122) |
Polypeptide molecular weights are from the following sources: E.coli his operon DNA sequence (49) E.coli his DNA sequence (99); E.coli hisT DNA sequence (21).
Enzyme native molecular weights are from the following sources: HisG (34, 208); HisD (34, 107, 211); HisC (34, 112, 120); HisB (181), HisH-HisF (137); HisA (113, 142); HisI (69); HisRS (20, 74, 130); TruA (HisT) (94).
Crystal structures can be found in the following references. Some structures are from thermophilic species or fungi, rather than E. coli and S. typhimurium. HisG (56, 65, 150); HisD (26); HisC (111, 199); HisB (181, 197); HisH-HisF (11, 61, 80, 165); HisA (113, 142); HisI (200); HisRS (19, 20, 96, 110); TruA (HisT) (94, 124).
Enzyme assay protocols can be found in the references. Additional and updated assay methods can be found in the following references: all of the his operon encoded enzymes (154), HisG (134, 139, 208); HisD (67, 107, 213); HisC (120); HisB (phosphatase activity) (40, 83); HisS (76, 92, 97); HisT (68, 155). Full substrate names and structures corresponding to abbreviations are given in Fig. 1.
The first reaction in the pathway is catalyzed by the HisG protein and involves a displacement on C-1 of PRPP by N-1 of the purine ring of ATP (Fig. 1). This Mg+2 ion-dependent condensation releases a pyrophosphate molecule, inverts the ribose moiety derived from PRPP from the α to the β configuration, and although reversible, is strongly product inhibited by L-histidine. The role played by feedback inhibition of the HisG enzyme in controlling the flow of intermediates through the histidine pathway is discussed in another section. Recent genome-scale metabolic modeling of E. coli indicates that this first reaction in histidine biosynthesis is one of the rare (5 of 873) thermodynamically unfavorable reactions involved in biomass production (114). The conclusion suggests that product inhibition likely limits flux into the histidine pathway (below and (114)).
The next steps in the pathway involve irreversible, Mg+2 ion-dependent hydrolysis of the N′-5-phosphoribosyl-ATP to N′-5′-phosphoribosyl-AMP and pyrophosphate catalyzed by the carboxyl terminal domain of the HisI enzyme, followed by a ring-opening reaction catalyzed by the amino terminal domain of HisI (Fig. 1). An internal redox reaction, known as an Amadori rearrangement, follows and is catalyzed by the HisA gene product (Fig. 1). For some time, the order and number of the next biosynthetic steps were not unequivocally established, and it was thought that at least one intermediate of unknown structure existed (201). However, it is now established from biochemical and structural characterization (10, 11, 136, 137, 164, 165) and the phenotypes of hisH and hisF mutants (125, 128, 178) that HisH and HisF are subunits of a single imidazoleglycerol phosphate (IGP) synthase that produces IGP and AICAR directly. Besides being cycled back to purine biosynthesis, AICAR is a precursor of the unusual ribotriphosphate 5-amino-4-imidazole carboxamide riboside 5′-triphosphate (ZTP in Fig. 1) (36). For this and other reasons, mutation frequencies were measured in E. coli with altered AICAR pools; however, AICAR was found not to be an endogenous metabolic mutagen (93, 95).
The final steps in histidine biosynthesis include a Mn+2 ion-dependent dehydration catalyzed by the carboxyl terminal domain of the HisB protein followed by a ketonization of the resulting enol, a reversible transamination with a nitrogen atom from glutamate catalyzed by the pyridoxal 5′- phosphate-containing HisC enzyme, and a dephosphorylation of L-histidinol phosphate catalyzed by the amino terminal domain of HisB. The L-histidinol product is then converted via an unstable histidinaldehyde intermediate to histidine by the bifunctional Zn+2-ion dependent HisD dehydrogenase (26, 108). This conversion involves two consecutive oxidation steps linked to the reduction of two NAD+ molecules (26, 108). In summary, the atoms of histidine are derived from the following precursors through the biosynthetic pathway: three carbon atoms of the amino acid backbone and carbons 4 and 5 of the imidazole ring are from PRPP, the amino group is from glutamate, nitrogen 3 of the imidazole ring is from glutamine, and carbon 2 and nitrogen 1 of the imidazole ring are from ATP.
Regulation of the Pathway
Energy equivalent to about 41 ATP molecules is required to synthesize each histidine molecule (42). Consequently, a bacterium that completely lacks control of histidine biosynthesis will waste about 2.5% of its metabolic energy synthesizing excess histidine when growing with a doubling time of about 50 min (42). For this reason, it is not surprising that E. coli and S. typhimurium have evolved an elaborate network to control the rate of histidine biosynthesis. The two most important points of control are regulation of the flow of intermediates through the pathway and regulation of the amounts of histidine biosynthetic enzymes produced.
Regulation of the Flow of Intermediates through the Pathway
The flow of intermediates through the histidine biosynthetic pathway can be adjusted by varying the enzymatic activity of the HisG enzyme, which catalyzes the first reaction in the pathway (Fig. 1). Modulation of HisG enzyme activity is brought about by four interrelated forms of inhibition: (i) classical, noncompetitive feedback inhibition by histidine; (ii) inhibition by ppGpp in the presence of partially inhibiting concentrations of histidine; (iii) competitive inhibition by ADP and AMP in response to the overall energy status in the cell; and (iv) competitive product inhibition by phosphoribosyl-ATP (PR-ATP) (Fig. 1). In wild-type bacteria growing in minimal medium, the rate of histidine biosynthesis seems to be controlled primarily by regulation of HisG enzymatic activity. Several feedback-resistant and feedback-hypersensitive mutations were mapped in a region that encodes the carboxyl-terminal portion of the HisG protein (119, 206). Feedback-resistant mutants selected for their growth in the presence of the analog 2-thiazolealanine excrete histidine into the culture medium (194). This important observation indicates that feedback inhibition holds histidine biosynthesis far below its full capacity, even when histidine is not supplied as a supplement (42). Some feedback-hypersensitive mutants also have a distinct phenotype; they are growth restricted at 20°C because of severe inhibition of the mutant HisG enzyme by histidine at lower temperatures (170, 206).
Because of its crucial role, the enzymology of the HisG protein has been intensively studied (Table 1). One potential problem in interpreting HisG kinetic data is strong product inhibition by phosphoribosyl-ATP (PR-ATP) (Fig. 1) (71, 208). To minimize this product inhibition, HisG enzyme assays were coupled to HisI activity to convert PR-ATP to the next intermediate in the pathway (208). Stopped-flow kinetic analyses of the HisG reaction have also been performed (71). Another earlier issue about HisG enzymology was the aggregation state of the protein, which was influenced in a complex way by temperature, ionic strength, pH, and the presence of ligands and effectors (34, 71). The combined kinetic studies have led to the following picture of the regulation of HisG activity. (i) Active HisG is a dimer that is stabilized by the substrate PRPP (208, 209); (ii) HisG dimers are in equilibrium with an inactive hexamer form whose formation is favored by binding the PR-ATP product, AMP, ADP, or histidine (140, 208); (iii) AMP and ADP are competitive inhibitors for both of the substrates, PRPP and ATP (160), whereas histidine acts through an allosteric mechanism that does not overlap the enzyme active site (159); (iv) inhibition by AMP and histidine is synergistic (71), and histidine causes discrimination against ATP binding in favor of the co-inhibitors, AMP and ADP (160).
The apparent Km of the HisG enzyme for ATP is much lower than the intracellular ATP concentration, whereas the apparent Km for PRPP is probably closer to the intracellular PRPP concentration (Table 1 and footnote a to Table 2); therefore, the rate of histidine biosynthesis most likely is directly affected by variations in the intracellular PRPP pool size. Since ATP and PRPP are the first substrates in the pathway and considerable cellular energy is consumed in histidine biosynthesis, inhibition of HisG enzyme activity by AMP and ADP has frequently been cited as an example of an energy-utilizing system that responds to the overall energy status in the cell, as expressed by the Atkinson energy-charge formula (42, 138). The Ki of the HisG enzyme for histidine (Table 1) is comparable to the intracellular histidine concentration found in bacteria growing in minimal medium containing histidine (Table 2), an observation that implies substantial inhibition of the rate of biosynthesis through the pathway. In contrast, the Ki for histidine is considerably higher than the intracellular histidine concentration found in bacteria that must synthesize histidine.
TABLE 2.
Parameters of histidine biosynthesis in wild-type S. typhimurium and E. coli
| Parametera | Growth conditionb | Value | References |
|---|---|---|---|
| Intracellular free His concn | Minimal medium | 15 μM | (17) |
| Minimal medium + 50 μM His | 100 μM | ||
| Total residue composition of His in protoplasm | Minimal medium | 90 μmol/g of dried cells | (167) |
| % of RNAHis charged with His | Minimal medium | 77% | (146) |
| Minimal medium + 50 μM His | 88% | ||
| Relative his operon expression | Minimal medium | ≡1.0 | (42, 222) |
| Minimal medium + 50 μM His | 0.9 | ||
| Rich medium | 0.3 | ||
| Steady-state rate of His biosynthesis | Minimal medium | ≈ 1 μmol/g (dry wt)/min | (42) |
Additional relevant parameters: the Km for histidine transport is approximately 10−8 for the high affinity periplasmic-binding protein (HisJ-HisQ-HisM-HisP) permease (16). The intracellular concentrations of the following compounds were estimated for bacteria growing in minimal-glucose medium: 3,000 μM ATP; 31 μM ppGpp; 2 μM tRNAHis; 2 μM histidyl-tRNA synthetase (35, 73).
Glucose is the carbon source in the minimal medium. Rich medium is Luria-Bertani (LB) or nutrient broth
Three recent structural studies have provided the first insights into the mechanism of HisG allosteric control (56, 65, 150). The structure of HisG from Mycobacterium tuberculosis was determined without bound ligand and complexed with inhibitors AMP and histidine (65). The structure of HisG from E. coli was determined complexed with the inhibitor AMP or the PR-ATP product (150). Last, the structure of the HisG-HisZ complex from Lactococcus lactis was determined complexed with the substrate PRPP or with phosphate ion and ATP (56). From these combined crystal structures and previous biochemical studies (Table 1 and above), the following picture of HisG feedback control emerges. (i) The Mg+-dependent HisG ATP-phosphoribosyltransferase (ATP-PRTase) reaction proceeds by an ordered Bi-Bi kinetic mechanism by which ATP binds before PRPP and PPi leaves before PR-ARP (see (150)). (ii) HisG ATP-PRTases represent a separate class of PRTs, designated as type IV, that have a fold related to that in periplasmic binding proteins (56, 65, 150). Lack of amino acid sequence similarity and the differences in structure suggests that the different types of PRTs arose by convergent evolution (150). (iii) Each monomer of HisG consists of three domains; domains I and II form a crevice used as the catalytic core, and the carboxyl terminal domain III binds to histidine (56, 65, 150). (iv) ATP and PRPP substrate binding favors dimer formation and causes the enzyme to assume an activated state similar to that of glycosyltransferases (56, 150). This state involves a conformational change at the dimer interface that brings into the active site conserved amino acids that promote pyrophosphate group leaving (56) (v) The “long” form of HisG found in E. coli, S. typhimurium, M. tuberculosis, and related bacteria has a fundamentally different quaternary structure (dimers and hexamers) from that of the L. lactis “short” form of HisG (heterooctamer). The catalytic HisG subunit of the short form is missing about 100 amino acids from its carboxyl terminus compared to the long form and requires heterodimer formation with a separate HisZ subunit for activity (56, 198). The HisZ subunit is a paralog of amino acyl-tRNA synthetases (198), although it lacks this activity. The remaining properties of HisG allosteric control discussed here refer to the long form found in E. coli and S. typhimurium. Further information about the HisZ subunit is in references (39, 198), and the structures of the HisG short and long forms are compared in reference (56)
(vi) Competitive inhibition of ATP and PRPP binding by AMP is caused by binding of a monophosphate and the ribose ring of AMP to the PRPP binding site in domain II and binding of the adenine ring of AMP to the ATP binding site in domain I (150). In addition, AMP and the product PR-ATP cause the catalytically active ATP-PRT dimers to aggregate into inactive hexamers, which are formed by binding together the carboxyl terminal ends of three dimers (65, 150). Aggregation into hexamers structurally closes the active site of the enzyme (65, 150). (vii) Binding of AMP and histidine induces a large conformational change in HisG that causes steric hindrance of the active sites in domains I and II (65). Binding of AMP orients residues in the carboxyl terminal domains of the hexamer subunits and thereby increases the affinity for histidine (65, 150) Conversely, the conformation change that occurs upon histidine binding decreases the dissociation of AMP from the active site.
The basis for inhibition of HisG by the alarmone ppGpp awaits future structural elucidation. ppGpp, which is a positive effector of his operon transcription (see below), does not inhibit HisG enzyme activity by itself; however, in the presence of moderate histidine concentrations (≥25 μM), physiologically significant concentrations of ppGpp (≥200 μM) strongly inhibit HisG enzyme activity in a positively cooperative manner (161). The synergistic inhibition of HisG enzyme by ppGpp and histidine might play a physiological role (25). Starvation of bacteria for amino acids elicits ppGpp accumulation as part of the stringent response (see Chapter XX). If bacteria are starved for an amino acid in the presence of histidine, then the synergistic inhibition of HisG enzyme by intracellular ppGpp (≥200 μM) and histidine (≈100 μM) will completely inhibit histidine biosynthesis. In contrast, ppGpp accumulation in bacteria starved for histidine will not inhibit the HisG enzyme. In addition, HisG enzyme activity will not be strongly inhibited by the intracellular pools of histidine (≈15 μM) and ppGpp (≈30 μM) present in bacteria growing exponentially in minimal medium lacking histidine.
In summary, the HisG ATP-PRTase is a complicated protein that is present in multiple aggregation states in response to its substrate and combinations of the inhibitors histidine, ppGpp, AMP, ADP, and PR-ATP. This multivalent inhibition allows sensitive control of the rate of histidine biosynthesis in response to a variety of cellular metabolic states. This complex, multivalent control is required because histidine biosynthesis is regulated chiefly by modulating the flow of intermediates through the pathway in wild-type bacteria growing under common culture conditions, as discussed in the next section. Recent structural studies can account for most of the catalytic, inhibition, and aggregation properties of the HisG ATP-PRTase. This combination of biochemical and structural information provides powerful information about the mechanism of allosteric feedback inhibition of the critical HisG enzyme.
Regulation of the Amounts of Histidine Biosynthetic Enzymes
In effect, noncompetitive inhibition by histidine lowers the apparent Vmax of the HisG enzyme reaction and makes it appear as if less total enzyme were present (152). Another way to control the rate of histidine biosynthesis is to adjust the intracellular concentrations of the histidine biosynthetic enzymes in response to histidine and other metabolites. The structure and regulation of the his operon, which encodes all of the histidine biosynthetic enzymes (Fig. 2 and Table 3), have been subjects of investigation for over fifty years. Results from many studies show that two mechanisms regulate his operon expression at the level of transcription: (i) transcription initiation at the his operon primary promoter (hisp1) is positively regulated by increasing ppGpp concentrations up to the ppGpp concentration found in cells growing in minimal-glucose medium; and (ii) transcription of the his structural genes is regulated by an attenuation mechanism that responds to the intracellular concentration of His-tRNAHis. The concentration of His-tRNAHis is, in turn, determined by cellular histidine concentration, histidyl-tRNA synthetase activity and amount, and chromosomal DNA supercoiling levels in response to anaerobiosis and osmolarity (Tables 2 and 3; see below).
FIGURE 2.
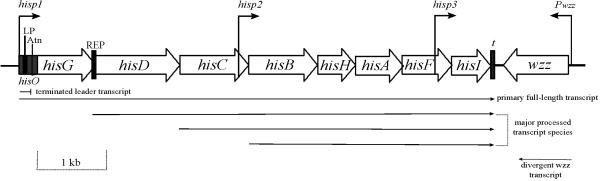
Diagram of the his operon in E. coli and S. typhimurium drawn to scale. hisp1, his operon primary promoter; LP, leader peptide; Atn, his attenuator; hisp2 and hisp3, his operon internal promoters; t, terminator at the end of operon; REP is a repetitive extragenic palindromic element that is present between hisG and hisD in S. typhimurium but not in E. coli; Pwzz, promoter of wzz gene. Arrows at the bottom of the figure show the length of the transcripts.
TABLE 3.
Parameters of the his operon structure and regulation
| Parametera | Comments | Organismb | Reference(s) |
|---|---|---|---|
| Gene order; length | OGDCBHAFI; 7,389 nt OG-(REPb)-DCBHAFI; 7,438 nt |
E.coli
S. typhimurium |
(49) |
| Size of full-length primary transcript: hisp1 to t | 7,337 nt (DNA sequence) 7,390 nt (DNA sequence) ≈7,300 (Northern blot) |
E.coli S. typhimurium E.coli and S. typhimurium |
(5, 49) |
| Primary transcript half-life | ≈4 min | S. typhimurium | |
| Sizes of major processed transcripts on Northern blots | ≈6,300 (hisDCBHAFI) ≈5,000 (hisCBHAFI) ≈3,900 (hisBHAFI)c |
S. typhimurium | (5) |
| Size of major terminated leader transcript: hisp1 to A | 180 nt 177 nt |
E.coli
S. typhimurium |
(49, 59, 99, 100) |
| Sizes of secondary transcriptd |
hisp2 to t 3,943 nt 3,888 nt hisp3 to t 809 nt 763 nt |
E.coli
S. typhimurium E.coli S. typhimurium |
(5, 6, 49) |
| % of transcripts from hisp2 relative to hisp1 in wild-type bacteria | ≤10% (and hisp2 is stronger than hisp3) | S. typhimurium | (5) |
| Established modes of operon regulation | ppGpp stimulation of transcription initiation from hisp1 Attenuation control in response to His-tRNAHis concentration, which changes in response to histidine supply and chromosomal DNA supercoiling density (see text) |
E. coli and S. typhimurium (but mainly S. typhimurium) | See text |
| Type of terminator (t) at end of operon | Bidirectional, Rho factor independent | E. coli and S. typhimurium | (31, 52) |
hisp1, his operon primary promoter; A, his attenuator; hisp2 and hisp3, his operon internal promoters; t, terminator at the end of operon (see Fig. 2)
REP is a repetitive extragenic palindromic element that is present between hisG and hisD in S. typhimurium but not in E. coli (123, 207).
