Abstract
ERp44 is a pH-regulated chaperone of the secretory pathway. In the acidic milieu of the Golgi, its C-terminal tail changes conformation, simultaneously exposing the substrate-binding site for cargo capture and the RDEL motif for ER retrieval via interactions with cognate receptors. Protonation of cysteine 29 in the active site allows tail movements in vitro and in vivo. Here we show that also conserved histidines in the C-terminal tail regulate ERp44 in vivo. Mutants lacking these histidines are hyperactive in retaining substrates. Surprisingly, they are also O-glycosylated and partially secreted. Co-expression of client proteins prevents secretion of the histidine mutants, forcing tail opening and RDEL accessibility. Client-induced RDEL exposure allows retrieval of proteins from distinct stations along the secretory pathway, as indicated by the changes in O-glycosylation patterns upon over-expression of different partners. The ensuing gradients may help optimising folding and assembly of different cargoes. Endogenous ERp44 is O-glycosylated and secreted by human primary endometrial cells, suggesting possible pathophysiological roles of these processes.
Introduction
ERp44 is a multifunctional chaperone of the PDI-family that regulates Ca2+ signalling, redox homeostasis and thiol-dependent protein quality control at the endoplasmic reticulum (ER)-Golgi interface (Anelli et al., 2012; Cortini and Sitia, 2010). ERp44 is key for the retrieval of orphan subunits of disulfide-linked oligomers, like IgM and adiponectin (Anelli et al., 2003; Anelli et al., 2007; Qiang et al., 2007) and for the intracellular localization of Ero1 oxidases, Sumf1 and peroxiredoxin 4 (Prx4) (Fraldi et al., 2008; Kakihana et al., 2013; Otsu et al., 2006). It associates with its client proteins covalently via cysteine 29 (C29) and non-covalently, likely through the surrounding hydrophobic patches in the substrate-binding site (SBS). In the available crystal structure, the SBS is shileded by a C-terminal tail (C-tail) (Wang et al., 2008). Tail movements simultaneously expose the SBS and the C-terminal RDEL, allowing interaction with the KDELR and the retrieval to the ER of the chaperone and its client proteins (Anelli et al., 2003; Cortini and Sitia, 2010; Vavassori et al., 2013).
We demonstrated recently that the pH gradient existing along the early secretory compartment (ESC), a term used herein to define the ER, ERGIC and Golgi complex, regulates C29 protonation and, consequently, ERp44 tail opening (Vavassori et al., 2013). In the closed state, C29 is concealed in the interface between domain a and the C-tail, forming the hydrogen bonds with the side chains of T369 and S32 and the main chain amides of F31 and S32. At lower pH, however, the hydrogen bonds are significantly weakened probably due to the protonation of the C29 thiol group, leading to the release of the C-tail. Such pH-dependent conformational change is advantageous in the regulated Golgi to ER transport of client proteins by ERp44. Besides C29, T369 and S32, we also noted that five histidines residues located at the border between the domain b′ and the C-tail are highly conserved in the ERp44 family proteins (Fig. 1A). To test the possibility that these residues could also contribute in modulating ERp44 activity, we here generated deletion and replacement mutants and analyzed their activity in vitro and in vivo. Our data reveal a novel mechanism controlling the binding of both clients and KDELR by ERp44 at ER-Golgi interface. We also show that O-glycosylated ERp44 can be secreted by certain cell types.
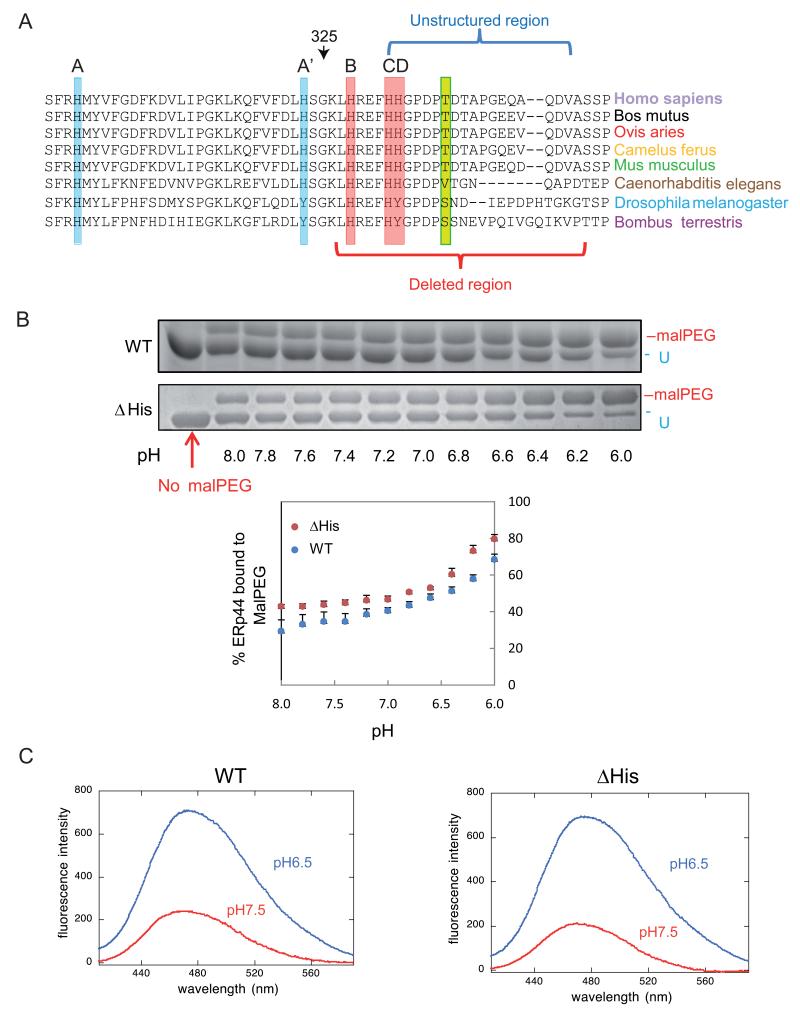
Figure 1. pH dependent conformational changes of ΔHis ERp44 in vitro.