Determined from the DNA sequences and by Northern blotting and SI mapping.
When E. coli and S. typhimurium are growing in a nutrient-rich medium, there apparently is an advantage in decreasing the amounts of the histidine biosynthetic enzymes by about four fold (Table 2). This metabolic regulation, which gears histidine biosynthesis to cellular growth rate, appears to be mediated by ppGpp and is independent of His-tRNAHis-specific attenuation control (see below) (184, 193, 205, 223). Surprisingly, in vivo his operon expression is largely unaffected by the presence of histidine in the growth medium (Table 2), despite the potential for a wide range of control by His-tRNAHis-specific attenuation. This unusual feature of his operon expression partly reflects the fact that histidine addition does not greatly increase the percentage of tRNAHis molecules charged with histidine (Table 2). Consequently, even when exogenous histidine is absent from growth media, the amount of charged tRNAHisis still relatively high. Another surprising feature of his operon attenuation is that even when 77 to 88% of tRNAHis molecules are charged with histidine, there is significant readthrough transcription beyond the his attenuator. The high basal level of wild-type his operon expression is most readily apparent in bacteria containing mutations that increase his operon attenuation. These mutations, which prevent translation of the his leader peptide or formation of the antiterminator RNA secondary structure (see below), completely prevent transcription of the his operon structural genes and cause histidine auxotrophy (126, 127). Clearly, there is a much greater potential to limit his operon expression by attenuation than is actually used in the bacterium growing under the laboratory conditions tested so far. A mechanism that might contribute to setting the basal level of his attenuation is described in the section on regulation of the his operon. The need to maintain relatively high cellular concentrations of the histidine biosynthetic enzymes may be related to the high affinity of the histidine periplasmic transport system and is discussed near the end of this review.
The attenuation mechanism also modulates his operon expression in response to physiological conditions that change chromosomal DNA supercoiling density, such as anaerobiosis and osmolarity (169). his operon expression is found to increase when the bacterial chromosome relaxes (i.e., is less negatively supercoiled), as occurs in DNA gyrase mutants or in wild-type bacteria during growth in the presence of oxygen and low osmolarity or upon addition of DNA gyrase inhibitors, such as novobiocin (169). For example, in stationary-phase cells, novobiocin addition increases his operon expression about 10-fold, and this effect is reversed by salt addition or anaerobiosis. Even without novobiocin addition, his operon expression is reduced by high osmolarity or anaerobiosis (169). This experimental result does not support a prediction from a recent metabolic network model that his operon expression should increase during oxygen deprivation (190). Other experiments with mutants lacking the his attenuator region (Δhis01242) or defective in ppGpp synthesis (relA) indicated that supercoiling control of his operon expression occurs mostly by changes in attenuation rather than by changes in the frequency of transcription initiation from the primary promoter hisp1 (see below). Several lines of investigation support the idea that supercoiling control of his operon attenuation is mediated by changes in the total cellular content of tRNAHis molecules, encoded by the HisR gene (90, 169, 187). The regulation of hisR by DNA supercoiling and the identification of the early his regulatory loci, hisW and hisU(I), as mutant alleles of gyrA and gyrB, respectively, are considered later in this review.
The Histidine Biosynthetic Enzymes
Since the last version of this review, there has been an explosion of knowledge about the structure and biochemical mechanisms of most of the histidine biosynthetic enzymes. Table 1 compiles these recent references and some of the older ones whose results have held up well over time. The biochemistry and structural basis for allosteric feedback inhibition of the HisG ATP-PRTase is presented above. In this section, I briefly summarize some of the most important conclusions about the other histidine biosynthetic enzymes from these recent structure and mechanism papers. Where possible, I focus the scope of this section on enzymes from E. coli and S. typhimurium. Some definitive structural and biochemical papers using enzymes from thermo-stable prokaryotes are discussed for enzymes that have proven difficult in the enteric bacteria. The references cited and the NCBI Entrez site should be consulted for the structures and additional details.
Bifunctional HisI
The carboxyl and amino terminal domains of the HisI enzyme catalyze the second PR-ATP pyrophosphohydrolase (sometimes called HisE) and third PR-AMP-cyclohydrolase steps, respectively, of histidine biosynthesis (Fig. 1 and above). Bifunctional HisI has been biochemically intractable (69). The protein tends to be unstable and has ill-defined metal coenzyme requirements. For this reason, the monofunctional PR-AMP-cyclohydrolase from Methanococcus vannielii was characterized biochemically (69), and the structure of HisI from Methanobacterium thermoautotrophicum was determined (200). The PR-ATP pyrophosphohydrolase (step 2) remains the only enzyme in the histidine biosynthesis pathway whose structure remains to be determined (V. J. Davisson, personal communication). The HisI PR-AMP-cyclohydrolase exists as a dimer that requires both Zn+2 and Mg+2 ions for catalysis (69, 200), although Cd+2 can replace Zn+2 with some change in catalytic properties. The active site of the PR-AMP-cyclohydrolase is formed by the dimer interface, and each Zn+2 is bound by three conserved Cys residues, one from one subunit and the other two from the other subunit (200) The Zn+2 ions bind with high affinity and likely play a direct role in catalysis by activating a water molecule, similar to the mechanism in nucleoside/nucleotide hydrolases (200). The Mg+2 ion is freely exchangeable, and there may be co-binding of Mg+2 and the substrate (69) Mg+2 is bound by conserved aspartate residues in the cleft of the active site, and Mg+2 may also play a role in the catalytic mechanism (200).
HisA
The fourth step of histidine biosynthesis is carried out by the HisA enzyme (Fig. 1). HisA catalyzes an Amadori rearrangement that leads to the irreversible isomerization of the aminoaldose Pro-FAR substrate to a more stable aminoketose product (see above and Fig. 1) (113). The structure of HisA is a (βα)8 barrel structure, which is a common fold shared by many enzymes, including other sugar isomerases, such as the TrpF enzyme involved in tryptophan biosynthesis (see (113, 129, 142, 145)). HisA and TrpF share almost no amino acid similarity, but remarkably, a single amino acid change in HisA (Asp127Val) imparts TrpF enzymatic activity (129). Like other (βα)8 barrel enzymes, the active site of HisA is located on the C-terminal ends of β strands, which form an inner parallel β-sheet. In HisA, aspartate residues at the C-terminal ends of β-sheets 1 and 5 are required for catalytic activity (113, 145). The Asp127Val mutation in HisA is thought to change the catalytic specificity by allowing the more negatively charged TrpF substrate to bind to the HisA active site (145). Kinetic and mutagenesis analyses have been performed on the HisA and TrpF enzymes of Thermotoga maritima and E. coli (113). These combined biochemical and structural studies suggest that the HisA and TrpF isomerases share a common catalytic mechanism that involves general acid-base catalysis (113). In addition, both enzymes seem to have high catalytic turnover that allows rapid consumption of their relatively unstable aminoaldose substrates. Finally, the extraordinary plasticity of (βα)8 barrel enzymes was reiterated by the demonstration that a single mutation (Asp130Val) caused the T. maritima IGP synthase HisF subunit, which is folded into a (βα)8 barrel structure, to acquire TrpF isomerase activity (145). These finding supports the notion that the reaction specificities of current (βα)8 barrel enzymes evolved from an ancestral enzyme of broad substrate specificity (129, 145). An important corollary of this idea is that (βα)8 barrel enzymes might be engineered to acquire new activities (129).
HisH-HisF heterodimer
The HisH-HisF IGP synthase catalyzes the fifth step of histidine biosynthesis (Fig. 1; (10, 11, 136, 137, 164, 165)). HisH-HisF is a class I glutamine amidotransferase, which carries out two concerted reactions. The HisH subunit contains a flavodoxin-like fold and is a glutaminase that produces a protected ammonia molecule from glutamate (see (164)). The HisF subunit, which is folded into an (αβ)8-barrel structure, uses this ammonia in a carbon-nitrogen ligation and cyclization reaction that produces IGP and AICAR (Fig. 1) (164). The HisH-HisF IGP synthase was the last step worked out in the histidine biosynthesis pathway (137) and is noteworthy, because it carries out its reactions by a concerted allosteric mechanism that channels the ammonia product formed by the HisH subunit to the PRFAR substrate in the active site of the HisF subunit (see (11, 164)). No additional intermediates are synthesized by HisH in the absence of HisF, as was initially proposed (see (137)).
The concerted allosteric regulation and metabolic channeling prevent waste of glutamine and protect the ammonia molecule for the second step in the reaction (see (11, 164)). The current model for the mechanisms of catalysis, regulation, and channeling are based on the structures of the enzyme from yeast (61, 165) and Thermotoga maritima (80), biochemical analyses, and extensive modeling approaches (see (11, 164)). Briefly, glutamine is thought to bind to the HisH active site that contains a conserved cyteine-histidine-glutamate triad that catalyzes glutamine hydrolysis (164). Yet, the HisH glutaminase remains essentially inactive until the other substrate, PRFAR, binds to the active site of the HisF barrel, which is about 30 Å away. PRFAR binding to HisF stimulates the rate of the distant HisH glutaminase by about 5,000-fold to a robust kcat ≈ 7 s-1 (11, 165). PRFAR binding occurs by electrostatic interactions to the C-terminal face over the hole of the HisF beta barrel. Binding of PRFAR induces an ordering of an unstructured loop in HisF that is communicated directly to the interface between the HisH and HisF subunits by a network of evolutionarily conserved residues. Subsequent formation of a key salt bridge leads to tightening of the interface between the subunits into a closed conformation, which opens an internal channel and increases the rigidity of the HisH active site, thereby stimulating cleavage of glutamine. Released ammonia is relayed from the HisH active site across the interface to the core of the HisF-beta barrel by the network of conserved residues, which exclude water. Following the cyclo-ligase reaction catalyzed by HisF, release of the IGP and AICAR products is thought to disrupt salt-bridges and hydrogen bonds in the subunit interface and loosen the subunit interface. Glutamate can then exchange from the HisH active site for a new glutamine substrate molecule, and the enzyme is set for another round of concerted reactions.
Recently, it was reported that acivicin, which is a natural product analog of glutamine, prevents the growth of E. coli in minimal medium (202). Acivicin is produced by Streptomyces sviceus and is being tested as an antitumor compound based on its inhibition of mammalian amidotransferases (see (116)). Growth of E. coli was restored by the addition of purines or histidine, which suggested that acivicin was inhibiting the HisH-HisF IGP synthase. Enzyme assays containing purified HisH-HisF confirmed that acivin is a potent inhibitor of IGP synthase (202). Other studies showed that acivicin inhibits HisH-HisF activity by a mechanism-based formation of a covalent bond between acivicin and the active-site Cys residue of HisH (64). Interestingly, inactivation is enhanced by binding of the PRFAR substrate to HisF, presumably by the same allosteric mechanism that activates the glutamine cleavage (64). Microarray analyses of transcripts levels in bacteria treated with the drug confirmed a block in histidine biosynthesis, including induction of stringent response, but also showed a complex pattern of transcript changes, indicative of other targets (202).
There is one other long-standing, curious observation concerning the HisH-HisF IGP synthase. Overexpression of HisH-HisF is highly pleiotropic and inhibits cell division of E. coli and S. typhimurium, particularly in the presence of high concentrations of sugar sources (102, 163). Induction of the SOS response or production of AICAR cannot account for these defects (93, 102). It was suggested that HisH-HisF overproduction may cause a partial block in cell septum formation (98). This idea was supported by the observation that D-cycloserine suppresses the filamentation caused by HisH-HisF overexpression (47). D-cycloserine inhibits D-alanine ligase and racemase, thereby curtailing production of complete five amino acid stem peptides. Therefore, peptidoglycan precursor amount is somehow linked to the filamentation of HisH-HisF mutants, but the mechanism remains unclear. Finally, it had been noted that some strains of S. typhimurium form filaments inside of host cells, such as macrophages (115). Strains which have these growth defects contain a non-polar mutation in hisG and the growth-arrest phenotype depends on HisH-HisF expression (115). Likewise, some mutations outside of the his operon that lead to filamentation inside of host cells, such as ΔtrpC insertions, also depend on possible HisH-HisF over-expression. However, it is not clear whether triggering the his operon to overexpress HisH-HisF and inhibit bacterial cell division plays any role in host defense.
Bifunctional HisB
The C-terminal and N-terminal domains of HisB protein catalyze the sixth (IGP dehydratase) and eighth (Hol-P phosphatase) steps, respectively, in the histidine pathway (Fig. 1 and above). The two domains seem to function independently and are found as separate enzymes in many other species (see (181)). Fusion to form bifunctional HisB has been proposed to be a recent evolutionary event (45). The structure of the monofunctional IGP dehydratase was determined from the fungus Filobasidiella neoformans (197), and the structure of the N-terminal domain of HisB (HisB-N) was determined for E. coli (181).
The IGP dehydratase removes a non-acidic hydrogen atom from IGP (Fig. 1), which contrasts to the leaving hydrogen in most other dehydratase reactions (197). IGP dehydratase does not show any amino acid similarity with other known proteins and has the unusual property that it requires Mn+2 ion for activity and formation of higher 24-mer oligomeric structures (197). In the absence of Mn+2, IGP dehydratase crystallizes as a trimer in which each monomer contains a compact domain that is formed by half-domains of an unusual topology (197). Each half-domain contains a histidine-rich motif that is highly conserved in IGP dehydratases. The two half domains likely arose by a duplication event. Modeling of the trimer into a 24-mer places the conserved sites into one site that likely is the Mn+2 binding site and possibly the active site (197). The structure of IGP dehydratase has allowed a better understanding of the aggregation state of bifunctional HisB in E. coli and S. typhimurium (see below). In addition, plant IGP dehydratase is a target for herbicides, and the structure will further efforts to improve the potency and specificity of these chemicals (103, 191).
In contrast to monofunctional IGP dehydratase, the HisB-N domain, which catalyzes HolP phosphatase in the presence of divalent cations, forms a dimer in solution and crystals (181). However, the intact bifunctional HisB protein forms large aggregates in the presence of Mn+2 due to oligomerization of the HisB-C IGP dehydratase domain (181, 197). Importantly, oligomerization of the intact HisB protein does not affect the Hol-P phosphatase activity. A model based on the structures of the individual HisB-N and HisB-C domains was constructed (181). In this model, bifunctional HisB forms a 24-mer in which the HisB-N and HisB-C domains are matched, and hexamers are the basic unit within the structure.
The structure of the dimeric His-N phosphatase domain was determined in the presence of Mg+2 ion, Ca+2 ion, Mg+ + histidinol, Ca+2 + pAsp, or Mg+2 + sulfate ion. These different structures represent “snapshots” of various intermediates in the phosphatase reaction pathway. Each monomer of HisB-N folds into a single domain with the structure of a haloacid dehalogenase (HAD) superfamily enzyme (45, 181). Each monomer contains the four invariant aspartate residues (DDDD) involved in catalysis, which proceeds through a phosphoaspartate intermediate. Zn+2 ion stabilizes an extended loop in the HisB-N structure. The active site contains two divalent cation binding sites, and the phosphatase is active in the presence of Mg+2, Mn+2, Co+2, or Zn+2, but is inhibited completely by Ca+2. The structures suggest that the second bound divalent cation participates in the reaction mechanism, which is different from other characterized HAD enzymes (181). The proposed mechanism also suggests how Ca+2ion, which has a relatively large atomic radius, binds to the two metal binding sites, distorts the active site, and arrests the reaction after formation of the phosphoaspartate intermediate.
HisC
Aminotransferases homologous to HisC are widespread in amino acid biosynthetic pathways and catalyze the transfer from glutamate of an amino group that eventually becomes the α-amino group in the amino acid end product of the pathway (Fig. 1 and above). Aminotransferases use the vitamin B6-derived coenzyme pyridoxal 5’-phosphate in a reaction mechanism involving formation of a Schiff’s base (see (111, 158, 199) and Chapter XX). The structure of the E. coli HisC aminotransferase, which catalyzes the seventh step of histidine biosynthesis (Fig. 1), was determined complexed with pyridoxamine 5’-phosphate, as an internal covalent aldimine with pyridoxal 5’-phosphate, or as a covalent complex with pyridoxal 5’-phosphate and HolP (199) or complexed with HolP or N-(5’-phosphopyridoxyl)-L-glutamate (111). HisC exists as a dimer, and each monomer consists of a larger α/ β/ α domain that binds pyridoxal 5’-phosphate, a smaller domain, and an N-terminal dimerization arm (111, 199). The residue forming the Schiff’s base with PLP is Lys214 or Lys217 in E. coli or S. typhimurium HisC, respectively, and the HolP substrate binds in the dimer interface at a site that partially overlaps the glutamate binding site (199). The spectroscopic properties of wild-type and mutant HisC are consistent with the idea that bond strain within the Shiff’s base lowers its pKa and contributes to catalysis (158).
The structure of HisC is similar to that of other aminotransferases, despite weak homology at the amino acid level, and amino acids involved in pyridoxal 5’-phosphate binding, substrate binding, catalysis, and dimerization are conserved in different aminotransferases (111, 199). Binding of Hol-P, pyridoxamine 5’-phosphate, or pyridoxal 5’-phosphate to HisC does not cause large structural changes in the protein or ligands (199). The covalent tetrahedral complex formed between HisC, pyridoxal 5’-phosphate, and HolP resembles a gem-diamine structure, which is an expected intermediate in the reaction (199). This is the first report for any aminotransferase of the structure of this normally transient intermediate, which may have been stabilized by crystal packing forces (199). Finally, amino acid sequence and protein structure comparisons suggest that the HisC aminotransferase is closely related to the PLP-dependent CobD decarboxylase from the cobalamin biosynthetic pathway of S. typhimurium, suggesting a common evolutionary origin (62, 63). In addition, the HisC HolP aminotransferase from Thermotoga maritima has evolved the ability to bind aromatic amino acids and functions both as a HolP and aromatic amino acid aminotransferase (88).
Bifunctional HisD
HisD catalyzes the final two steps of histidine biosynthesis (Fig. 1 and above). The crystal structure of the HisD homodimer unbound and bound to the substrate histidinol and cofactors NAD+ and Zn+ has been recently reported (26). This paper also reviews the proposed reaction mechanism of the four-electron oxidation reaction catalyzed by HisD (26, 210). Each monomer of the HisD dimer consists of four domains; two make up a globule structure containing incomplete Rossmann folds, and two make up a tail-like structure. The dimer is formed by domain swapping in the tail structures. The incomplete Rossmann folds, some sequence similarity, and tandem arrangement suggest that the two domains in the globule structure arose by a duplication event. The active site of the enzyme is formed by amino acids from both of the monomers that form the dimer. One of the Rossmann folds is involved in NAD+, binding, whereas the other Rossmann fold binds Zn+2 ion and the substrate histidinol. Zn+ functions in substrate binding, but not catalysis, and is required for histidinol substrate binding. NAD+ binds relatively weakly to the Rossmann fold in an unusual way compared to a canonical NAD+-Rossmann fold structure (26). The structures of HisD are consistent with a mechanism for HisD catalysis that was proposed earlier based on kinetic and biochemical studies of wild-type and mutant HisD proteins (210). This acid-base catalysis mechanism involves four bases, including conserved histidine residues (26, 210). The crystal structure also refines the stereochemistry of the NAD+ reactions.
STRUCTURE AND REGULATION OF THE his OPERON
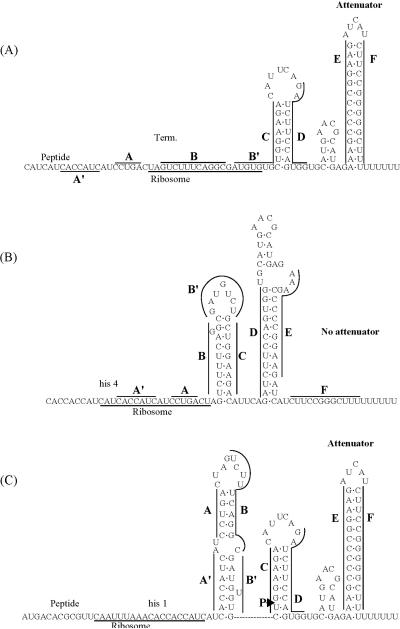
The his operon located at 42 min in the S. typhimurium chromosome is depicted in Fig. 2. The S. typhimurium and E. coli his DNA sequences are 81% identical, and the structure of the E. coli his operon at 44 min (2) is essentially the same as that of S. typhimurium, with the exception of a repetitive extragenic palindromic (REP) sequence between S. typhimurium hisG and hisD and several other minor differences (49). The locations of some his regulatory loci genes are also listed in Table 1. Parameters relevant to his operon structure and control are collected in Table 3 and have not changed since the last version of this review. They give the following general picture of the operon. The eight structural genes are transcribed into a single, polycistronic mRNA molecule (Table 3) (49, 153), which extends from the primary promoter (hisp1) to the strong, bidirectional Rho-independent terminator (Fig. 2). The frequency of transcription initiations at hisp1 is positively regulated by a limited range of intracellular ppGpp concentrations. New results indicated that this stimulation by ppGpp occurs by a direct mechanism (66, 175). An RNA polymerase molecule that initiates transcription at hisp1 first transcribes a leader region and must continue transcribing past the attenuator control site if it is to enter the first structural gene (hisG) of the operon. The hisp1 promoter, leader region, and attenuator are contained in a genetic locus which traditionally has been designated the hisO region, even though the operon is not controlled by a classical repression mechanism (Fig. 2 and 3). Transcription termination or readthrough at the his attenuator will occur when the percentage of tRNAHis charged with histidine is high (88%) or low (≈12%), respectively (126, 132, 146). In addition, the percentage of transcription termination may be modulated indirectly by chromosome DNA supercoiling density, which is set by the total cellular concentration of tRNAHis molecules (90, 169, 187). Transcription termination at the his attenuator produces a terminated leader transcript of approximately 180 nucleotides (nt), which extends from hisp1 to a site in the attenuator region (59, 99). In some instances, transcription initiation can occur at two internal promoters (hisp2 and hisp3, Fig. 2) located before the start of the hisB and hisI genes; however, transcription from hisp1 occludes expression from hisp2 in vivo (5). The polycistronic hisOGDCBHAFI primary transcript is processed in several discrete endonucleolytic steps (Fig. 2 and Table 3) (5). The processed transcript containing hisBHAFI has a chemical half-life of about 15 min and is much more stable than the hisOGDCBHAFI primary transcript, whose half-life is about 4 min (5). These features of his operon structure and regulation are described in detail in the next sections.
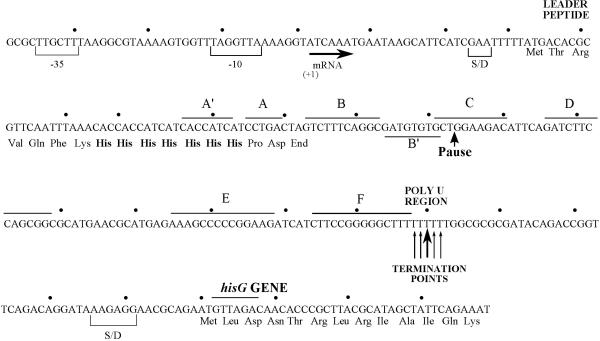
FIGURE 3.
DNA sequence of the hisO region in S. typhimurium. The -35 and -10 regions of the hisp1 promoter and the start point of transcription are indicated. S/D designates the weak and strong Shine-Dalgarno sequences in the translation initiation regions for the his leader peptide and hisG polypeptide, respectively. A′, A, B, B′, C, D, E, and F refer to segments of the his leader transcript that can base pair to form the alternative, mutually exclusive RNA secondary structures shown in Fig. 4. The site of pausing by RNA polymerase during in vitro transcription of the his leader region is indicated (59). The major (heavy arrow) and minor (light arrows) are points of transcription termination in the his attenuator region. Figure is redrawn from reference (25).
Structure of the his Operon
The Primary Promoter hisp1
Figure 3 presents the nucleotide sequence of the S. typhimurium hisO region (25, 29, 49, 184). The start of transcription determined in vitro and in vivo and the corresponding -35 and -10 regions for binding of σ70 RNA polymerase are indicated (29, 49, 78, 99, 100). There is an AT-rich “discriminator” sequence between the -10 box and the +1 start of transcription that is required for ppGpp stimulation of transcription from hisp1 (70, 184). The nucleotide sequence of the E. coli hisp1 promoter is identical to that of the S. typhimurium promoter in its -35 and -10 regions and is similar in the 18-nt -30 to -14 spacer and in the -6 to +1 discriminator region. The start of transcription at the E. coli hisp1 promoter in vivo and in vitro is analogous to the position indicated in Figure 3. his-lac fusions were used to identify mutations that decrease the efficiency of the hisp1 promoter by 4- to 400-fold compared with the wild-type promoter. These down mutations alter the sequences of the -35 or -10 regions or the spacing between these two regions and thereby confirm the position of hisp1 (25). The hisp1 promoter is unusually strong in vivo and in vitro on supercoiled templates (218). The E. coli hisp1 promoter was cloned into a galK expression vector and found to be about four times stronger than the gal promoter in vivo (180). The strength of hisp1 in vivo is further indicated by the result that the histidine biosynthetic enzymes amount to at least 4% of the total cellular proteins in bacteria deleted for the his attenuator (174). Results from in vitro experiments confirm that hisp1 is stronger than a pBR322 promoter and show that transcription from hisp1 is about 20-fold stronger on supercoiled templates than on linear templates (218). However, his-lac expression from a mutant lacking the his attenuator is decreased slightly by high osmolarity and anaerobiosis, which are thought to increase chromosomal negative supercoiling (169). The inference from this result is that transcription from hisp1 is not strongly affected by the changes in chromosomal DNA supercoiling density that occurs in vivo in response to physiological conditions. The effects of ppGpp on transcription from the hisp1 promoter are discussed below.
The Internal Promoters hisp2 and hisp3
The positions of two internal promoters have been evolutionarily conserved in the his operons of E. coli and S. typhimurium (49, 106, 189). By using Tn10 insertions to block transcription from upstream regions in the S. typhimurium his operon, hisp2 was initially mapped near the end of the hisC coding region before the start of hisB and hisp3 mapped near the end of the hisF coding region before the start of hisI (189). The S. typhimurium and E. coli hisp2 promoters were S1 mapped at about 100 bp upstream from the end of the hisC coding region (106), very close to a major processing site of the his primary transcript (5). Experiments in which transcription from hisp2 was measured in mutants lacking transcription from hisp1 or in which transcription from hisp1 or hisp2 was measured in galK expression vectors showed that hisp2 is about half as strong as hisp1 (84, 106). This relatively strong expression contrasts with the low (<10%) expression from hisp2 observed in vivo compared with the amount of his primary transcript initiated at hisp1 and transcribed through the attenuator (5). An indirect method based on ratios of enzyme activities suggested that transcription from hisp2 is insignificant when transcription of the his structural genes is equal to or greater than the level found in wild-type bacteria (84). Together, these results imply that transcription of hisp2 is occluded by transcription from hisp1. This hypothesis has been confirmed by studies showing that the level of hisp2 transcription is inversely correlated with the amount of readthrough transcription beyond the his attenuator (5).
The internal promoter in the trp operon is thought to maintain sufficient levels of the TrpC and TrpB polypeptides, each of which contains multiple tryptophan residues. The TrpE and TrpD polypeptides, which are encoded by genes upstream from the internal promoter, contain few tryptophan residues and would not be seriously depleted by sudden tryptophan starvation (225). An analogous function for the hisp2 and hisp3 internal promoters seems unlikely because the HisG polypeptide, which is encoded by the first gene in the operon, contains multiple histidine residues (49, 177). In both E. coli and S. typhimurium, the hisp2 promoter is metabolically regulated (106, 222); however, the physiological functions of the his internal promoters remain unknown. This mystery is compounded by the fact that the promoter-distal segment of the primary his transcript is processed into a long-lived hisBHAFI transcript that contains the same intact genes as transcripts initiated at the hisp2 promoter (5). It would seem that for some unknown physiological reason, E. coli and S. typhimurium evolved mechanisms to maintain expression of the last five genes of the his operon (5).
his operon structural genes and evolution
The complete DNA sequences of the E. coli and S. typhimurium his operons have been determined and compared (25, 29, 46, 48, 49, 105, 183). The his operons of both species are extremely compact, except for the expected space between the end of the his leader peptide and hisG and in the hisG-hisD intercistronic region. In fact, there is only one small 5-bp intercistronic region between hisG and hisD in the entire 7,379-nt E. coli his operon; for all the other gene pairs, the translation stop codon of the upstream gene overlaps the translation initiation codon of the next downstream gene (49). Likewise, except for hisG and hisD, the reading frames of all of the S. typhimurium his genes overlap. The ≈100-nt S. typhimurium hisG-hisD border contains a REP DNA sequence that is found throughout the bacterial chromosome (49, 117, 207). The absence of a REP sequence in the E. coli hisG-hisD intercistronic region supports the idea that such sequences arose through transposition (117). The S. typhimurium hisG-hisD REP sequence has been shown to act as a joining point for chromosomal rearrangements, including the generation of tandem duplications in recA+ bacteria (195).
Overlapping of translational signals raises the possibility that extensive translational coupling occurs during the expression of the his operon. In the trp operon, translational coupling between the trpE and trpD genes is thought to guarantee equimolar synthesis of the corresponding gene products, which interact to form a multimeric enzyme (225). In S. typhimurium, the HisG, HisD, HisC, and HisA polypeptides are expressed in molar ratios of 3:1:1:1 (221). Because of the large intercistronic region, direct translational coupling would not be expected between hisG and hisD. Some level of translational coupling between other genes in the operon could produce the equimolar synthesis of the HisD, HisC, and HisA polypeptides detected in vivo. Translational coupling might also influence the kinetics of his operon expression after the onset of histidine limitation; perhaps the sequential appearance of the histidine biosynthetic enzymes in cells with low formylating capacity reflects translational coupling, whereas the simultaneous appearance of the histidine biosynthetic enzymes in cells with high formylating capacity indicates a mode of uncoupled, independent translation (32). Unfortunately, whether translational coupling occurs in the his operon expression remains unresolved (49).
Numerous recent studies have analyzed the distribution and evolution of the his operon and histidine biosynthetic genes. A couple of generalizations can be drawn from these reports that are relevant to this review. Compact attenuation-regulated his operons similar to those of E. coli and S. typhimurium occur in γ-proteobacteria, including pathogenic Yersinia and Vibrio species (219); however, there are orders of γ-proteobacteria, such as Pseudomonas species, whose histidine biosynthetic genes are scattered around the chromosome and that lack obvious attenuation control (219). This variety of structures and regulation is also evident in gram-positive bacteria, which contain different patterns of his gene arrangements and regulation, including histidine-specific T-boxes (219). Moreover, some species within an order synthesize histidine (e.g., Streptococcus mutans), whereas other species do not contain histidine biosynthetic genes (e.g., Streptococcus pneumoniae).
Two scenarios have emerged for the evolution of unified, compact his operons, such as those of E. coli and S. typhimurium. One analysis was based only on his gene arrangements in proteobacterial species (86). According to this scenario, ancestral his genes encoded monofunctional enzymes and were scattered throughout the chromosome. These separate his genes then underwent a series of joining steps that built highly compact his operons, such as those present in E. coli and S. typhimurium. These compact his operons then transferred horizontally among proteobacterial species (86). A later study included the organization of his genes from phyla outside of the Proteobacteria (179). The major conclusion from this analysis was that the compact his operon is widely distributed and ancient (179). According to this scenario, the unified his operon emerged early, was transferred horizontally, and was subsequently broken up in some bacterial species. Beyond this difference in conclusions about the origin of the unified operon, other analyses persuasively argue that gene elongation events (HisA), gene duplications (HisA and HisF), and gene fusions (leading to biofunctional HisI and HisB proteins) occurred during the evolution and assembly of the histidine biosynthetic pathway in E. coli and S. typhimurium (see (85)).
his operon expression and intracellular formylating capacity
A limited number of studies suggest that intracellular formylating capacity and its modulation of initiator fMet-tRNA pools may influence his operon expression (see (7)). Premature transcript 3′ ends were mapped in the hisC region in response to the drugs trimethoprim and kasugamycin, which decrease the intracellular fMet-tRNA pool and the binding of fMet-tRNA to ribosomes, respectively, and thereby uncouple transcription and translation (7). Both antibiotics lead to premature his transcript release, possibly by increasing Rho factor-dependent transcription termination within the operon (see below) and RNA polymerase pausing (7). Trimethoprim, and to a lesser extent kasugamycin, also led to an increase in his transcript processing in the hisC-hisB region (7), possibly by exposing a transcript processing site within hisC or by fostering ribosome stalling at the start of hisB (see below). Another link between histidine biosynthesis and intracellular formylation involves production of AICAR (Fig. 1), which must be formylated to reenter the nucleotide pool (7, 36). Increased AICAR production may reduce intracellular formylating capacity (7, 32), and this reduction may account for the increased his transcript release and processing observed in his constitutive and nonpolar hisD and hisC mutants (7).
his transcript processing by RNaseE and RNaseP
Three major processed his transcripts were detected by Northern (RNA) blotting (Fig. 2 and Table 3). Processing of longer transcripts to form the 3,900-nt hisBHAFI transcript was studied in detail and shows some remarkable features (5, 8). The processed hisBHAFI transcript is extraordinarily stable (t1/2≈ 15 min) compared with the full-length his operon transcript (t1/2≈ 4 min) (Table 2) (5). A cis element in hisC upstream from the processing point is required for efficient transcript cleavage (5, 8). This cis element was recognized specifically by an RNA-binding protein in gel shift assays (5) and probably binds RNaseE or an endoribonuclease associated with RNaseE (8). In addition, RNaseE probably cuts the full-length transcript at several other sites within the hisC region (8). Ribosome binding to the translation start site of hisB, which is the first complete gene in the processed transcript, also was required for efficient processing and transcript stability (5, 8). Recent data support the model that the initial RNaseE cut in hisC and ribosome binding at the start of hisB cause the processed transcript to assume a specific secondary structure that is further cut by RNaseP (8). This is the first report implicating the RNaseP ribozyme, which mainly cuts pre-tRNA, in mRNA processing. The resulting hisBHAFI transcript has a 5′-end hairpin structure, which together with translation of hisB may enhance the stability of the 3,990-nt mRNA fragment. The processing pattern of the his primary transcript needs to be reconciled with the 3:1:1:1 stoichiometry of the HisG, HisD, HisC, and HisA polypeptides. The greater stability of the hisBHAFI processed transcript than of transcripts that contain other his genes, such as hisD, raises the issue of how equimolar amounts of HisD, HisC, and HisA polypepties are maintained, if the hisBHAFI transcript is indeed translated. Finally, the processing site between hisG and hisD (Fig. 2) and the presence of a REP sequence that can stabilize mRNA from 3′→ 5′ endonucleolytic attack (123) suggest one mechanism that could account for the higher amount of HisG protein than other his proteins in S. typhimurium (221).