A) Sequence conservation in the proximal part of the ERp44 C-tail. ClustalW2 alignment of the histidine-rich region and of the first part of the C-terminal tail of ERp44 reveals high sequence conservation in different organisms. Three histidines reside in the tail (B, C and D) and two in the b′ domain (A and A′). A, B and C (highlighted in red) are the most conserved histidines, while A′ and D (in pale blue) are absent in arthropods. The unstructured region in the crystal and the region deleted in the ΔHis mutant are indicated by blue and red brackets.
B) pH-dependent accessibility of ERp44 cysteine 29. Upon in vitro alkylation with MalPEG (10 minutes at room temperature), ERp44 undergoes an easily detectable mobility shift in SDS-PAGE, indicating the accessibility of C29 to MalPEG (upper two panels; (Vavassori et al., 2013)). The percentage of MalPEG-bound ERp44 was plotted as a function of pH (lower panel). Clearly, lowering the pH increases the accessibility of C29 to MalPEG in the ΔHis mutant as well as in wt ERp44.
C) pH-dependent tail movements in wt and ΔHis ERp44 compared by ANS binding in vitro. ANS fluorescence spectra were measured for ERp44 wt or ΔHis. The exposure of hydrophobic surfaces correlates with increased and blue-shifted ANS fluorescence (Vavassori et al., 2013). Also with this assay, ERp44 wt and ΔHis show similar pH dependent changes.
Results
In the available ERp44 crystals, part of the C-tail (residues 332-350) is not resolved due to the lack of electron density (Wang et al., 2008). Two histidines at the beginning of this unstructured fragment (332-333) and additional ones in the upstream helices (299, 323 and 328) are highly conserved in ERp44 (Fig. 1 A), as are the surrounding sequences. To investigate the potential regulatory role of this histidine-rich region, we first replaced residues 326-350 with a short spacer (SGSG) to generate the ΔHis mutant (see supplementary Fig. 1A for details) and investigated its in vitro reactivity with polyethylene glycol 2000-modified maleimide (MalPEG, Fig. 1B) or 1-anilinonaphthalene-8-sulfonate (ANS Fig.1C) as a function of pH. ERp44 contains two unpaired cysteines (C29 and C63) and four cysteines engaged in structural disulfide bonds (C160-C212 and C272-C289 in domains b and b′, respectively) (Wang et al., 2008). In a preceding study, we found that C29 was the primary reactor of MalPEG, while C63 was also modifiable, though with much lower efficiency (Vavassori et al., 2013). In the ΔHis mutant, the C-tail was still long enough to shield C29 and the surrounding hydrophobic patches at pH >7.0 (Fig. 1B and 1C). Even after deletion of the His-rich loop, however, more MalPEG was bound to ERp44 and the ANS fluorescence peak was blue-shifted and enhanced upon lowering the pH. These data suggested that in vitro C29 and the hydrophobic regions in ΔHis became exposed at pH values encountered in the Golgi (6.5), as we previously demonstrated for wild type (wt) ERp44 (Vavassori et al., 2013).
The histidine rich loop regulates the accessibility of the active site and RDEL motif in vivo
Despite ΔHis behaved similar to wt ERp44 in vitro, the mutant did show evident phenotypes in vivo. Indeed, ΔHis was partially secreted by HeLa cells (Fig. 2A, lane 6, black arrowhead), suggesting lower RDEL accessibility. Accordingly, the double mutant ΔHis/ΔRDEL was secreted at levels similar to ERp44 ΔRDEL (Fig. 2B). Pulse chase assays (Supplementary Figure 2A) confirmed that the ΔHis mutant was secreted by HeLa transfectants, and not degraded to a significant extent. Taken together, these findings indicate that in the absence of the His loop the RDEL motif interacts less efficiently with its cognate receptors.
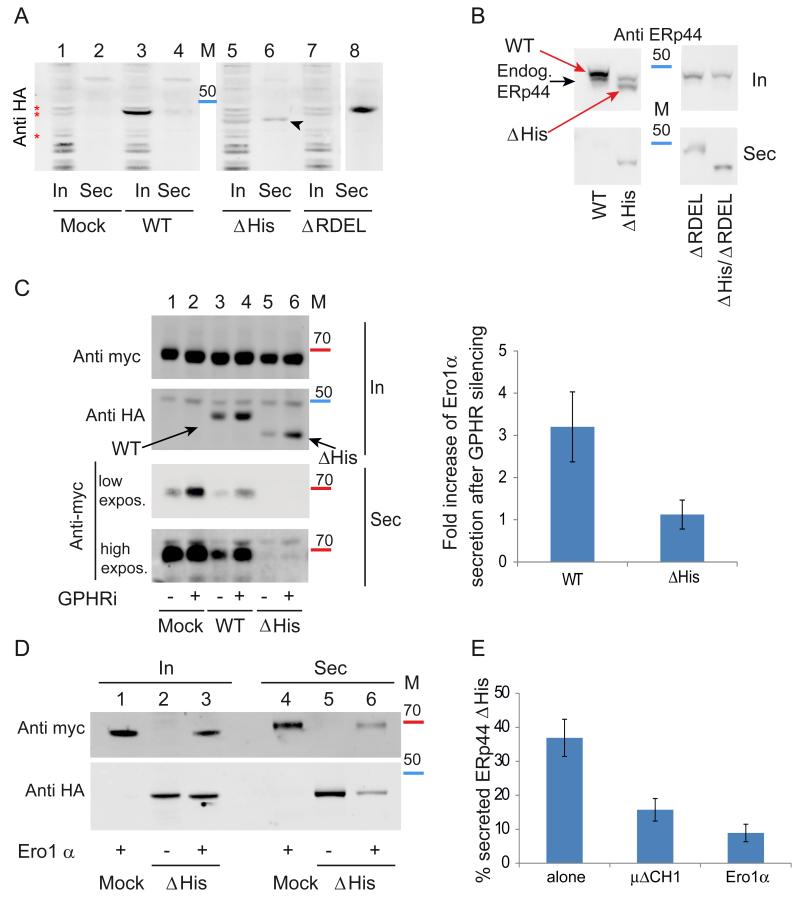
Figure 2. The histidine-rich loop regulates accessibility of the active site and RDEL motif in vivo.