A recent report suggests another role for RNaseP endonuclease processing of the his primary transcript (147). At a nonpermissive temperature, a temperature-sensitive mutant of RNaseP accumulated his mRNA extending from a region in front of hisG to the end of the operon (Fig. 1). However, transcripts corresponding to the hisO leader region did not accumulate in the mutant, and an RNaseP cleavage site was mapped to the intercistronic region between the hisO leader region and the start of hisG (147). This study also confirmed the RNaseP cleavage site near hisC mentioned above (8). Analogous RNaseP cleavage sites were detected in other operons (147) suggesting that RNaseP cleavage may control the stability of transcripts downstream from cut sites. Further studies of this phenomenon are needed for the his operon, especially direct demonstration that the accumulation is independent of tRNAHis amount and attenuation control.
Rho factor-dependent “classical” polarity within the his operon and roles of NusA and NusB
Mapping of cryptic transcription termination points within the his operon has provided an explanation for the strong gradient of classical polarity observed for hisG, but not for other his genes, such as hisD and hisA (3, 6). Nonsense mutations early, but not later in hisG, strongly reduce expression of downstream genes in the his operon; by contrast, nonsense mutations located throughout hisD and hisA strongly reduce downstream his operon expression (3, 6, 91). Premature translation termination within a coding region exposes potential mRNA targets to entry of Rho factor, which causes the premature transcription termination that underlies the decrease in downstream gene expression (3, 6). These Rho factor-dependent sites have been designated as transcription termination elements (TTEs) for the his operon (50). According to this model, the TTEs lie downstream of the premature translation stop codon and contain a transcription pause site which becomes the 3′ transcript end produced by Rho-dependent transcription termination. Each TTE in the his operon also contains a cytosine-rich and guanosine-poor region upstream from the pause site that serves as an entry for binding of the Rho factor hexamer. The locations of these TTEs correspond to the polarity patterns observed for his operon nonsense mutations. The only TTEs in hisG are located early in the gene; therefore, only nonsense mutations upstream of these entry sites are strongly polar. In contrast, TTEs are located throughout hisD and only at the end of hisA; therefore, nonsense mutations essentially anywhere within hisD or hisA are upstream of a TTE (6). The polarity pattern within hisC is complicated by the transcript processing mentioned above. It is not clear whether the putative translational coupling described above might also contribute to the strong polarity gradient observed in hisG in contrast to the uniform polarity observed in the internal genes of the his operon (3, 6, 91).
Further study of transcription termination at TTE1 and TTE2 from hisG in the absence of translation revealed that the NusA and NusB transcription elongation factors modulate polarity at these sites. TTE2 is the stronger Rho-dependent termination site and functions independently of the NusB factor in vitro (50). In contrast, the suboptimal TTE1 site is preceded by a BoxA sequence, which binds to NusB and enhances transcription termination at TTE1 by Rho factor in vivo and in vitro (50). This finding was unexpected, because the NusB factor often acts as an antitermination factor. Further studies suggested an involvement of NusA and NusE along with NusB on efficient termination at TTE1 (51). This study also showed that NusA increases the efficiency of termination at TTE2 by increasing transcription pausing. A last interesting result from this study is that NusA is required even when there is normal translation of the his operon. Mutants deficient in NusA function showed increased transcription termination at the TTE2 site suggesting that NusA modulation of transcription elongation may be important in maintaining tight coupling between transcription and translation (51). Finally, the his TTE1 and TTE2 sites have been used in screens of potential antibiotics that inhibit Rho factor (53, 54).
The terminator at the end of the his operon
Transcription termination at the end of the E. coli and S. typhimurium his operons occurs at strong Rho factor-independent terminators (48, 52). The S. typhimurium terminator is located only a few nucleotides downstream from the hisI stop codon, whereas the E. coli terminator is preceded by a nontranslated region of 40 nt (49). The S. typhimurium his terminator is a symmetrical, mirror-image structure; each strand contains (reading 5 ′ to 3′) a G+C-rich inverted repeat followed by several T residues. This mirror-image structure suggested that the terminator might function in both orientations. This prediction was confirmed both in vivo and in vitro (52). Analysis of in vivo transcription termination points by S1 mapping and Northern hybridizations demonstrated that this structure terminates his operon mRNA initiated at the his primary and internal promoters and, at the same time, terminates a 1,200-nt-long transcript synthesized from the DNA strand opposite to the one copied into his mRNA. The gene convergent to the his operon was identified as rol, which regulates the size distribution of the O-antigen moiety of lipopolysaccaride (Fig. 2) (31). Both the his and wzz (rol) transcripts synthesized in vivo end with polyuridylate residues, as expected for Rho factor- independent transcription termination. The his/wzz terminator functions at greater than 90% efficiency in either orientation in vivo, and it does not seem likely that there is any regulatory connection between the his operon and wzz gene through this shared terminator. The Rho factor-independent nature and high efficiency of the his/wzz terminator in both orientations were confirmed in a purified in vitro transcription system.
Regulation of his Operon
Metabolic regulation
As noted above, his operon expression is about four fold greater in bacteria growing in minimal-glucose medium than bacteria growing in rich medium (Table 2). This inverse relationship between his operon expression and cellular growth rate is a form of metabolic regulation that adjusts his operon expression in response to the general amino acid supply in the cell (176, 205, 222). Because ppGpp levels were known to vary inversely with growth rate (see Chapter XX), ppGpp was examined as a possible positive effector of his operon expression. In an in vitro coupled transcription-translation system prepared from a relA- mutant defective in ppGpp synthesis (see Chapter XX), addition of 100 μM ppGpp caused a 10-fold increase in expression of the wild-type his operon contained on a linear, transducing phage template (205). Equal levels of ppGpp-mediated stimulation were detected from the wild-type template and from a mutant template deleted for the his attenuator, which showed that ppGpp acts independently of attenuation. By uncoupling transcription and translation in the in vitro system, it was possible to show that ppGpp stimulates his operon transcription but not translation. From these results, ppGpp was postulated to be the effector molecule that directly mediates metabolic regulation of his operon expression (205).
Results from physiological experiments strongly support the model for the role of ppGpp as a positive effector of his operon expression. The increase in expression of the his operon in bacteria subjected to sudden histidine starvation in amino acid-rich medium is markedly less in relA- mutants than that in relA+ strains (205). There is a positive correlation between in vivo his operon expression and intracellular ppGpp concentrations, up to the ppGpp level found in bacteria growing in minimal-glucose medium (186, 205, 222, 223); increases in in vivo ppGpp concentrations beyond this level fail to increase his operon expression (222). These results support the notion that his operon transcription is maximally stimulated at lower than maximum in vivo ppGpp concentrations. Positive control of his operon expression by ppGpp was strongly confirmed by genetic schemes in which mutants defective in ppGpp metabolism were selected on the basis of growth characteristics in the presence of histidine analogs that inhibit histidine biosynthesis (see below) (186). Experiments were performed in which intracellular ppGpp was drastically reduced below the level found in relA+ bacteria growing in rich medium (193). Results from this study show that attenuator-independent his operon expression decreases about 15-fold in a relA- mutant and increases about 2-fold in a relA+ strain in response to the decreased and increased ppGpp levels, respectively, induced by addition of the analog serine hydroxamate to the amino acid-rich growth medium. Thus, the full range of the ppGpp-mediated metabolic regulation of in vivo his operon expression is at least 30-fold. Finally, a number of physiological experiments have confirmed the conclusion that ppGpp-mediated metabolic regulation and attenuation are independent mechanisms for the control of his operon transcription (182, 184, 186, 193, 222, 223). A corollary of this conclusion, that starvation for amino acids other than histidine increases transcription of the his operon, has been confirmed (222). In these experiments, the amino acid limitation should not have interfered directly with leader peptide synthesis in a way that would increase read-through transcription of the his attenuator (see below). ppGpp-mediated metabolic regulation induces the expression of several other amino acid biosynthetic operons besides the his operon (55, 205). These earlier results were recently confirmed by microarray analyses of the E. coli stringent response, which activated the transcription of leader transcripts of several amino acid biosynthetic operons, including hisL (81). In this regard, it is particularly noteworthy that relA spoT double null mutants show a complex requirement for amino acids that can be met by high concentrations of Casamino Acids (224). The effect of ppGpp accumulation on HisG enzyme activity, which controls the flow of intermediates through the histidine biosynthetic pathway, was described above.
One of the most interesting questions about his operon metabolic regulation concerns the mechanism by which ppGpp stimulates transcription. In early studies, strains containing putative mutations in the hisp1 promoter showed altered levels of attenuator-independent his operon expression in response to amino acid downshifts (222). These results suggested that ppGpp affects transcription initiation frequencies at the hisp1 promoter; but, more rigorous interpretations were not possible at that time because the base changes of the mutations were unknown. Two lines of investigation have served as prelude to recent definitive experiments on the mechanism of ppGpp stimulation of his operon transcription. Systematic studies by Artz and coworkers of his promoter mutations in vitro in a coupled transcription-translation system and in vivo in the bacterial chromosome showed similar results and strongly suggested that stimulation of his operon expression by ppGpp occurs at or near the hisp1 promoter (70, 184). In addition, stimulation of ppGpp of hisP1 transcription depends on the sequence of the -10 region in the promoter and an AT-rich discriminator region between the -10 box and the start of transcription. For example, site-directed mutations that increase the matches of the -10 region of the hisp1 promoter to the consensus for a σ70RNA polymerase promoter increased transcription from hisp1 in the absence of ppGpp and largely abolished the stimulation measured from the wild-type promoter in response to ppGpp both in vitro and in vivo (70, 184).
Paradoxically, earlier attempts to detect activation of transcription by ppGpp in purified in vitro transcription systems proved to be largely unsuccessful (27, 28). Therefore, it was proposed that positive regulation of certain promoters in response to ppGpp could occur by an indirect, passive mechanism in which RNA polymerase is limiting and decreased transcription of the strong rRNA promoters in response to ppGpp makes RNA polymerase available for transcription of other promoters (27, 28). The model of indirect stimulation was tested by Choy in a coupled in vitro transcription-translation system using positively (hisp1) and negatively (leuVP) regulated promoters (66). Mixed template and titration experiments demonstrated that there was no inverse correlation between hisp1 activation and leuVP repression under conditions where RNA polymerase amount was made limiting. Furthermore, the extracts seemed to contain titratable factors that could become limiting for promoter stimulation or repression. Together, these results strongly suggested that in the extract system using supercoiled DNA templates, ppGpp-dependent activation and repression are independent, which does not support the passive model of his metabolic control (66).
Advances in understanding auxiliary factors and subunits used by RNA polymerase prompted Gourse and coworkers to reprise experiments on the effects of ppGpp on in vitro transcription from hisp1 (175). They showed that ppGpp can indeed significantly increase transcription from the hisp1 promoter when the potentiator factor DksA is added to the reaction mixtures containing purified RNA polymerase. Other factors, such as the omega subunit of RNA polymerase, also are important for proper function of ppGpp in purified in vitro transcription systems (220). Consistent with the in vitro transcription results, a ΔdksA mutant did not show stimulation of his operon transcription following amino acid limitation that increase ppGpp levels (175). Kinetic analyses suggest that the synergistic positive effect of ppGpp and DksA on the transcription at some promoters, such hisp1, may occur by increasing the rate at which RNA polymerase isomerizes to an open complex. Together, these combined results show that ppGpp positively stimulates transcription from the hisp1 promoter by a direct mechanism, although indirect effects of RNA polymerase concentration might still contribute to his metabolic control (175).
Attenuation Control
Histidine-specific and supercoiling-responsive control of his operon expression in E. coli and S. typhimurium is exerted through an attenuation mechanism. The analysis of mutations indicates that attenuation can potentially regulate his operon expression over a ≈200-fold range (42, 83, 127). Therefore, the combination of metabolic regulation (≈30-fold) and attenuation (≈200-fold) gives a rather extraordinary total potential range of 6,000 for his operon regulation. The discovery of his operon attenuation paralleled the work of Yanofsky and his associates on trp operon attenuation (see Chapter XX), and results from the two systems led to a rapid elucidation of the mechanism underlying attenuation. Histories of the discovery of his operon attenuation were published previously (25, 34). To avoid redundancy with other chapters on attenuation, only the specific aspects of his operon attenuation are presented here.
Model for his attenuation
The attenuation model proposes that transcription termination at the his attenuator is modulated by synthesis of a peptide encoded by the his leader region (29, 30, 34, 59, 78, 126, 127, 132). Except for details, the mechanisms that bring about the coupling of transcription termination and translation are formally similar for his and trp attenuation. The his mechanism relies on two critical features of the his leader transcript, which precedes the start of the hisG coding region (Fig. 3). First, the his leader transcript encodes a 16-amino-acid peptide that contains seven consecutive histidine residues. The paper by Barnes reporting that the his leader peptide contains seven tandem histidines (29) had considerable impact, because it dramatically confirmed the generality of the attenuation hypothesis proposed by Lee and Yanofsky (144). Second, a series of mutually exclusive, alternative secondary structures can form in the his leader transcript (Fig. 3 and 4). These stem-and-loop structures are formed by pairing between bases in segments A′ and B′, A and B, B and C, C and D, D and E, and E and F in the his leader transcript and are designated as A′A:B′B, B:C, C:D, D:E, and E:F, respectively (Fig. 3 and 4). Secondary structure E:F together with the polyuridylate residues downstream from it constitutes a strong Rho factor-independent terminator. If E:F forms, transcription termination occurs at one of the uridylate residues and produces a terminated leader transcript (Fig. 3, Fig. 4, and Table 3). In essence, control by attenuation amounts to varying the frequency at which the E:F terminator structure forms. Any factor, condition, or mutation that prevents E:F terminator formation acts as an antiterminator and allows an RNA polymerase molecule to continue transcription into the his operon structural genes. In wild-type attenuation, the antiterminator is an alternative RNA secondary structure D:E (Fig. 4) whose frequency of formation is determined by translation of the leader transcript.
FIGURE 4.
Mutually exclusive, alternative stem-and-loop structures that form in the his leader transcript as part of the attenuation process. Letters designate segments of the leader transcript that base pair to form secondary structures. Ribosomes labeled lines indicate the nucleotides masked by a ribosome stopped at a codon. The model for his attenuation is described in the text. (A) Transcription termination configuration caused by translation of the leader transcript to the UAG stop codon; (B) readthrough transcription configuration caused by ribosome stalling at the fourth histidine codon in the leader transcript; (C) transcription termination configuration caused by ribosome stalling at the Gln codon in the leader transcript. Configuration C will also occur in the absence of translation of the leader transcript or after rapid dissociation of the ribosome at the translation stop codon. Configuration C also indicates the pause site (arrowhead marked with P) where RNA polymerase temporarily halts elongation following synthesis of A:B (57, 59, 146). The role of pausing in synchronizing the attenuation mechanism is described in the text. Figure modified from reference (127).
Translation of the his leader transcript and formation of alternative secondary structures that signal transcription antitermination or termination are synchronized by pausing of RNA polymerase after formation of the A:B perfect stem-loop structure (Fig. 3 and 4) (57-60, 143). Formation of the upper A:B hairpin is part of the pause signal, and most of RNA segment B ′ probably remains base paired to the DNA template (59, 60, 143). In addition, extending the perfect A:B hairpin by one G:C bp reduces pausing, possibly by interfering with an interaction between the hairpin and RNA polymerase (60). Landick and co-workers have used the his pause site as the primary model to dissect the mechanism of transcription pausing (reviewed in (141)). Studies of the his pause site have led to the model of the elemental pause state, which involves an off-line rearrangement of the active-site of RNA polymerase that prevents nucleotide substrate addition and does not involve translocation (see (141, 214)). The his elemental pause is further stabilized by a multipartite signal that includes the A:B RNA hairpin, which is spaced 2-3 nt from the RNA:DNA hybrid, sequences in the DNA downstream from the pause site in the RNA polymerase active site and sequences in the RNA:DNA hybrid itself (57, 60, 141, 214) Using a combination of biochemical approaches, including single-molecule studies, crosslinking, chemical probing of complexes, and analyses of specific mutations, Landick and co-workers have recently proposed a model for the rearrangements in the active site of RNA polymerase that lead to formation of the elemental pause state and its stabilization by the signals listed above (214). In this model, the “trigger loop” in the active site of RNA polymerase plays roles in NTP loading, translocation, and catalysis during elongation. During pausing, the trigger loop is rearranged to fray the RNA 3’ template away from the DNA template (214). Finally, the in vitro half-life of paused RNA polymerase in the his leader is unaffected by ppGpp addition and is moderately increased by NusA protein, although no boxA motif is present in or near the A:B structure (59). Enhanced pausing in response to NusA protein seems to require the signals in the his leader transcript, including the A:B pause hairpin, but not the downstream DNA signal (59). Whether NusA affects pausing in vivo has not been reported.
To see how the attenuation mechanism works in vivo, consider bacteria growing in minimal-glucose medium containing histidine. Under these growth conditions, the intracellular concentration of histidine will approach 100 μM, and about 90% of the tRNAHis molecules will be charged with histidine (Table 2). An RNA polymerase molecule that initiates transcription at the hisp1 promoter will start to transcribe the his leader region (Fig. 3). A transcribing RNA polymerase molecule will pause at position 102 in the leader transcript partly as a result of A:B hairpin formation (Fig. 4C). Meanwhile, a ribosome will begin to translate the leader transcript and produce the leader peptide. Because the intracellular concentration of His-tRNAHis is high, the entire leader peptide will be synthesized, and the translating ribosome will move all the way to the stop codon at position 80 in the transcript. Ribosome movement to this position will disrupt A:B, thereby releasing the RNA polymerase molecule from its paused state, and will mask segments B and B′ in the transcript. When transcription resumes, structure C:D will have an opportunity to form first, preclude formation of structure D:E, and allow the E:F terminator to form (Fig. 4A). Once transcription termination occurs, the RNA polymerase and terminated leader transcript are released spontaneously from the DNA template.