A) ERp44 mutants lacking the histidine-rich loop are secreted. The indicated HeLa transfectants were cultured for 4 hours in fresh medium, and aliquots from lysates (In) and supernatants (Sec) analyzed under reducing conditions by western blotting with anti-HA antibodies. The first two lanes (Mock) show cells transfected with an empty plasmid. The arrowhead points at secreted ΔHis ERp44. Red asterisks point at background bands recognized by anti-HA antibodies. Albeit not elegant, these unwanted bands provide god loading controls. In all Western blot analyses, molecular weight markers were used (see also Fig. 5): the red and blue bands indicate the migration of the 70 and 50 kDa markers.
B) Residual retention of ERp44 ΔHis is due to KDELR activity. Aliquots from lysates (In) and spent media (Sec) were collected from HeLa transfectants (treated as in A) and analyzed under reducing conditions by WB with monoclonal anti-ERp44 antibodies (36C9) that recognize also endogenous ERp44. The black and red arrows point to endogenous and HA-tagged over-expressed ERp44 molecules, respectively. Note that deletion of the C-terminal RDEL allows complete ΔHis secretion, while no endogenous ERp44 is detectable extracellularly.
C) ERp44 ΔHis is hyperactive in retaining Ero1α in a pH-independent manner. HeLa cells were co-transfected with ERp44 variants and Ero1α as indicated and the pH gradient in the ESC was altered by GPHR silencing (− lanes show cells treated with irrelevant siRNA) as described previously (Vavassori et al., 2013). Aliquots from lysates (In) and culture media (Sec) were collected and analyzed by WB by sequential staining of the same filter with anti-HA and anti-myc antibodies to detect ERp44 or Ero1α respectively (as indicated on the left hand margin). Note that less Ero1 is secreted out of cells expressing the ΔHis mutant although its intracellular level is lower than that of wt ERp44. A higher exposure of the secreted samples is shown to detect the small amounts of Ero1α secreted out of cells expressing the ΔHis mutant. Intracellular and secreted proteins were quantified by densitometric analyses, and the mean fold of induction of Ero1α secretion upon GPHR silencing calculated relative to controls (right panel). Data represent the average of ≥ 8 experiments like the one shown in the left panel ± s.d.
D) Client-induced retention of ΔHis. Aliquots of lysates (In) and supernatants (Sec) of the indicated HeLa transfectants were analyzed as above. Note that much less ERp44 ΔHis is secreted in the presence of Ero1α, and vice versa.
E) Secretion of ERp44 ΔHis is inhibited by client proteins (Ero1α and μΔCH1) Secretion of ΔHis in the presence or absence of over-expressed Ero1α or μΔCH1 was analyzed as above. To quantify secretion, we calculated the ratios between the ΔHis bands detected extra- and intracellularly after 4 hours cultivation in fresh medium. To facilitate the comparison amongst different transfectants, the values obtained for wt ERp44 and ΔRDEL were arbitrarily assumed as zero and 100, respectively. Histograms show the average ≥3 experiments +/− s.d.
Next, we investigated whether the ΔHis mutant maintained the ability to prevent the secretion of Ero1α. Like peroxiredoxin 4 (Prx4), also Ero1α lacks a KDEL motif and is retained inside cells through interactions with PDI or ERp44 (Kakihana et al., 2013; Otsu et al., 2006). Secretion assays clearly demonstrated that: i) ΔHis was in part secreted (Fig. 2A), ii) ΔHis was more efficient than wt ERp44 in preventing Ero1α secretion (Fig. 2C, lanes 3 and 5), and iii) paradoxically, the co-expression of a client (Ero1α) inhibited the secretion of the retainer (ΔHis, Fig. 2D, lanes 5 and 6). These findings suggested that client binding could favour RDEL exposure in ΔHis. The ‘client-induced retention’ of ΔHis was evident upon co-expressing not only Ero1α (Fig. 2E) but also Ig-μΔCH1, another known ERp44 substrate (Anelli et al., 2007;Ronzoni et al., 2010;Vavassori et al., 2013).
To analyze the pH dependency of ΔHis in living cells, we silenced the Golgi pH regulator (GPHR (Maeda et al., 2008)), specifically raising the pH in the Golgi (Vavassori et al., 2013). Due to the unavailability of anti-GPHR antibodies, the efficiency of silencing was monitored by RT-PCR assays (Supplementary Figure 2B). As previously observed (Vavassori et al., 2013), neutralizing the ER-Golgi pH gradient allowed Ero1α secretion (Fig.2C, lanes 1-2), also in cells over-expressing wt ERp44 (lanes 3-4). In contrast, basification of the cisGolgi did not affect the capability of ΔHis to retain Ero1α (Fig. 2C, lanes 5-6).
Thus, deletion of histidine rich loop allowed secretion of ERp44 unless high-affinity client proteins were co-expressed. At the same time, it increased its ability to retain Ero1α (Fig. 2C) and Ig-μΔCH1 (not shown) and limited its pH dependency (Fig. 2C). Secreted Ero1α could be detected only upon overexposure of the Western blot, such was the efficiency of ΔHis in retaining the oxidase.