If the translating ribosome rapidly dissociates from the stop codon, A′A:B′B will have an opportunity to form, prevent structure B:C formation, and still allow C:D and E:F to form (Fig. 4C). On the other hand, rapid dissociation of the ribosome at the instant the RNA polymerase molecule is released from pausing could allow synthesis of segment C before A′A:B′B has a chance to form. This situation would allow B:C formation, which will lead to readthrough transcription. B:C might also have a chance to form by re-equilibration of secondary structures if the transcribing RNA polymerase molecule pauses again in response to C:D formation. However, pausing was detected in a purified in vitro transcription system only after A:B hairpin synthesis (59, 143). In either instance, rapid release of ribosomes at the stop codon could account for the relatively high basal level of his operon expression detected in vivo in the presence of histidine.
Consider next bacteria moderately starved for histidine. Omission of histidine from minimal-glucose medium does not reduce the concentration of His-tRNAHis sufficiently (≈80% charged; Table 2) to cause significant read through of the wild-type his attenuator. However, mild histidine starvation can be induced genetically (e.g., reference (25)) or by adding the analog 3-amino-1,2,4-triazole (AT), which inhibits one of the HisB enzyme activities (Table 1). Under these conditions, a ribosome translating the leader transcript will stall at the tandem histidine codons, because the cellular concentration of His-tRNAHis is low (≤ 12% charged [112]). If a ribosome prevents secondary structure formation in a transcript for about 16 nt downstream from a codon in the aminoacyl site (25, 127), then ribosome stalling at histidine codons 3 to 5 will mask segments A and A′ and release the RNA polymerase molecule from its paused state. Segment C will then be synthesized, and subsequent B:C formation will preclude C:D formation, allow the antiterminator D:E to form, and allow transcription to continue past the his attenuator (Fig. 4B).
Antitermination can also be caused by reducing the concentration of His-tRNAHis molecules by decreasing the total cellular content of tRNAHis molecules (90, 146, 169, 187). In wild-type cells, changes in the absolute cellular concentration of tRNAHis molecules may be brought about by modulating the expression of hisR, the structural gene for tRNAHis (see below). Physiological conditions (e.g., aerobiosis or low osmolarity), antibiotics (e.g., novobiocin), or mutations (e.g., gyrA (hisW) or gyrB [hisU(I)]) that decrease negative chromosomal supercoiling are thought to reduce hisR expression. This, in turn, reduces the total cellular concentration of tRNAHis molecules, thereby decreasing the amount of His-tRNAHis and leading to increased his operon expression by readthrough of the his attenuator (90, 146, 169, 188). In contrast to control by histidine, regulation of his operon attenuation by supercoiling should be adaptive and slow, because changes in the total cellular content of tRNAHis molecules would require turnover or dilution of stable tRNA molecules by cell growth and division. Finally, if translation of the leader peptide is prevented by a mutation in the initiation codon, super-attenuation occurs. An RNA polymerase molecule transcribes the leader region, pauses after formation of A:B, eventually resumes transcription without assistance from a ribosome, and synthesizes segment C and then segment D of the leader transcript. Formation of C:D prevents D:E formation as the RNA polymerase molecule continues transcription, and the absence of D:E allows formation of the E:F terminator. Super-attenuation should also occur if a translating ribosome stalls upstream of the third histidine codon in the leader transcript, because a ribosome stalled so far upstream in the leader transcript will fail to disrupt A′A:B′B formation (Fig. 4C). It has been reported that in vivo attenuator-dependent his operon expression is less during severe histidine starvation than during mild histidine starvation (46). This observation can be explained by super-attenuation which results from occasional stalling of ribosomes at the first two histidine codons in response to severe histidine limitation (25).
Evidence and recent developments
The following features have been well established experimentally for the model of his attenuation: (i) response to the intracellular amount of His-tRNAHis rather than free histidine (146); (ii) nearly complete evolutionary conservation of the DNA sequence of the his leader regions of E. coli and S. typhimurium (126); (iii) existence of his terminated leader and his readthrough transcripts in vivo and in vitro (59, 99, 100, 132); (iv) formation of mutually exclusive, alternative RNA secondary structures in the his leader transcript (30, 34, 59, 83, 126, 127, 143); (v) translation of the his leader to form the encoded peptide (24, 25, 126, 127); (vi) masking of transcript segments by stalled ribosomes (25, 127); (vii) lack of function of the leader peptide other than to be translated as part of the attenuation mechanism (127); (viii) transcriptional pausing in vitro after A:B synthesis (57-60, 141, 143, 214); and (ix) the structure of the paused RNA polymerase complex following A:B synthesis (see (141, 214)).
Some of the most innovative work done on his attenuation since the last version of this review concerns the mechanism of pausing (see above). The setting of basal levels of his attenuation by rapid release of ribosomes and spontaneous release of the termination complex at the his attenuator are based on the model for trp attenuation (see Chapter XX) and have not been experimentally verified for his attenuation. A recent probabilistic model for attenuation predicts that attenuation is hypersensitive to changes in amino acid supply because of the synchronized start of this mechanism and the small frequencies of usage of the regulatory codons that encode leader peptides (82). The model predicts high basal expression and low sensitivity for the trp attenuator and large dynamic range and high sensitivity for his attenuation, which includes extra stem- loop structures in the his leader compared to the trp leader (Fig. 4). However, this model assumes that the basal level of his expression is low, which does not match the experimentally results described above. Nevertheless, this model suggests how the extra stem-loop structures in the his attenuator might influence the frequency of attenuation (82).
One issue that has been partially resolved is the postulated role of the HisG enzyme as a regulator of his operon expression. Suggestive results from a number of experimental approaches led to the conclusion that the HisG enzyme, which binds histidine and His-tRNAHis molecules, acts as an autogenous corepressor of his operon expression (157). Although this conclusion was instrumental in the formulation of the model of autogenous regulation (104), subsequent genetic and physiological experiments showed conclusively that HisG protein is not an essential component of the mechanism for his operon regulation (192). Therefore, the only known forms of his operon control are metabolic regulation and attenuation. Results from physiological experiments that implicated HisG protein as a putative corepressor probably reflect indirect effects on intracellular PRPP, ppGpp, or His-tRNAHis concentrations. Nevertheless, the HisG enzyme may play a direct but ancillary role in controlling his attenuation by acting as a regulated reservoir for His-tRNAHis molecules (135). It has also been noted, but not experimentally verified, that the his leader transcript has the potential to fold into a cloverleaf secondary structure that resembles the folded tRNAHis molecule (14), and this folded his leader might bind HisG or other proteins. The question of whether the HisG enzyme plays some auxiliary role in his operon regulation remains unanswered and still needs to be readdressed experimentally.
Role of the his Regulatory Loci
One of the early schemes devised for the selection of S. typhimurium mutants with high his operon expression relied on resistance to a combination of the analogs AT and 1,2,4-triazole-3-alanine (TRA) (185). AT inhibits the HisB dehydratase activity (Table 1) and reduces histidine biosynthesis, whereas TRA is mistaken for histidine in the cell and is charged onto tRNAHis and incorporated into proteins. Growth of wild-type cells is inhibited by a combination of analogs, because AT causes reduction of the histidine supply below a critical level and TRA incorporation presumably inactivates proteins. High expression of the his operon caused by mutations can overcome the effect of the analogs and allow the mutant bacteria to grow. Because the absolute amount of His-tRNAHis controls the level of his attenuation, mutants containing defects in tRNAHis biosynthesis were selected by resistance to a combination of AT and TRA. The genes affected in these mutants include: hisR (structural gene for tRNAHis), gyrA (hisW) and gyrB (hisU(I)) (subunits of DNA gyrase), hisS (aminoacylation of tRNAHis with histidine), truA (hisT) (pseudouridine modification of the anticocodon stem-loop of tRNAHis), and rnpA (hisU(II)) (RNaseP) (processing of tRNAHis and the his mRNA transcript). Accounting for the high his operon expression in these mutants played an important role in working out the mechanism of attenuation control. It was also found that relA mutants, which are deficient in ppGpp biosynthesis, are sensitive to AT, and spoT mutations, which are defective in ppGpp breakdown, suppress the AT sensitivity of relA mutants (186). Moreover, spoT mutants, which contain increased ppGpp levels, were found to be resistant to TRA and low concentrations of AT, provided that amino acids other than histidine were added to minimal-glucose media (186).
Genetic schemes were also developed to select for decreases in high-level his operon expression. Selection for decreased his operon expression has been particularly useful in locating suppressors of mutations in the his regulatory loci (e.g., see reference (90). These schemes rely on the complicated phenotypes caused by increased his operon expression, including changes in colony morphology and growth inhibition by temperature, adenine, or high salt (90). As mentioned above, some of these phenotypes may be partially attributable to overexpression of the HisH and HisF proteins (163); however, in most cases, the phenotypes remain incompletely understood.
It is beyond the scope of this chapter to describe in detail the molecular genetics of each his regulatory locus; however, some features of the his regulatory loci that are relevant or unique to the topic of histidine biosynthesis are described. Additional information about the his regulatory loci and relA, relC, and spoT is found in other chapters.
hisR
The hisR gene codes for the single cellular species of tRNAHis in E. coli and S. typhimurium (37, 42, 126). The tRNAHis molecule is unique among tRNAs in that it contains one additional 5′-end nucleotide, which is critical for aminoacylation (118). The single-copy hisR gene is part of a tRNA gene cluster whose order is tRNAArg tRNAHis-tRNALeu-tRNAPro in both E. coli and S. typhimurium (37, 121). Recently, it was shown that the RNaseE endonuclease acts to process the precursor transcript that contains tRNAArg-tRNAHis-tRNALeu-tRNAPro (172) This cleavage by RNaseE precedes processing by RNaseP and 3’ → 5’ exonucleases and accounts for the reason that RNaseE activity is essential for cell viability (172). The genes in the tRNAArg-tRNAHis tRNALeu-tRNAPro operon are bounded by a promoter, which has an upstream activating sequences in its -70 region, a stringency-control discriminator sequence between the -10 region and the start point of transcription, and a Rho-independent terminator (37, 38, 121).
Mutations in the S. typhimurium hisR promoter reduce the total cellular content of tRNAHis molecules by about 50% and thereby cause increased read-through transcription of the his attenuator (38). Likewise, mutations or antibiotics that decrease negative supercoiling, such as in gyrAB or quinolone-family antibiotics, reduce expression of the hisR promoter, which leads to decreased cellular amounts of HistRNAHis, increased read-through of the his attenuator, and increased his operon expression (42, 72, 89, 90, 187). As noted above, this effect of supercoiling is restricted to his attenuation rather than on transcription from the hisp1 promoter (169), even though transcription from hisp1 does depend on supercoiling in an in vitro coupled transcription-translation system (184). Consistent with this conclusion, normal his operon expression is restored in a gyrA (hisW) mutant by an episome carrying a single copy of the hisR gene (42). The dependence of the hisR promoter on supercoiling seems to depend, in part, on the stringency discriminator sequence (89, 90). Another interesting recent finding is that temperature-sensitive dnaA mutants show increased read-through of the his attenuator and increased his operon expression when grown in rich media (33). This effect is suppressed by simply increasing the copy number of hisR. In this case, an imbalance between the gene dosage may occur between the hisR locus, which is close to oriC, and the his operon, which is far from oriC, leading to a decrease in the hisR to his operon gene ratio (33).
hisS
The hisS gene encodes histidyl-tRNA synthetase (HisRS), which aminoacylates tRNAHis molecules with histidine (Table 1). The histidyl-tRNA synthetase is a class II synthetase (188). Francklyn and coworkers have determined the structure of HisRS bound to several substrates and amino acid analogues (see (96)). They have also used HisRS as a model to study the mechanism of aminoacylation of a class II synthetase and the basis of the discrimination between cognate and noncognate tRNA substrates (see (110)). Because attenuation responds to the amount of His-tRNAHis, the activity of the histidyl-tRNA synthetase affects histidine biosynthesis. In wild-type cells growing in minimal-glucose medium without histidine, the Km of histidyl-tRNA synthetase for histidine is comparable to the histidine concentration (Tables 1 and 2). In this concentration range, the rate of aminoacylation should be strongly affected by fluctuations in the histidine concentration (42). In addition, aminoacylation of tRNAHis is noncompetitively inhibited by AMP (77), competitively inhibited by ADP or adenosine (77), very strongly inhibited by AMP in the presence of pyrophosphate (44), and strongly product inhibited by His-tRNAHis (44). This pattern of inhibition has three consequences that could affect the intracellular amount of His-tRNAHis and thereby influence the rate of histidine biosynthesis: (i) the activity of histidyl-tRNA synthetase is subject to control by cellular energy charge, like the HisG enzyme (see above and reference (43)); (ii) product inhibition probably plays a role in setting the percentage of tRNAHis molecules that are charged at a given histidine concentration (146); and (iii) only a small fraction of the His-tRNA synthetase molecules will be active in bacteria growing under normal conditions because of product inhibition by the high percentage of charged tRNAHis molecules (Table 2 and reference (44)).
Mutations in the hisS gene that affect the level of his attenuation act by reducing the percentage of tRNAHis molecules charged with histidine (146). These mutations generally lower the catalytic activity of HisRS or decrease the affinity of HisRS for histidine, tRNAHis, or ATP (75). This conclusion was confirmed by recent detailed biochemical analysis of five of these earlier hisS mutants (96). Furthermore, histidine biosynthesis seems to be linked directly to the control of the hisS gene, since limitation for histidine causes increased expression of hisS in vivo (156). However, the regulation of hisS is not well understood.
truA) hisT
The truA (hisT) gene encodes PSUI, which catalyzes formation of pseudouridine residues at positions 38, 39, and 40 in the anti-codon stem and loop of at least 30 tRNA isoaccepting species, including tRNAHis (Table 1) (196, 216). The structure of the PSUI homodimer was recently reported unbound and bound to tRNA (94, 124), and the ability of PSUI to modify three different position in different tRNA molecules was studied by combining the structural studies with kinetic studies and computer simulations (124). Transcription termination at the his attenuator is greatly decreased in truA (hisT) mutants even though the undermodified tRNAHis molecules are charged with histidine to the same extent as in wild-type strains (148). To explain this observation, it was postulated that undermodification of His-tRNAHis molecules in truA (hisT) mutants slows translation of the seven histidine codons contained in the his leader transcript and thereby mimics ribosome stalling induced by low concentrations of His-tRNAHis in wild-type bacteria moderately starved for histidine (126). Slow translation or ribosome stalling will result in increased transcription read-through of the his attenuator (Fig. 4B). Support for this conjecture has come from kinetic experiments which show that the absence of pseudouridine modifications in truA (hisT) mutants reduces the general rate of translation elongation by at least 25% and severely reduces translation of mRNA molecules and leader transcripts containing runs of codons for the same amino acid (23, 173). In addition, in some codon contexts, truA (hisT)-mediated tRNA modification is important for reading frame maintenance during translation (217).
Starvation for amino acids results in preferential undermodification for pseudouridine residues at positions 38, 39, and 40 in tRNA molecules in some genetic backgrounds (133, 212). This tRNA undermodification parallels the effect of the truA (hisT) mutation and may lead to increased his operon expression in response to general amino acid deprivation, although further study is needed. A related issue concerns the possibility that histidine biosynthesis is affected by fluctuations in the level of expression of the truA (hisT) gene itself. The truA (hisT) gene is in a complex superoperon with at least three other genes, including the pdxB gene required for the biosynthesis of the essential coenzyme, pyridoxal 5′-phosphate (21, 22, 155). truA (hisT) expression is positively regulated by growth rate in a way that would coordinate the amounts of PSUI and tRNA molecules (215). PSUI amounts may also be regulated by translational coupling to the expression of an upstream dehydrogenase gene (22, 155). The number of PSUI molecules in the cell seems limited (131), and so parallel regulation of PSUI and tRNA amounts may be important for maintaining full tRNA modification levels.
HISTIDINE TRANSPORT
Histidine uptake and its regulation is reviewed elsewhere (see Chapter XX), but there are a couple of considerations about uptake that impact histidine biosynthesis. Histidine can enter Salmonella typhimurium by at least five uptake system ((12); Chapter XX on Amino Acid Utilization), including the high-affinity histidine periplasmic-binding-protein (HisJ-HisQ-HisM-HisP) system (Table 2). The histidine periplasmic-binding-protein permease has been studied extensively by G. F. Ames and coworkers (see references (15, 18, 79, 149, 168) and is one of the best characterized ABC ATP-dependent transport systems (see Chapter XX). The Km of the HisJ-HisQ-HisM-HisP permease for histidine is about 10-8 M and the Kd (histidine) of the HisJ periplasmic binding protein is about 0.1 μM, which means that its affinity for histidine is orders of magnitude greater than that of some of the other histidine transport systems (12); Chapter XX). The HisJ-HisQ-HisM-HisP permease can concentrate histidine against a very high concentration gradient and scavenge histidine at very low concentrations (15, 17, 79). The S. typhimurium histidine limit concentration, which is the lowest external concentration required by the HisJ-HisQ-HisM-HisP permease to supply the cell with enough histidine for protein synthesis without endogenous histidine biosynthesis, is only 0.15 μM (Table 2) (12, 15). It has been suggested that bacterial cells need to maintain such an efficient and complex histidine permease because there is a certain amount of leakage of histidine from bacterial cells (15, 17, 79). Since the biosynthesis of each histidine molecule requires so much energy (see above), it may be advantageous for bacteria to recover any histidine lost to the growth medium. It has also been pointed out that the high affinity of amino acid permeases makes bioassays possible (15). Finally, the high affinity of the HisJ-HisQ-HisM-HisP permease may explain why bacterial cells need to maintain high basal concentrations of the histidine biosynthetic enzymes even in the presence of exogenous histidine (Table 2) (42). Since transport of histidine is so effective at low concentrations, the internal pool of histidine will not decrease significantly until the external histidine supply is almost completely gone. If attenuation greatly lowered the amounts of the histidine biosynthetic enzymes in response to external histidine supply, then it is conceivable that there would not be enough time for adequate transcription and translation of the his structural genes between when the internal histidine pool starts to fall and when the total histidine supply is exhausted. Hence, a system may have evolved that favors histidine transport at the relatively small energy expense of maintaining a constant supply of the histidine biosynthetic enzymes (42).