Deletion of histidine rich loop favours accumulation of ERp44 in distal ESC stations
How could ΔHis be so efficient in retaining Ero1α? Endogenous ERp44 is mainly localized in the ERGIC and cisGolgi, partly due to interactions with the lectin ERGIC-53 (Anelli et al., 2007). In contrast, over-expressed ERp44 accumulates in the ER (Anelli et al., 2002; Anelli et al., 2003). The ΔHis mutant localized more distally than wt ERp44 when expressed in the secretory HepG2 cell line (Fig. 3A). Organelle fractionation assays confirmed that ΔHis accumulated in ERGIC and cisGolgi, a localization that would favour capture of clients destined to be retrieved (data not shown).
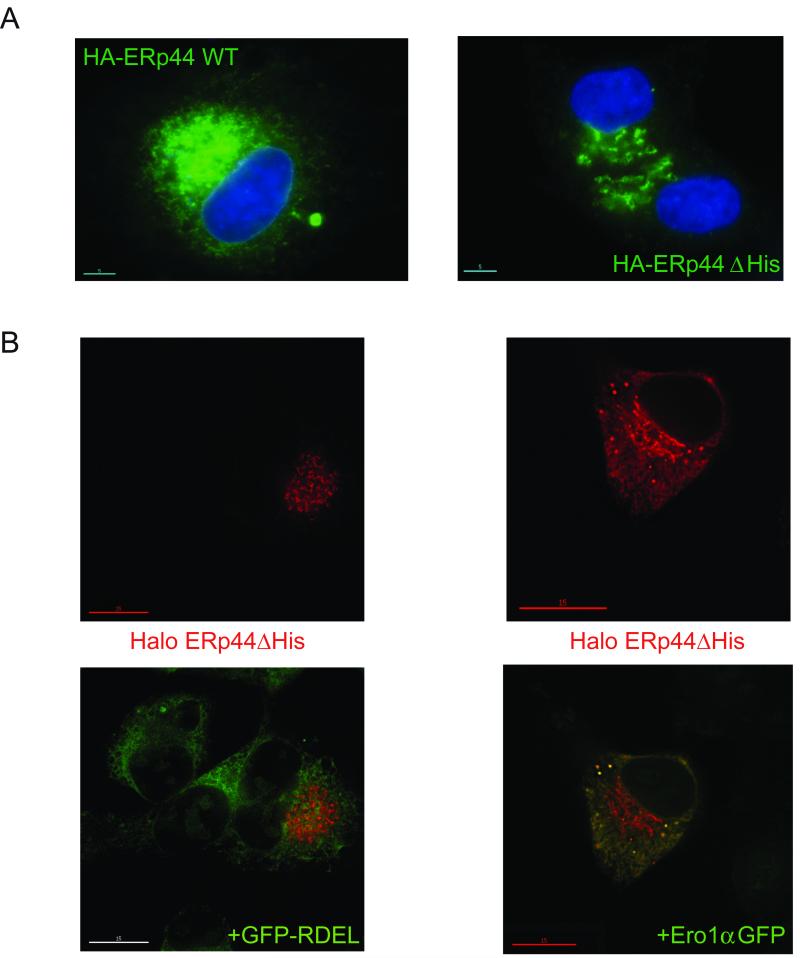
Figure 3. Deletion of histidine rich loop favours accumulation of ERp44 in distal ESC stations.
A) Steady state localization. HepG2 transfectants expressing HA-tagged wt and ΔHis ERp44 were analyzed by immunofluorescence with anti-HA (green). The blue staining (DAPI) was used to detect nuclei. Note that much less green staining is detected in the nuclear membrane or peripheral ER in ΔHis transfectants, as compared to cells expressing wt ERp44.
B) Client-induced relocalisation of ΔHis to the ER. Co-expressing Ero1α-GFP, but not sGFP-RDEL, causes the re-localization of ΔHis in the ER. HepG2 cells were co-transfected with Halo-tagged ERp44 ΔHis and sGFP-RDEL or Ero1α-GFP and then decorated with a Halo ligand (red signal). Clearly, no co-localization is detectable between sGFP-RDEL (green) and ERp44 ΔHis (red) while expression of Ero1α-GFP causes ΔHis to localize also in the ER yielding a reticular yellow staining (right panel).
Based on the earlier observation that overexpression of high-affinity client proteins prevented secretion of ΔHis, we investigated whether the presence of Ero1α-GFP could alter the subcellular localization of the mutant. An ER-resident protein that does not bind ERp44 (sGFP-RDEL) was used as a control (Fig. 3B). Clearly, the co-expression of Ero1α-GFP, but not sGFP-RDEL, caused re-localization of ΔHis into the ER. These data reinforced the notion that interactions with high affinity clients (i.e. Ero1α) keep the tail of ERp44 in an open conformation, favouring RDEL exposure and KDELR-dependent retrieval of the complex to the ER.
Conserved histidines regulate C-tail movements in vivo
Considering their conservation from Homo sapiens to Caenorhabditis elegans (Fig. 1A), we replaced histidines 299, 323, 328, 332 and 333 (called hereafter A, A′, B, C or D, respectively) with alanines, singularly or in combinations. The former two are part of the domain b′, whilst B, C and D are located in the C-tail (Wang et al., 2008). Histidines A, A′ and B are part of structured 310 helices while histidines C and D reside in a flexible loop (Fig. 4A) and are hence most likely exposed to the solvent. . A, B and C are conserved throughout evolution. Single substitution of each of them allowed partial secretion. In contrast, replacing the less conserved histidines, A′ or D, had little if any effect (Table I and data not shown). Clearly, the more histidines were substituted, the more ERp44 was secreted (Fig. 4B-C). Deleting the entire loop had even stronger effects (Fig. 2A). Altogether, these results suggest that in living cells the flexible His-rich loop is important to favour tail movements leading to RDEL exposure in the absence of high affinity clients. Without this loop, the RDEL motif likely remains less accessible to cognate receptors and as a consequence the mutant is secreted. By contrast, co-expression of Ero1α strongly inhibited secretion of the mutants (Fig. 4C), as described earlier for ΔHis (Fig.2 D). Since ERp44 is a pH-regulated chaperone, it is not surprising that the histidines (whose intrinsic pKa is near neutral) contained in or immediately upstream the loop contribute in controlling RDEL exposure in vivo. In line with their high evolutionary conservation, histidines A, B and C appear to be particularly important in regulating tail movements.