SUMMARY
In all major respects, histidine biosynthesis seems to be the same in E. coli and S. typhimurium. Histidine biosynthesis in E. coli and S. typhimurium has continued to provide a wealth of fundamental information about biochemical and cellular regulation that go far beyond the topic of histidine biosynthesis. Some of these advances are highlighted in this review, including insights into the mechanisms of allosteric feedback inhibition, cooperative control of enzyme function, unique enzymatic reactions, positive metabolic control by ppGpp, pausing of RNA polymerase, attenuation, and pathway evolution. The progress in these areas will continue as sophisticated genomic, metabolomic, proteomic, and structural approaches converge in studies of the histidine biosynthetic pathway and mechanisms of control of the his operon. In addition, the new methods that have emerged recently to construct directed chromosomal mutations efficiently combined with whole-genome methods to identify mutations that arise in selections rapidly at reasonable cost should build on the more classical studies done so far on the physiological consequences of biochemical changes on histidine biosynthesis and his operon control.
ACKNOWLEDGMENTS
I thank the many scientists who have worked or continue to work on the biosynthesis of histidine in E. coli and S. typhimurium for helpful discussions, information, and insights into this topic and Krystyna M. Kazmierczak for comments on the manuscript. Preparation of this review was supported by grants AI060744 from the National Institutes of Health and 0543289 from the National Science Foundation of the U.S.A. to M. E. W., and by funds from the Indiana METACyt Initiative of Indiana University Bloomington, funded in part through a major grant from the Lilly Endowment, Inc. to M.E.W.
This chapter is dedicated to the memory of Philip E. Hartman and Zlata Demerec Hartman (Johns Hopkins University), who were pioneers of bacterial genetics and education, made numerous fundamental discoveries about the his operon, bacterial metabolism, and mutagenesis, and were outstanding mentors and role models to many in the field.
LITERATURE CITED
- 1.Albritton WL, Levin AP. Some comparative kinetic data on the enzyme imidazoleacetol phosphate:L-glutamate aminotransferase derived from mutant strains of Salmonella typhimurium. J Biol Chem. 1970;245:2525–8. [PubMed] [Google Scholar]
- 2.Alifano P, Carlomagno MS, Bruni CB. Location of the hisGDCBHAFI operon on the physical map of Escherichia coli. J Bacteriol. 1992;174:3830–1. doi: 10.1128/jb.174.11.3830-3831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alifano P, Ciampi MS, Nappo AG, Bruni CB, Carlomagno MS. In vivo analysis of the mechanisms responsible for strong transcriptional polarity in a “sense” mutant within an intercistronic region. Cell. 1988;55:351–60. doi: 10.1016/0092-8674(88)90058-x. [DOI] [PubMed] [Google Scholar]
- 4.Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alifano P, Piscitelli C, Blasi V, Rivellini F, Nappo AG, Bruni CB, Carlomagno MS. Processing of a polycistronic mRNA requires a 5′ cis element and active translation. Mol Microbiol. 1992;6:787–98. doi: 10.1111/j.1365-2958.1992.tb01529.x. [DOI] [PubMed] [Google Scholar]
- 6.Alifano P, Rivellini F, Limauro D, Bruni CB, Carlomagno MS. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991;64:553–63. doi: 10.1016/0092-8674(91)90239-u. [DOI] [PubMed] [Google Scholar]
- 7.Alifano P, Rivellini F, Nappo AG, Bruni CB, Carlomagno MS. Alternative patterns of his operon transcription and mRNA processing generated by metabolic perturbation. Gene. 1994;146:15–21. doi: 10.1016/0378-1119(94)90828-1. [DOI] [PubMed] [Google Scholar]
- 8.Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994;8:3021–31. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- 9.Allen S, Zilles JL, Downs DM. Metabolic flux in both the purine mononucleotide and histidine biosynthetic pathways can influence synthesis of the hydroxymethyl pyrimidine moiety of thiamine in Salmonella enterica. J Bacteriol. 2002;184:6130–7. doi: 10.1128/JB.184.22.6130-6137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amaro RE, Myers RS, Davisson VJ, Luthey-Schulten ZA. Structural elements in IGP synthase exclude water to optimize ammonia transfer. Biophys J. 2005;89:475–87. doi: 10.1529/biophysj.104.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amaro RE, Sethi A, Myers RS, Davisson VJ, Luthey-Schulten ZA. A network of conserved interactions regulates the allosteric signal in a glutamine amidotransferase. Biochemistry. 2007;46:2156–73. doi: 10.1021/bi061708e. [DOI] [PubMed] [Google Scholar]
- 12.Ames BN. A bacterial system for detecting mutagens and carcinomas. In: Sutton HE, Harris MI, editors. Mutagenic Effects of Environmental Contaminants. Academic Press, Inc.; New York: 1972. pp. 57–66. [Google Scholar]
- 13.Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA. 1973;70:2281–5. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ames BN, Tsang TH, Buck M, Christman MF. The leader mRNA of the histidine attenuator region resembles tRNAHis: possible general regulatory implications. Proc Natl Acad Sci USA. 1983;80:5240–2. doi: 10.1073/pnas.80.17.5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ames GF. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 16.Ames GF. Components of histidine transport. In: Fox CF, editor. Membrane Research. Academic Press, Inc.; New York: 1972. pp. 409–426. [Google Scholar]
- 17.Ames GF. Uptake of Amino Acids by Salmonella typhimurium. Arch Biochem Biophys. 1964;104:1–18. doi: 10.1016/s0003-9861(64)80028-x. [DOI] [PubMed] [Google Scholar]
- 18.Ames GF, Nikaido K, Wang IX, Liu PQ, Liu CE, Hu C. Purification and characterization of the membrane-bound complex of an ABC transporter, the histidine permease. J Bioenerg Biomembr. 2001;33:79–92. doi: 10.1023/a:1010797029183. [DOI] [PubMed] [Google Scholar]
- 19.Arnez JG, Augustine JG, Moras D, Francklyn CS. The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci USA. 1997;94:7144–9. doi: 10.1073/pnas.94.14.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnez JG, Harris DC, Mitschler A, Rees B, Francklyn CS, Moras D. Crystal structure of histidyl-tRNA synthetase from Escherichia coli complexed with histidyl-adenylate. EMBO J. 1995;14:4143–55. doi: 10.1002/j.1460-2075.1995.tb00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arps PJ, Marvel CC, Rubin BC, Tolan DA, Penhoet EE, Winkler ME. Structural features of the hisT operon of Escherichia coli K-12. Nucleic Acids Res. 1985;13:5297–315. doi: 10.1093/nar/13.14.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arps PJ, Winkler ME. An unusual genetic link between vitamin B6 biosynthesis and tRNA pseudouridine modification in Escherichia coli K-12. J Bacteriol. 1987;169:1071–9. doi: 10.1128/jb.169.3.1071-1079.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Artz S, Holzschu D, Blum P, Shand R. Use of M13mp phages to study gene regulation, structure and function: cloning and recombinational analysis of genes of the Salmonella typhimurium histidine operon. Gene. 1983;26:147–58. doi: 10.1016/0378-1119(83)90184-1. [DOI] [PubMed] [Google Scholar]
- 24.Artz SW, Broach JR. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci USA. 1975;72:3453–7. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Artz SW, Holzschu D. Histidine biosynthesis and its regulation. In: Herrmann KM, Somerville RL, editors. Amino Acids: Biosynthesis and Genetic Regulation. Addison-Wesley Publishing Co.; Rading, Massachusetts: 1983. pp. 379–404. [Google Scholar]
- 26.Barbosa JA, Sivaraman J, Li Y, Larocque R, Matte A, Schrag JD, Cygler M. Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase. Proc Natl Acad Sci USA. 2002;99:1859–64. doi: 10.1073/pnas.022476199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 28.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–88. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 29.Barnes WM. DNA sequence from the histidine operon control region: seven histidine codons in a row. Proc Natl Acad Sci USA. 1978;75:4281–5. doi: 10.1073/pnas.75.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes WM, Tuley E. DNA sequence changes of mutations in the histidine operon control region that decrease attenuation. J Mol Biol. 1983;165:443–59. doi: 10.1016/s0022-2836(83)80212-5. [DOI] [PubMed] [Google Scholar]
- 31.Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol. 1992;174:5228–36. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berberich MA, Kovach JS, Goldberger RF. Chain initiation in a polycistronic message: sequential versus simultaneous derepression of the enzymes for histidine biosynthesis in Salmonella typhimurium. Proc Natl Acad Sci USA. 1967;57:1857–64. doi: 10.1073/pnas.57.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanc-Potard AB, Figueroa-Bossi N, Bossi L. Histidine operon deattenuation in dnaA mutants of Salmonella typhimurium correlates with a decrease in the gene dosage ratio between tRNA(His) and histidine biosynthetic loci. J Bacteriol. 1999;181:2938–41. doi: 10.1128/jb.181.9.2938-2941.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blasi F, Bruni CB. Regulation of the histidine operon: translation-controlled transcription termination (a mechanism common to several biosynthetic operons). Curr Top Cell Regul. 1981;19:1–45. doi: 10.1016/b978-0-12-152819-5.50018-x. [DOI] [PubMed] [Google Scholar]
- 35.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–69. [PubMed] [Google Scholar]
- 36.Bochner BR, Ames BN. ZTP (5-amino 4-imidazole carboxamide riboside 5′-triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell. 1982;29:929–37. doi: 10.1016/0092-8674(82)90455-x. [DOI] [PubMed] [Google Scholar]
- 37.Bossi L. The hisR locus of Salmonella: nucleotide sequence and expression. Mol Gen Genet. 1983;192:163–70. doi: 10.1007/BF00327662. [DOI] [PubMed] [Google Scholar]
- 38.Bossi L, Smith DM. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984;39:643–52. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- 39.Bovee ML, Champagne KS, Demeler B, Francklyn CS. The quaternary structure of the HisZ-HisG N-1-(5′-phosphoribosyl)-ATP transferase from Lactococcus lactis. Biochemistry. 2002;41:11838–46. doi: 10.1021/bi020243z. [DOI] [PubMed] [Google Scholar]
- 40.Brady DR, Houston LL. New assay for histidinol phosphate phosphatase using a coupled reaction. Anal Biochem. 1972;48:480–2. doi: 10.1016/0003-2697(72)90101-7. [DOI] [PubMed] [Google Scholar]
- 41.Brady DR, Houston LL. Some properties of the catalytic sites of imidazoleglycerol phosphate dehydratase-histidinol phosphate phosphatase, a bifunctional enzyme from Salmonella typhimurium. J Biol Chem. 1973;248:2588–92. [PubMed] [Google Scholar]
- 42.Brenner M, Ames BN. The histidine operon and its regulation. In: Vogel HJ, editor. Metabolic Pathways. Vol. 5. Academic Press; New York: 1971. pp. 349–387. [Google Scholar]
- 43.Brenner M, De Lorenzo F, Ames BN. Energy charge and protein synthesis. Control of aminoacyl transfer ribonucleic acid synthetases. J Biol Chem. 1970;245:450–2. [PubMed] [Google Scholar]
- 44.Brenner M, Lewis JA, Straus DS, De Lorenzo F, Ames BN. Histidine regulation in Salmonella typhimurium. XIV. Interaction of the histidyl transfer ribonucleic acid synthetase with histidine transfer ribonucleic acid. J Biol Chem. 1972;247:4333–9. [PubMed] [Google Scholar]
- 45.Brilli M, Fani R. Molecular evolution of hisB genes. J Mol Evol. 2004;58:225–37. doi: 10.1007/s00239-003-2547-x. [DOI] [PubMed] [Google Scholar]
- 46.Bruni CB, Musti AM, Frunzio R, Blasi F. Structural and physiological studies of the Escherichia coli histidine operon inserted into plasmid vectors. J Bacteriol. 1980;142:32–42. doi: 10.1128/jb.142.1.32-42.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cano DA, Mouslim C, Ayala JA, Garcia-del Portillo F, Casadesus J. Cell division inhibition in Salmonella typhimurium histidine-constitutive strains: an ftsI-like defect in the presence of wild-type penicillin-binding protein 3 levels. J Bacteriol. 1998;180:5231–4. doi: 10.1128/jb.180.19.5231-5234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlomagno MS, Blasi F, Bruni CB. Gene organization in the distal part of the Salmonella typhimurium histidine operon and determination and sequence of the operon transcription terminator. Mol Gen Genet. 1983;191:413–20. doi: 10.1007/BF00425756. [DOI] [PubMed] [Google Scholar]
- 49.Carlomagno MS, Chiariotti L, Alifano P, Nappo AG, Bruni CB. Structure and function of the Salmonella typhimurium and Escherichia coli K-12 histidine operons. J Mol Biol. 1988;203:585–606. doi: 10.1016/0022-2836(88)90194-5. [DOI] [PubMed] [Google Scholar]
- 50.Carlomagno MS, Nappo A. The antiterminator NusB enhances termination at a sub-optimal Rho site. J Mol Biol. 2001;309:19–28. doi: 10.1006/jmbi.2001.4678. [DOI] [PubMed] [Google Scholar]
- 51.Carlomagno MS, Nappo A. NusA modulates intragenic termination by different pathways. Gene. 2003;308:115–28. doi: 10.1016/s0378-1119(03)00472-4. [DOI] [PubMed] [Google Scholar]
- 52.Carlomagno MS, Riccio A, Bruni CB. Convergently functional, Rho-independent terminator in Salmonella typhimurium. J Bacteriol. 1985;163:362–8. doi: 10.1128/jb.163.1.362-368.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrano L, Alifano P, Corti E, Bucci C, Donadio S. A new inhibitor of the transcription-termination factor Rho. Biochem Biophys Res Commun. 2003;302:219–25. doi: 10.1016/s0006-291x(03)00131-1. [DOI] [PubMed] [Google Scholar]
- 54.Carrano L, Bucci C, De Pascalis R, Lavitola A, Manna F, Corti E, Bruni CB, Alifano P. Effects of bicyclomycin on RNA- and ATP-binding activities of transcription termination factor Rho. Antimicrob Agents Chemother. 1998;42:571–8. doi: 10.1128/aac.42.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cashel M, Rudd KE. Stringent response. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology; Washington, D.C.: 1987. pp. 1410–1438. [Google Scholar]
- 56.Champagne KS, Sissler M, Larrabee Y, Doublie S, Francklyn CS. Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog. J Biol Chem. 2005;280:34096–104. doi: 10.1074/jbc.M505041200. [DOI] [PubMed] [Google Scholar]
- 57.Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 58.Chan CL, Landick R. Effects of neutral salts on RNA chain elongation and pausing by Escherichia coli RNA polymerase. J Mol Biol. 1997;268:37–53. doi: 10.1006/jmbi.1997.0934. [DOI] [PubMed] [Google Scholar]
- 59.Chan CL, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–804. [PubMed] [Google Scholar]
- 60.Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3′ end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- 61.Chaudhuri BN, Lange SC, Myers RS, Davisson VJ, Smith JL. Toward understanding the mechanism of the complex cyclization reaction catalyzed by imidazole glycerolphosphate synthase: crystal structures of a ternary complex and the free enzyme. Biochemistry. 2003;42:7003–12. doi: 10.1021/bi034320h. [DOI] [PubMed] [Google Scholar]
- 62.Cheong CG, Bauer CB, Brushaber KR, Escalante-Semerena JC, Rayment I. Three-dimensional structure of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica. Biochemistry. 2002;41:4798–808. doi: 10.1021/bi012111w. [DOI] [PubMed] [Google Scholar]
- 63.Cheong CG, Escalante-Semerena JC, Rayment I. Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes. Biochemistry. 2002;41:9079–89. doi: 10.1021/bi020294w. [DOI] [PubMed] [Google Scholar]
- 64.Chittur SV, Klem TJ, Shafer CM, Davisson VJ. Mechanism for acivicin inactivation of triad glutamine amidotransferases. Biochemistry. 2001;40:876–87. doi: 10.1021/bi0014047. [DOI] [PubMed] [Google Scholar]
- 65.Cho Y, Sharma V, Sacchettini JC. Crystal structure of ATP phosphoribosyltransferase from Mycobacterium tuberculosis. J Biol Chem. 2003;278:8333–9. doi: 10.1074/jbc.M212124200. [DOI] [PubMed] [Google Scholar]
- 66.Choy HE. The study of guanosine 5′-diphosphate 3′-diphosphate-mediated transcription regulation in vitro using a coupled transcription-translation system. J Biol Chem. 2000;275:6783–9. doi: 10.1074/jbc.275.10.6783. [DOI] [PubMed] [Google Scholar]
- 67.Ciesla Z, Salvatore F, Broach JR, Artz SW, Ames BN. Histidine regulation in Salmonella typhimurium. XVI. A sensitive radiochemical assay for histidinol dehydrogenase. Anal Biochem. 1975;63:44–55. doi: 10.1016/0003-2697(75)90187-6. [DOI] [PubMed] [Google Scholar]
- 68.Cortese R, Kammen HO, Spengler SJ, Ames BN. Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem. 1974;249:1103–8. [PubMed] [Google Scholar]
- 69.D'Ordine RL, Klem TJ, Davisson VJ. N1-(5′-phosphoribosyl) adenosine-5′-monophosphate cyclohydrolase: purification and characterization of a unique metalloenzyme. Biochemistry. 1999;38:1537–46. doi: 10.1021/bi982475x. [DOI] [PubMed] [Google Scholar]