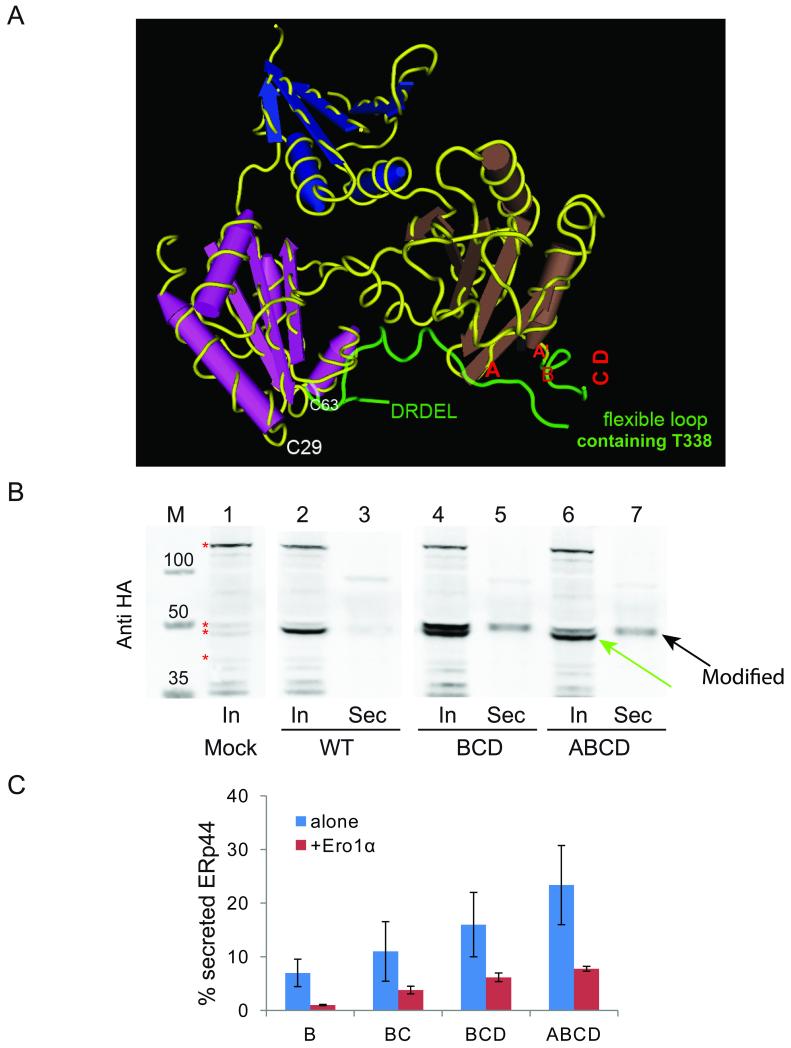
Figure 4. Replacing conserved histidines causes O-glycosylation and secretion of ERp44.
A) 3D localization of conserved histidine residues. The panel highlights the 3D crystal structure of ERp44 with the C-tail in green. Red letters (A, A′, B, C and D) point at the position of the five histidine residues. Histidines C and D are located at the beginning of an unstructured histidine loop (332-350) which, as the five C-terminal DRDEL residues, is not resolved in the crystal.
B) Replacement of histidine residues induces secretion of modified ERp44. Aliquots from lysates (In) and culture media (Sec) were collected from HeLa transfectants expressing the indicated ERp44 variants and blots decorated with anti-HA. Similar experiments were performed for other histidine mutants (data not shown). Note that anti-HA antibodies specifically detect two bands in the lysates of the BCD and ABCD mutants, of which only the upper one is secreted (black arrow). The green arrow points at the only intracellular band recognized in transfectants expressing wt ERp44. Red asterisks point at some background bands recognized by anti-HA antibodies, which provide useful loading controls.
C) His replacement mutants are retrieved more efficiently upon Ero1α co-expression. ERp44 secretion was quantified as described in Figure 2E in HeLa cells expressing the indicated ERp44 mutants alone or with Ero1α. The histograms show the % of secreted ERp44 for the different mutants. The data represent the average of 3 or more experiments ± s.d. in the presence of Ero1α. Secretion efficiency was calculated as described in legend to Figure 2E.
Table I. Phenotypic characterization of ERp44 histidine mutants.
The data summarize the main features of the mutants described in this paper, including the capability of retaining overexpressed Ero1α, and of being secreted in different conditions, and their basal subcellular localization.
| ERp44 variant | Erolα retentiona | ERp44 secretionb | O- glyc. | Subcellular localizationc | ||
|---|---|---|---|---|---|---|
| Untreated | GPHRi | alone | Ero1α | |||
| WT | ++ | + | undetectable | undetectable | − | ER-ERGIC |
| ΔRDEL | − | − | 100%c | 100% | + | Barely detectable |
| ΔHis | ++++ | ++++ | 36% | 9% | − | ERGIC -cisGolgi |
| A | ++ | ND | <10% | undetectable | + | ND |
| A′ | ND | <5% | undetectable | + | ND | |
| B | ++ | ND | <10% | − | + | ER-ERGIC |
| C | ND | ND | <10% | ND | + | ER-ERGIC |
| D | + | ND | <5% | undetectable | + | ER-ERGIC |
| BC | ++ | ND | ≈10% | <5% | + | ERGIC |
| BD | + | ND | <10% | undetectable | + | ND |
| CD | +++ | ++ | <10% | undetectable | + | ERGIC |
| BCD | +++ | ND | 15-20% | ≈5% | + | ERGIC-cisGolgi |
| A′BCD | +++ | ND | ≈10% | ≈5% | + | ND |
| ABCD | +++ | ND | 20-25% | <10% | + | ERGIC-cisGolgi |
Scoring is relative the mutants with the highest (δHis ++++) and lowest (δRDEL -) efficiency. The former efficiently retains Erol even when expressed at rather low levels (see figure 2C).
For ERp44 secretion, scoring is relative to a maximum (100%, δRDEL) and a minimum (0%, wt ERp44, which is undetectable in the spent media (4 hours) of our HeLa transfectants. Precise values are given only for δHis, for which the experiment has been repeated over 10 times.
Subcellular localization was determined by immunofluorescence, as described in Figure 3. Owing to its rapid secretion (Anelli et al., 2007), the δRDEL mutant is barely detectable inside cells.
Secreted ERp44 is O-glycosylated at threonine 338
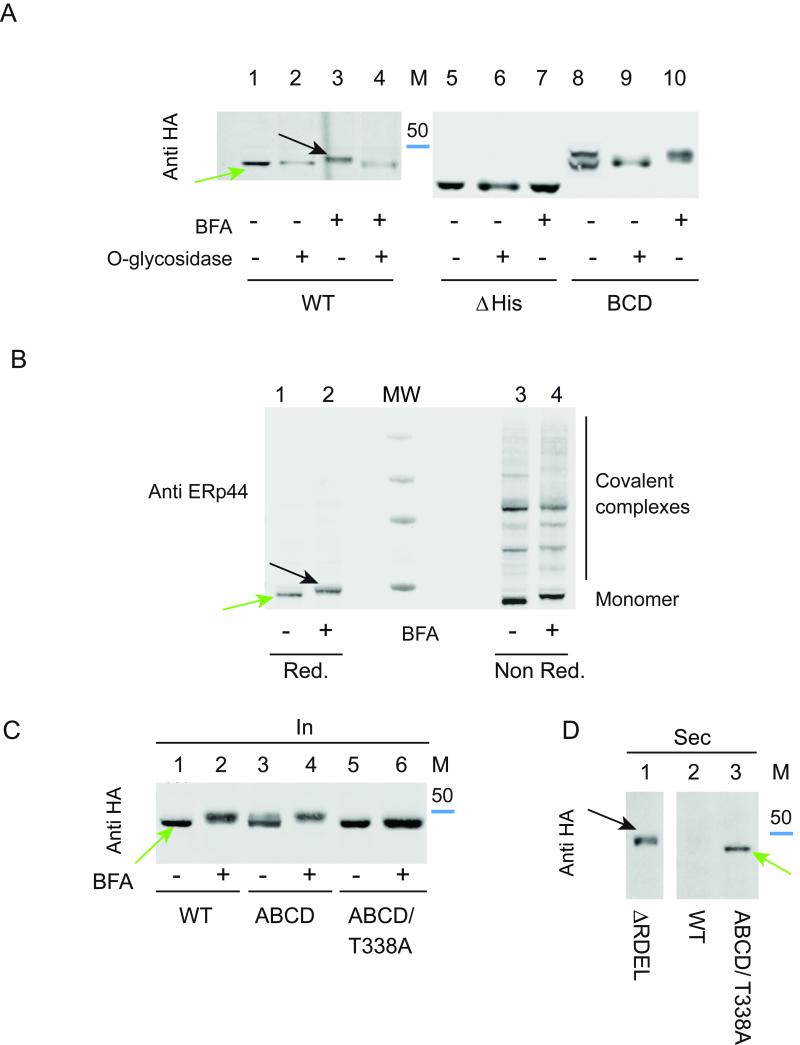
Western blotting analyses revealed the presence of two bands in the lysates of the BCD and ABCD mutants, of which only the upper was secreted (Fig. 4B, see arrows). Although ERp44 has no potential N-glycosylation sites, mutants that escaped KDELR-dependent retrieval could undergo O-glycosylation in the Golgi. To ascertain whether this was indeed the case, we treated HeLa transfectants with brefeldin A (BFA), a drug known to induce the retrograde transport of Golgi enzymes to the ER (Lippincott-Schwartz et al., 1989). The electrophoretic mobility of wt and BCD ERp44, but not of ΔHis, was lower after BFA-treatment (Fig. 5A, compare lanes 3, 7, 10). Treatment with O-glycosidase restored faster migration (lane 4) and abolished the doublet observed in the BCD mutant (lane 9), demonstrating that ERp44 can undergo O-glycosylation before being secreted. In contrast, ΔHis ERp44 was not modified in the presence of BFA, indicating that either the target residue was deleted in this mutant or the replacement caused its inaccessibility to O-glycosyl transferases.
Figure 5. Mapping client-induced retrieval of His replacement mutants with respect to O-glycosylation.
A) Secreted ERp44 is O-glycosylated. HeLa cells transfected as indicated were treated with (lanes 3, 4, 7, 10) or without brefeldin A (BFA) for 4 hours to determine whether retrieval of Golgi enzymes to the ER caused processing of ERp44. Aliquots from the lysates were digested with (lanes 2, 4, 6, 9) or without O-glycosidase (see Materials and methods), resolved by gel electrophoresis and blots decorated with anti-HA. The blue band indicates the migration of the 50 kDa marker.
B) Endogenous ERp44 can also be O-glycosylated and form covalent complexes with client proteins. Untransfected HeLa cells were treated with BFA as above and resolved under reducing (lanes 1-2) and non-reducing (lanes 3-4) conditions before blotting and decoration with 36C9 monoclonal anti ERp44 antibodies. Molecular weight markers (250, 130, 70 and 50 kDa) are shown in the center lane (MW).
C) ERp44 is O-glycosylated on threonine 338. Aliquots from the lysates of the indicated HeLa transfectants treated with or without BFA were analyzed by WB with anti-HA.