- 70.Da Costa XJ, Artz SW. Mutations that render the promoter of the histidine operon of Salmonella typhimurium insensitive to nutrient-rich medium repression and amino acid downshift. J Bacteriol. 1997;179:5211–7. doi: 10.1128/jb.179.16.5211-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dall-Larsen T. Stopped flow kinetic studies of adenosine triphosphate phosphoribosyl transferase, the first enzyme in the histidine biosynthesis of Escherichia coli. Int J Biochem. 1988;20:811–5. doi: 10.1016/0020-711x(88)90069-9. [DOI] [PubMed] [Google Scholar]
- 72.Davis L, Williams LS. Characterization of a cold-sensitive hisW mutant of Salmonella typhimurium. J Bacteriol. 1982;151:867–78. doi: 10.1128/jb.151.2.867-878.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Lorenzo F, Ames BN. Histidine regulation in Salmonella typhimurium. VII. Purification and general properties of the histidyl transfer ribonucleic acid synthetase. J Biol Chem. 1970;245:1710–6. [PubMed] [Google Scholar]
- 74.De Lorenzo F, Di Natale P, Schechter AN. Chemical and physical studies on the structure of the histidyl transfer ribonucleic acid synthetase from Salmonella typhimurium. J Biol Chem. 1974;249:908–13. [PubMed] [Google Scholar]
- 75.De Lorenzo F, Straus DS, Ames BN. Histidine regulation in Salmonella typhimurium. X. Kinetic studies of mutant histidyl transfer ribonucleic acid synthetases. J Biol Chem. 1972;247:2302–7. [PubMed] [Google Scholar]
- 76.Di Natale P, Cimino F, De Lorenzo F. The pyrophosphate exchange reaction of histidyl-tRNA synthetase from Salmonella typhimurium: reaction parameters and inhibition by transfer ribonucleic acid. FEBS Lett. 1974;46:175–9. doi: 10.1016/0014-5793(74)80362-5. [DOI] [PubMed] [Google Scholar]
- 77.Di Natale P, Schechter AN, Castronuovo Lepore G, De Lorenzo F. Histidyl transfer ribonucleic acid synthetase from Salmonella typhimurium. Interaction with substrates and ATP analogues. Eur J Biochem. 1976;62:293–8. doi: 10.1111/j.1432-1033.1976.tb10160.x. [DOI] [PubMed] [Google Scholar]
- 78.Di Nocera PP, Blasi F, Di Lauro R, Frunzio R, Bruni CB. Nucleotide sequence of the attenuator region of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci USA. 1978;75:4276–80. doi: 10.1073/pnas.75.9.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doige CA, Ames GF. ATP-dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug resistance. Annu Rev Microbiol. 1993;47:291–319. doi: 10.1146/annurev.mi.47.100193.001451. [DOI] [PubMed] [Google Scholar]
- 80.Douangamath A, Walker M, Beismann-Driemeyer S, Vega-Fernandez MC, Sterner R, Wilmanns M. Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex. Structure. 2002;10:185–93. doi: 10.1016/s0969-2126(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 81.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–96. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elf J, Ehrenberg M. What makes ribosome-mediated transcriptional attenuation sensitive to amino acid limitation? PLoS Comput Biol. 2005;1:e2. doi: 10.1371/journal.pcbi.0010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ely B. Physiological studies of Salmonella histidine operator-promoter mutants. Genetics. 1974;78:593–606. doi: 10.1093/genetics/78.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ely B, Ciesla Z. Internal promoter P2 of the histidine operon of Salmonella typhimurium. J Bacteriol. 1974;120:984–6. doi: 10.1128/jb.120.2.984-986.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fani R, Brilli M, Fondi M, Lio P. The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol Biol 7 Suppl. 2007;2:S4. doi: 10.1186/1471-2148-7-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fani R, Brilli M, Lio P. The origin and evolution of operons: the piecewise building of the proteobacterial histidine operon. J Mol Evol. 2005;60:378–90. doi: 10.1007/s00239-004-0198-1. [DOI] [PubMed] [Google Scholar]
- 87.Fani R, Mori E, Tamburini E, Lazcano A. Evolution of the structure and chromosomal distribution of histidine biosynthetic genes. Orig Life Evol Biosph. 1998;28:555–70. doi: 10.1023/a:1006531526299. [DOI] [PubMed] [Google Scholar]
- 88.Fernandez FJ, Vega MC, Lehmann F, Sandmeier E, Gehring H, Christen P, Wilmanns M. Structural studies of the catalytic reaction pathway of a hyperthermophilic histidinol-phosphate aminotransferase. J Biol Chem. 2004;279:21478–88. doi: 10.1074/jbc.M400291200. [DOI] [PubMed] [Google Scholar]
- 89.Figueroa-Bossi N, Guerin M, Rahmouni R, Leng M, Bossi L. The supercoiling sensitivity of a bacterial tRNA promoter parallels its responsiveness to stringent control. Embo J. 1998;17:2359–67. doi: 10.1093/emboj/17.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Figueroa N, Wills N, Bossi L. Common sequence determinants of the response of a prokaryotic promoter to DNA bending and supercoiling. Embo J. 1991;10:941–9. doi: 10.1002/j.1460-2075.1991.tb08028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fink GR, Martin RG. Translation and polarity in the histidine operon. II. Polarity in the histidine operon. J Mol Biol. 1967;30:97–107. doi: 10.1016/0022-2836(67)90246-x. [DOI] [PubMed] [Google Scholar]
- 92.Fishman SE, Kerchief KR, Parker J. Specialized lambda transducing bacteriophage which carries hisS, the structural gene for histidyl-transfer ribonucleic acid synthetase. J Bacteriol. 1979;139:404–10. doi: 10.1128/jb.139.2.404-410.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Flores A, Fox M, Casadesus J. The pleiotropic effects of his overexpression in Salmonella typhimurium do not involve AICAR-induced mutagenesis. Mol Gen Genet. 1993;240:360–4. doi: 10.1007/BF00280387. [DOI] [PubMed] [Google Scholar]
- 94.Foster PG, Huang L, Santi DV, Stroud RM. The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nat Struct Biol. 2000;7:23–7. doi: 10.1038/71219. [DOI] [PubMed] [Google Scholar]
- 95.Fox M, Frandsen N, D'Ari R. AICAR is not an endogenous mutagen in Escherichia coli. Mol Gen Genet. 1993;240:355–9. doi: 10.1007/BF00280386. [DOI] [PubMed] [Google Scholar]
- 96.Francklyn C, Adams J, Augustine J. Catalytic defects in mutants of class II histidyl-tRNA synthetase from Salmonella typhimurium previously linked to decreased control of histidine biosynthesis regulation. J Mol Biol. 1998;280:847–58. doi: 10.1006/jmbi.1998.1902. [DOI] [PubMed] [Google Scholar]
- 97.Francklyn C, Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci USA. 1990;87:8655–9. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frandsen N, D'Ari R. Excess histidine enzymes cause AICAR-independent filamentation in Escherichia coli. Mol Gen Genet. 1993;240:348–54. doi: 10.1007/BF00280385. [DOI] [PubMed] [Google Scholar]
- 99.Freedman R, Schimmel P. In vitro transcription of the histidine operon. Identification of the his promoter and leader and readthrough transcripts. J Biol Chem. 1981;256:10747–50. [PubMed] [Google Scholar]
- 100.Frunzio R, Bruni CB, Blasi F. In vivo and in vitro detection of the leader RNA of the histidine operon of Escherichia coli K-12. Proc Natl Acad Sci USA. 1981;78:2767–71. doi: 10.1073/pnas.78.5.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gee P, Maron DM, Ames BN. Detection and classification of mutagens: a set of base-specific Salmonella tester strains. Proc Natl Acad Sci USA. 1994;91:11606–10. doi: 10.1073/pnas.91.24.11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gibert I, Casadesus J. sulA-independent division inhibition in his-constitutive strains of Salmonella typhimurium. FEMS Microbiol Lett. 1990;57:205–10. doi: 10.1016/0378-1097(90)90066-y. [DOI] [PubMed] [Google Scholar]
- 103.Glynn SE, Baker PJ, Sedelnikova SE, Davies CL, Eadsforth TC, Levy CW, Rodgers HF, Blackburn GM, Hawkes TR, Viner R, Rice DW. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure. 2005;13:1809–17. doi: 10.1016/j.str.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 104.Goldberger RF. Autogenous regulation of gene expression. Science. 1974;183:810–6. doi: 10.1126/science.183.4127.810. [DOI] [PubMed] [Google Scholar]
- 105.Grisolia V, Carlomagno MS, Nappo AG, Bruni CB. Cloning, structure, and expression of the Escherichia coli K-12 hisC gene. J Bacteriol. 1985;164:1317–23. doi: 10.1128/jb.164.3.1317-1323.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grisolia V, Riccio A, Bruni CB. Structure and function of the internal promoter (hisBp) of the Escherichia coli K-12 histidine operon. J Bacteriol. 1983;155:1288–96. doi: 10.1128/jb.155.3.1288-1296.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grubmeyer C, Skiadopoulos M, Senior AE. L-histidinol dehydrogenase, a Zn2+-metalloenzyme. Arch Biochem Biophys. 1989;272:311–7. doi: 10.1016/0003-9861(89)90224-5. [DOI] [PubMed] [Google Scholar]
- 108.Grubmeyer C, Teng H. Mechanism of Salmonella typhimurium histidinol dehydrogenase: kinetic isotope effects and pH profiles. Biochemistry. 1999;38:7355–62. doi: 10.1021/bi982757x. [DOI] [PubMed] [Google Scholar]
- 109.Grubmeyer CT, Gray WR. A cysteine residue (cysteine-116) in the histidinol binding site of histidinol dehydrogenase. Biochemistry. 1986;25:4778–84. doi: 10.1021/bi00365a009. [DOI] [PubMed] [Google Scholar]
- 110.Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–42. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haruyama K, Nakai T, Miyahara I, Hirotsu K, Mizuguchi H, Hayashi H, Kagamiyama H. Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5′-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme. Biochemistry. 2001;40:4633–44. doi: 10.1021/bi002769u. [DOI] [PubMed] [Google Scholar]
- 112.Henderson GB, Snell EE. Crystalline L-histidinol phosphate aminotransferase from Salmonella typhimurium. Purification and subunit structure. J Biol Chem. 1973;248:1906–11. [PubMed] [Google Scholar]
- 113.Henn-Sax M, Thoma R, Schmidt S, Hennig M, Kirschner K, Sterner R. Two (betaalpha)(8)-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms and common strategies for protecting their labile substrates. Biochemistry. 2002;41:12032–42. doi: 10.1021/bi026092h. [DOI] [PubMed] [Google Scholar]
- 114.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90:1453–61. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henry T, Garcia-Del Portillo F, Gorvel JP. Identification of Salmonella functions critical for bacterial cell division within eukaryotic cells. Mol Microbiol. 2005;56:252–67. doi: 10.1111/j.1365-2958.2005.04540.x. [DOI] [PubMed] [Google Scholar]
- 116.Hidalgo M, Rodriguez G, Kuhn JG, Brown T, Weiss G, MacGovren JP, Von Hoff DD, Rowinsky EK. A Phase I and pharmacological study of the glutamine antagonist acivicin with the amino acid solution aminosyn in patients with advanced solid malignancies. Clin Cancer Res. 1998;4:2763–70. [PubMed] [Google Scholar]
- 117.Higgins CF, Ames GF, Barnes WM, Clement JM, Hofnung M. A novel intercistronic regulatory element of prokaryotic operons. Nature. 1982;298:760–2. doi: 10.1038/298760a0. [DOI] [PubMed] [Google Scholar]
- 118.Himeno H, Hasegawa T, Ueda T, Watanabe K, Miura K, Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989;17:7855–63. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoppe I, Johnston HM, Biek D, Roth JR. A refined map of the hisG gene of Salmonella typhimurium. Genetics. 1979;92:17–26. doi: 10.1093/genetics/92.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsu LC, Okamoto M, Snell EE. L-Histidinol phosphate aminotransferase from Salmonella typhimurium. Kinetic behavior and sequence at the pyridoxal-P binding site. Biochimie. 1989;71:477–89. doi: 10.1016/0300-9084(89)90178-8. [DOI] [PubMed] [Google Scholar]
- 121.Hsu LM, Klee HJ, Zagorski J, Fournier MJ. Structure of an Escherichia coli tRNA operon containing linked genes for arginine, histidine, leucine, and proline tRNAs. J Bacteriol. 1984;158:934–42. doi: 10.1128/jb.158.3.934-942.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang L, Pookanjanatavip M, Gu X, Santi DV. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–51. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- 123.Hulton CS, Higgins CF, Sharp PM. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:825–34. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 124.Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson MS, Taylor BL. Comparison of methods for specific depletion of ATP in Salmonella typhimurium. Appl Environ Microbiol. 1993;59:3509–12. doi: 10.1128/aem.59.10.3509-3512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnston HM, Barnes WM, Chumley FG, Bossi L, Roth JR. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci USA. 1980;77:508–12. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Johnston HM, Roth JR. DNA sequence changes of mutations altering attenuation control of the histidine operon of Salmonella typhimurium. J Mol Biol. 1981;145:735–56. doi: 10.1016/0022-2836(81)90312-0. [DOI] [PubMed] [Google Scholar]
- 128.Johnston HM, Roth JR. Histidine mutants requiring adenine: selection of mutants with reduced hisG expression in Salmonella typhimurium. Genetics. 1979;92:1–15. doi: 10.1093/genetics/92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jurgens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R. Directed evolution of a (beta alpha)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc Natl Acad Sci USA. 2000;97:9925–30. doi: 10.1073/pnas.160255397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kalousek F, Konigsberg WH. Purification and characterization of histidyl transfer ribonucleic acid synthetase of Escherichia coli. Biochemistry. 1974;13:999–1006. doi: 10.1021/bi00702a026. [DOI] [PubMed] [Google Scholar]
- 131.Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J Biol Chem. 1988;263:2255–63. [PubMed] [Google Scholar]
- 132.Kasai T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature. 1974;249:523–7. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- 133.Kitchingman GR, Fournier MJ. Modification-deficient transfer ribonucleic acids from relaxed control Escherichia coli: structures of the major undermodified phenylalanine and leucine transfer RNAs produced during leucine starvation. Biochemistry. 1977;16:2213–20. doi: 10.1021/bi00629a027. [DOI] [PubMed] [Google Scholar]
- 134.Kleeman J, Parsons SM. A sensitive assay for the reverse reaction of the first histidine biosynthetic enzyme. Anal Biochem. 1975;68:236–41. doi: 10.1016/0003-2697(75)90700-9. [DOI] [PubMed] [Google Scholar]
- 135.Kleeman JE, Parsons SM. Inhibition of histidyl-tRNA-adenosine triphosphate phosphoribosyltransferase complex formation by histidine and by guanosine tetraphosphate. Proc Natl Acad Sci USA. 1977;74:1535–7. doi: 10.1073/pnas.74.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Klem TJ, Chen Y, Davisson VJ. Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase. J Bacteriol. 2001;183:989–96. doi: 10.1128/JB.182.3.989-996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Klem TJ, Davisson VJ. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–86. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- 138.Klungsoyr L, Hagemen JH, Fall L, Atkinson DE. Interaction between energy charge and product feedback in the regulation of biosynthetic enzymes. Aspartokinase, phosphoribosyladenosine triphosphate synthetase, and phosphoribosyl pyrophosphate synthetase. Biochemistry. 1968;7:4035–40. doi: 10.1021/bi00851a034. [DOI] [PubMed] [Google Scholar]
- 139.Kronenberg HM, Vogel T, Goldberger RF. A new and highly sensitive assay for the ATP phosphoribosyltransferase that catalyzes the first step of histidine biosynthesis. Anal Biochem. 1975;65:380–8. doi: 10.1016/0003-2697(75)90522-9. [DOI] [PubMed] [Google Scholar]
- 140.Kryvi H. Thermal stability of phosphoribosyladenosine triphosphate synthetase as reflected in its circular dichroism and activity properties. Effect of inhibitors. Biochim Biophys Acta. 1973;317:123–30. doi: 10.1016/0005-2795(73)90204-3. [DOI] [PubMed] [Google Scholar]
- 141.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 142.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–50. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 143.Lee DN, Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992;228:759–77. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- 144.Lee F, Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci USA. 1977;74:4365–9. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leopoldseder S, Claren J, Jurgens C, Sterner R. Interconverting the catalytic activities of (betaalpha)(8)-barrel enzymes from different metabolic pathways: sequence requirements and molecular analysis. J Mol Biol. 2004;337:871–9. doi: 10.1016/j.jmb.2004.01.062. [DOI] [PubMed] [Google Scholar]
- 146.Lewis JA, Ames BN. Histidine regulation in Salmonella typhimurium. XI. The percentage of transfer RNA His charged in vivo and its relation to the repression of the histidine operon. J Mol Biol. 1972;66:131–42. doi: 10.1016/s0022-2836(72)80011-1. [DOI] [PubMed] [Google Scholar]