D) Replacing threonine 338 does not impede secretion of the ABCD mutant. The supernatants of cells transfectants were resolved under reducing conditions and decorated with anti-HA. The green arrow points at the secreted product of the T338A. This mutant displays faster mobility than the O-glycosylated ΔRDEL secretory products (black arrow).
Noteworthy, also endogenous ERp44 was modified upon BFA treatment (Figure 5B, see black arrow). Non-reducing gels revealed that O-linked glycosylation did not prevent ERp44 from forming covalent complexes with its clients (see lanes 3-4). The fact that most wt ERp44 was not modified in normal conditions implied its retrieval before the modification occurs. In silico prediction programs (Hamby and Hirst, 2008; Steentoft et al., 2013) pointed at threonines 338 and 340 as possible substrates of O-glycosylation. Both are located within the loop deleted in ΔHis, which would explain why this mutant is not O-glycosylated. Replacing the most conserved one, T338, with alanine in a mutant that was in part secreted and modified (ABCD) was sufficient to prevent the processing (Fig. 5C, lanes 5-6). Failure to undergo O-glycosylation did not prevent secretion (Fig. 5D). Thus, ERp44 can be O-glycosylated at T338.
Despite in the experiment shown in Fig. 4B the BCD and ABCD mutants were secreted at similar levels, more O-glycosylated accumulated intracellularly in the former. These findings suggested that BCD could be more efficiently retrieved also from stations downstream the compartment where O-glycosylation takes place. The lower pH of these downstream stations could increase the exposure of RDEL (Vavassori et al., 2013) and/or the activity of KDELR (Wilson et al., 1993).
Exploiting O-glycosylation to map where ERp44 binds its clients
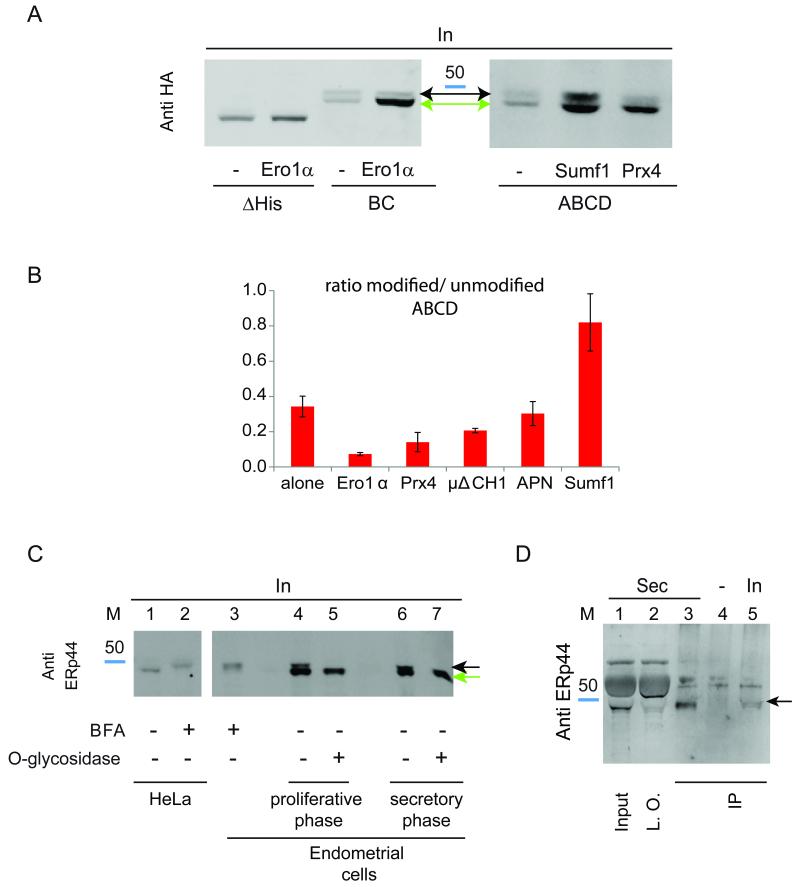
As described in Figs. 2D and 4C, overexpression of high affinity ERp44 clients prevented the secretion of histidine mutants, likely increasing RDEL accessibility. The notion that these histidine mutants could be O-glycosylated prompted us to determine whether the binding of different clients occurred before or after the compartment where this modification takes place. Substrates binding before encountering the glycosylating enzymes would increase the lower molecular weight band accumulating in cell lysates, while O-glycosylated forms would prevail if the interaction occurred in downstream stations. The ABCD mutant was hence expressed with Ero1α, Prx4, Ig-μΔCH1, adiponectin or Sumf1, and the unmodified/modified band ratios were calculated (Fig. 6A). Clearly, Ero1α and Prx4 favoured accumulation of the unglycosylated species. In contrast, Sumf1 induced retrieval of ABCD after it had been processed by O-glycosyl transferases. μΔCH1 and adiponectin had intermediate effects (Fig. 6B). These observations may reflect differences in the binding affinities or pH dependency of the interactions between ERp44 and its clients. Also other factors, including molecules binding to the histidine-rich region, could regulate the strength and location of the interactions between ERp44 and its clients.
Figure 6.
A) Sumf1 can bind ERp44 also after O-glycosylation. HeLa cells were co-transfected as indicated. Aliquots from the lysates were then resolved by SDS-PAGE and blots decorated with anti-HA. Note the accumulation of non-glycosylated histidine mutants in the lysates of cells co-expressing Ero1α or Prx4. Instead, Sumf1 expression allows significant accumulation of O-glycosylated ABCD ERp44.
B) Sequential client-induced retrieval of ERp44 histidine mutants. The histograms show the ratio between modified and unmodified ABCD ERp44 in the presence of Ero1α, Prx4, Sumf1, Ig-μΔCH1 or adiponectin (APN). Data represent the average of ≥3 experiments ± s.d.