- 147.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA. 2003;100:13213–8. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Limauro D, Avitabile A, Puglia AM, Bruni CB. Further characterization of the histidine gene cluster of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis of hisD. Res Microbiol. 1992;143:683–93. doi: 10.1016/0923-2508(92)90063-t. [DOI] [PubMed] [Google Scholar]
- 149.Liu CE, Ames GF. Characterization of transport through the periplasmic histidine permease using proteoliposomes reconstituted by dialysis. J Biol Chem. 1997;272:859–66. doi: 10.1074/jbc.272.2.859. [DOI] [PubMed] [Google Scholar]
- 150.Lohkamp B, McDermott G, Campbell SA, Coggins JR, Lapthorn AJ. The structure of Escherichia coli ATP-phosphoribosyltransferase: identification of substrate binding sites and mode of AMP inhibition. J Mol Biol. 2004;336:131–44. doi: 10.1016/j.jmb.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 151.Loper JC, Adams E. Purification and Properties of Histidinol Dehydrogenase from Salmonella typhimurium. J Biol Chem. 1965;240:788–95. [PubMed] [Google Scholar]
- 152.Martin RG. The first enzyme in histidine biosynthesis: the nature of feedback inhibition by histidine. The Journal of Biological Chemistry. 1963;238:257–268. [Google Scholar]
- 153.Martin RG. Presented at the Cold Spring Harbor Symp. Quant. Biol. 1963 [Google Scholar]
- 154.Martin RG, Berberich B, Ames BN, Davis WW, Goldberger RF, Yourno JD. Enzymes and intermediates of histidine biosynthesis in Salmonella typhimurium. Methods Enzymol. 1971;17B:3–44. [Google Scholar]
- 155.Marvel CC, Arps PJ, Rubin BC, Kammen HO, Penhoet EE, Winkler ME. hisT is part of a multigene operon in Escherichia coli K-12. J Bacteriol. 1985;161:60–71. doi: 10.1128/jb.161.1.60-71.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McGinnis E, Williams LS. Regulation of histidyl-transfer ribonucleic acid synthetase formation in a histidyl-transfer ribonucleic acid synthetase mutant of Salmonella typhimurium. J Bacteriol. 1972;111:739–44. doi: 10.1128/jb.111.3.739-744.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Meyers M, Levinthal M, Goldberger RF. trans-Recessive mutation in the first structural gene of the histidine operon that results in constitutive expression of the operon. J Bacteriol. 1975;124:1227–35. doi: 10.1128/jb.124.3.1227-1235.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mizuguchi H, Hayashi H, Miyahara I, Hirotsu K, Kagamiyama H. Characterization of histidinol phosphate aminotransferase from Escherichia coli. Biochim Biophys Acta. 2003;1647:321–4. doi: 10.1016/s1570-9639(03)00082-7. [DOI] [PubMed] [Google Scholar]
- 159.Morton DP, Parsons SM. Biosynthetic direction substrate kinetics and product inhibition studies on the first enzyme of histidine biosynthesis, adenosine triphosphate phosphoribosyltransferase. Arch Biochem Biophys. 175:677–86. doi: 10.1016/0003-9861(76)90559-2. [DOI] [PubMed] [Google Scholar]
- 160.Morton DP, Parsons SM. Inhibition of ATP phosphoribosyltransferase by AMP and ADP in the absence and presence of histidine. Arch Biochem Biophys. 1977;181:643–8. doi: 10.1016/0003-9861(77)90270-3. [DOI] [PubMed] [Google Scholar]
- 161.Morton DP, Parsons SM. Synergistic inhibition of ATP phosphoribosyltransferase by guanosine tetraphosphate and histidine. Biochem Biophys Res Commun. 1977;74:172–7. doi: 10.1016/0006-291x(77)91390-0. [DOI] [PubMed] [Google Scholar]
- 162.Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol. 2007;144:890–903. doi: 10.1104/pp.107.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Murray ML, Hartman PE. Overproduction of hisH and hisF gene products leads to inhibition of cell cell division in Salmonella. Can J Microbiol. 1972;18:671–81. doi: 10.1139/m72-105. [DOI] [PubMed] [Google Scholar]
- 164.Myers RS, Amaro RE, Luthey-Schulten ZA, Davisson VJ. Reaction coupling through interdomain contacts in imidazole glycerol phosphate synthase. Biochemistry. 2005;44:11974–85. doi: 10.1021/bi050706b. [DOI] [PubMed] [Google Scholar]
- 165.Myers RS, Jensen JR, Deras IL, Smith JL, Davisson VJ. Substrate-induced changes in the ammonia channel for imidazole glycerol phosphate synthase. Biochemistry. 2003;42:7013–22. doi: 10.1021/bi034314l. [DOI] [PubMed] [Google Scholar]
- 166.Nagai A, Ward E, Beck J, Tada S, Chang JY, Scheidegger A, Ryals J. Structural and functional conservation of histidinol dehydrogenase between plants and microbes. Proc Natl Acad Sci USA. 1991;88:4133–7. doi: 10.1073/pnas.88.10.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Neidhardt FC. Chemical composition of Escherichia coli. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology; Washington, DC.: 1987. [Google Scholar]
- 168.Nikaido K, Liu PQ, Ames GF. Purification and characterization of HisP, the ATP-binding subunit of a traffic ATPase (ABC transporter), the histidine permease of Salmonella typhimurium. Solubility, dimerization, and ATPase activity. J Biol Chem. 1997;272:27745–52. doi: 10.1074/jbc.272.44.27745. [DOI] [PubMed] [Google Scholar]
- 169.O'Byrne CP, Ni Bhriain N, Dorman CJ. The DNA supercoiling-sensitive expression of the Salmonella typhimurium his operon requires the his attenuator and is modulated by anaerobiosis and by osmolarity. Mol Microbiol. 1992;6:2467–76. doi: 10.1111/j.1365-2958.1992.tb01423.x. [DOI] [PubMed] [Google Scholar]
- 170.O'Donovan GA, Ingraham JL. Cold-sensitive mutants of Escherichia coli resulting from increased feedback inhibition. Proc Natl Acad Sci USA. 1965;54:451–7. doi: 10.1073/pnas.54.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst. 2008;4:135–47. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 172.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–15. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Palmer DT, Blum PH, Artz SW. Effects of the hisT mutation of Salmonella typhimurium on translation elongation rate. J Bacteriol. 1983;153:357–63. doi: 10.1128/jb.153.1.357-363.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Parsons SM, Koshland DE., Jr. A rapid isolation of phosphoribosyladenosine triphosphate synthetase and comparison to native enzyme. J Biol Chem. 1974;249:4104–9. [PubMed] [Google Scholar]
- 175.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–8. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pease AJ, Roa BR, Luo W, Winkler ME. Positive growth rate-dependent regulation of the pdxA, ksgA, and pdxB genes of Escherichia coli K-12. J Bacteriol. 2002;184:1359–69. doi: 10.1128/JB.184.5.1359-1369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Piszkiewicz D, Tilley BE, Rand-Meir T, Parsons SM. Amino acid sequence of ATP phosphoribosyltransferase of Salmonella typhimurium. Proc Natl Acad Sci USA. 1979;76:1589–92. doi: 10.1073/pnas.76.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pons FW, Neubert U, Muller P. Evidence for frameshift mutations in the hisH gene of Escherichia coli causing synthesis of a partially active glutamine amidotransferase. Genetics. 1988;120:657–65. doi: 10.1093/genetics/120.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Price MN, Alm EJ, Arkin AP. The histidine operon is ancient. J Mol Evol. 2006;62:807–8. doi: 10.1007/s00239-005-0191-3. [DOI] [PubMed] [Google Scholar]
- 180.Prival MJ, Cebula TA. Sequence analysis of mutations arising during prolonged starvation of Salmonella typhimurium. Genetics. 1992;132:303–10. doi: 10.1093/genetics/132.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rangarajan ES, Proteau A, Wagner J, Hung MN, Matte A, Cygler M. Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway. J Biol Chem. 2006;281:37930–41. doi: 10.1074/jbc.M604916200. [DOI] [PubMed] [Google Scholar]
- 182.Riccio A, Bruni CB, Rosenberg M, Gottesman M, McKenney K, Blasi F. Regulation of single and multicopy his operons in Escherichia coli. J Bacteriol. 1985;163:1172–9. doi: 10.1128/jb.163.3.1172-1179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Riggs D, Artz S. The hisD-hisC gene border of the Salmonella typhimurium histidine operon. Mol Gen Genet. 1984;196:526–9. doi: 10.1007/BF00436203. [DOI] [PubMed] [Google Scholar]
- 184.Riggs DL, Mueller RD, Kwan HS, Artz SW. Promoter domain mediates guanosine tetraphosphate activation of the histidine operon. Proc Natl Acad Sci USA. 1986;83:9333–7. doi: 10.1073/pnas.83.24.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Roth JR, Anton DN, Hartman PE. Histidine regulatory mutants in Salmonella typhimurium. I. Isolation and general properties. J Mol Biol. 1966;22:305–23. doi: 10.1016/0022-2836(66)90134-3. [DOI] [PubMed] [Google Scholar]
- 186.Rudd KE, Bochner BR, Cashel M, Roth JR. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J Bacteriol. 1985;163:534–42. doi: 10.1128/jb.163.2.534-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rudd KE, Menzel R. his operons of Escherichia coli and Salmonella typhimurium are regulated by DNA supercoiling. Proc Natl Acad Sci USA. 1987;84:517–21. doi: 10.1073/pnas.84.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schimmel P, Giege R, Moras D, Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA. 1993;90:8763–8. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schmid MB, Roth JR. Internal promoters of the his operon in Salmonella typhimurium. J Bacteriol. 1983;153:1114–9. doi: 10.1128/jb.153.2.1114-1119.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schramm G, Zapatka M, Eils R, Konig R. Using gene expression data and network topology to detect substantial pathways, clusters and switches during oxygen deprivation of Escherichia coli. BMC Bioinformatics. 2007;8:149. doi: 10.1186/1471-2105-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schweitzer BA, Loida PJ, CaJacob CA, Chott RC, Collantes EM, Hegde SG, Mosier PD, Profeta S. Discovery of imidazole glycerol phosphate dehydratase inhibitors through 3-D database searching. Bioorg Med Chem Lett. 2002;12:1743–6. doi: 10.1016/s0960-894x(02)00283-4. [DOI] [PubMed] [Google Scholar]
- 192.Scott JF, Roth JR, Artz SW. Regulation of histidine operon does not require hisG enzyme. Proc Natl Acad Sci USA. 1975;72:5021–5. doi: 10.1073/pnas.72.12.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shand RF, Blum PH, Mueller RD, Riggs DL, Artz SW. Correlation between histidine operon expression and guanosine 5′-diphosphate-3′-diphosphate levels during amino acid downshift in stringent and relaxed strains of Salmonella typhimurium. J Bacteriol. 1989;171:737–43. doi: 10.1128/jb.171.2.737-743.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sheppard DE. Mutants of Salmonella typhimurium Resistant to Feedback Inhibition by L-Histidine. Genetics. 1964;50:611–23. doi: 10.1093/genetics/50.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shyamala V, Schneider E, Ames GF. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. Embo J. 1990;9:939–46. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Singer CE, Smith GR, Cortese R, Ames BN. [Mutant tRNA His ineffective in repression and lacking two pseudouridine modifications.]. Nat New Biol. 1972;238:72–4. doi: 10.1038/newbio238072a0. [DOI] [PubMed] [Google Scholar]
- 197.Sinha SC, Chaudhuri BN, Burgner JW, Yakovleva G, Davisson VJ, Smith JL. Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold. J Biol Chem. 2004;279:15491–8. doi: 10.1074/jbc.M312733200. [DOI] [PubMed] [Google Scholar]
- 198.Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci USA. 1999;96:8985–90. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sivaraman J, Li Y, Larocque R, Schrag JD, Cygler M, Matte A. Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5′-phosphate and lhistidinol phosphate. J Mol Biol. 2001;311:761–76. doi: 10.1006/jmbi.2001.4882. [DOI] [PubMed] [Google Scholar]
- 200.Sivaraman J, Myers RS, Boju L, Sulea T, Cygler M, Jo Davisson V, Schrag JD. Crystal structure of Methanobacterium thermoautotrophicum phosphoribosyl-AMP cyclohydrolase HisI. Biochemistry. 2005;44:10071–80. doi: 10.1021/bi050472w. [DOI] [PubMed] [Google Scholar]
- 201.Smith DW, Ames BN. Intermediates in the early steps of histidine biosynthesis. J Biol Chem. 1964;239:1848–55. [PubMed] [Google Scholar]
- 202.Smulski DR, Huang LL, McCluskey MP, Reeve MJ, Vollmer AC, Van Dyk TK, LaRossa RA. Combined, functional genomic-biochemical approach to intermediary metabolism: interaction of acivicin, a glutamine amidotransferase inhibitor, with Escherichia coli K-12. J Bacteriol. 2001;183:3353–64. doi: 10.1128/JB.183.11.3353-3364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sonenshein AL. Introduction to metabolic pathways. In: Sosenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram Positive Bacteria. American Society for Microbiology; Washington, D.C.: 1993. pp. 127–132. [Google Scholar]
- 204.Stepansky A, Leustek T. Histidine biosynthesis in plants. Amino Acids. 2006;30:127–42. doi: 10.1007/s00726-005-0247-0. [DOI] [PubMed] [Google Scholar]
- 205.Stephens JC, Artz SW, Ames BN. Guanosine 5′-diphosphate 3′-diphosphate (ppGpp): positive effector for histidine operon transcription and general signal for amino-acid deficiency. Proc Natl Acad Sci U S A. 1975;72:4389–93. doi: 10.1073/pnas.72.11.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sterboul CC, Kleeman JE, Parsons SM. Purification and characterization of a mutant ATP phosphoribosyltransferase hypersensitive to histidine feedback inhibition. Arch Biochem Biophys. 1977;181:632–42. doi: 10.1016/0003-9861(77)90269-7. [DOI] [PubMed] [Google Scholar]
- 207.Stern MJ, Ames GF, Smith NH, Robinson EC, Higgins CF. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–26. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 208.Tebar AR, Ballesteros AO. Kinetic properties of ATP phosphoribosyltransferase of Escherichia coli. Mol Cell Biochem. 1976;11:131–6. doi: 10.1007/BF01744993. [DOI] [PubMed] [Google Scholar]
- 209.Tebar AR, Fernandez VM, Martin Del Rio R, Ballesteros AO. Studies on the quaternary structure of the first enzyme for histidine biosynthesis. Experientia. 1973;29:1477–9. doi: 10.1007/BF01943865. [DOI] [PubMed] [Google Scholar]
- 210.Teng H, Grubmeyer C. Mutagenesis of histidinol dehydrogenase reveals roles for conserved histidine residues. Biochemistry. 1999;38:7363–71. doi: 10.1021/bi982758p. [DOI] [PubMed] [Google Scholar]
- 211.Teng H, Segura E, Grubmeyer C. Conserved cysteine residues of histidinol dehydrogenase are not involved in catalysis. Novel chemistry required for enzymatic aldehyde oxidation. J Biol Chem. 1993;268:14182–8. [PubMed] [Google Scholar]
- 212.Thomale J, Nass G. Alteration of the intracellular concentration of aminoacyl-tRNA synthetases and isoaccepting tRNAs during amino-acid limited growth in Escherichia coli. Eur J Biochem. 1978;85:407–18. doi: 10.1111/j.1432-1033.1978.tb12253.x. [DOI] [PubMed] [Google Scholar]
- 213.Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 215.Tsui HC, Arps PJ, Connolly DM, Winkler ME. Absence of hisT-mediated tRNA pseudouridylation results in a uracil requirement that interferes with Escherichia coli K-12 cell division. J Bacteriol. 1991;173:7395–400. doi: 10.1128/jb.173.22.7395-7400.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Turnbough CL, Jr., Neill RJ, Landsberg R, Ames BN. Pseudouridylation of tRNAs and its role in regulation in Salmonella typhimurium. J Biol Chem. 1979;254:5111–9. [PubMed] [Google Scholar]
- 217.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J. 2001;20:4863–73. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Verde P, Frunzio R, di Nocera PP, Blasi F, Bruni CB. Identification, nucleotide sequence and expression of the regulatory region of the histidine operon of Escherichia coli K-12. Nucleic Acids Res. 1981;9:2075–86. doi: 10.1093/nar/9.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vitreschak AG, Lyubetskaya EV, Shirshin MA, Gelfand MS, Lyubetsky VA. Attenuation regulation of amino acid biosynthetic operons in proteobacteria: comparative genomics analysis. FEMS Microbiol Lett. 2004;234:357–70. doi: 10.1016/j.femsle.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 220.Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–87. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Whitfield HJ, Jr., Gutnick DL, Margolies MN, Martin RG, Rechler MM, Voll MJ. Relative translation frequencies of the cistrons of the histidine operon. J Mol Biol. 1970;49:245–9. doi: 10.1016/0022-2836(70)90391-8. [DOI] [PubMed] [Google Scholar]
- 222.Winkler ME, Roth DJ, Hartman PE. Promoter- and attenuator-related metabolic regulation of the Salmonella typhimurium histidine operon. J Bacteriol. 1978;133:830–43. doi: 10.1128/jb.133.2.830-843.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Winkler ME, Zawodny RV, Hartman PE. Mutation spoT of Escherichia coli increases expression of the histidine operon deleted for the attenuator. J Bacteriol. 1979;139:993–1000. doi: 10.1128/jb.139.3.993-1000.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–90. [PubMed] [Google Scholar]
- 225.Yanofsky C, Platt T, Crawford IP, Nichols BP, Christie GE, Horowitz H, VanCleemput M, Wu AM. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981;9:6647–68. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]