C) Endogenous ERp44 is O-glycosylated in primary endometrial cells Aliquots of the lysates obtained from endometrial cells in their proliferative (lanes 4-5) or secretory (lanes 3, 6-7) phase (Di Blasio et al., 1995) were digested with (lanes 5, 7) or without (lanes 3, 4, 6) O-glycosidase and resolved by gel electrophoresis under reducing conditions. Note that upon digestion, the doublet visible in untreated samples collapses into a single higher mobility band, which comprises un-glycosylated and de-glycosylated ERp44 (green arrow). The black arrow points at the O-glycosylated species, the main accumulating in HeLa (lane 2) or endometrial (lane 3) cells after treatment with BFA.
D) Endogenous ERp44 is secreted by endometrial stromal cells Aliquots from the lysates or spent media (48 h) corresponding to 8x104 or 7x105 endometrial cells in their secretory phase were immune-precipitated with 36C9 monoclonal anti ERp44 (IP), resolved under reducing conditions and decorated with rabbit anti ERp44 (JDA1). Lanes 1 and 2 show one third of the spent medium before (Input) or after (left over) immunoprecipitation. The band migrating above the 50 kDa marker is bovine serum albumin, most abundant in foetal calf serum. Cross-linked beads were used as a negative control (lane 4). Endogenous ERp44 is clearly detectable in the culture medium and migrates with a molecular weight similar to the O-glycosylated ERp44 species present intracellularly (black arrow).
The above data demonstrated that certain ERp44 mutants could be O-glycosylated. To verify the physiological relevance of ERp44 processing, we screened a wide panel of cell lines and tissues (E. Yoboue, TA, SS and RS, data not shown). ERp44 yielded a doublet band in primary endometrial cells obtained from normal volunteers (Figure 6C, lanes 4 and 6). The mobility of the upper band (highlighted by a black arrow) is comparable to the one accumulating upon treatment with BFA (lanes 2 and 3). Treatment with O-glycosidase collapsed the doublet into a single band (lanes 5-7, see green arrow), confirming that endogenous ERp44 can undergo O-glycosylation. Notably, the relative abundance of O-glycosylated ERp44 was higher in endometrial cells in the secretory than in the proliferative phase (Di Blasio et al., 1995). Processed ERp44 was detected in the spent medium of secretory endometrial cells (panel D, lanes 1 and 3).
Discussion
To promote productive folding, chaperones must bind and release their clients. Some are regulated by ATP, others by redox, while ERp44 exploits the pH gradient existing between the ER and the Golgi (Vavassori et al., 2013, and references therein). How does ERp44 sense pH? We show here that highly conserved histidines in the ERp44 C- tail provide a second sensing device that acts co-ordinately with the C29 protonation mechanism previously shown to regulate ERp44 cycling and activity within ESC (Vavassori et al., 2013).
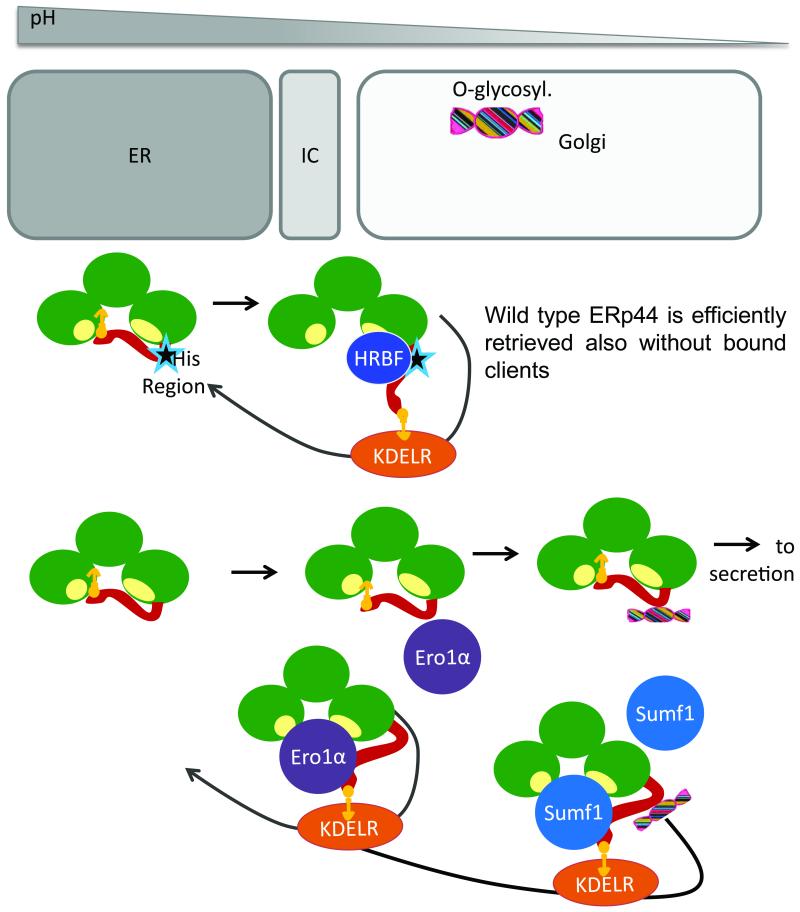
Deletion or replacement of the key conserved ERp44 histidines had notable consequences in vivo. In fact, it caused secretion of the histidine mutants by cells, likely owing to reduced accessibility of the RDEL motif to KDELR in the Golgi. Accordingly, co-expression of clients that would bind to the SBS and force RDEL exposure restored their retrieval (see Fig. 7). In contrast, deletion or mutation of the conserved histidines had marginal effects in vitro (Fig 1A and our unpublished results), indicating that additional ‘histidine region-binding factors’ regulate ERp44 in vivo (Fig. 7). Such factors could bind ERp44 in the ER and limit its forward transport, accounting for the enrichment of the histidine mutants in post-ER compartments (Fig. 3). Alternatively, the mutants may be present and/or be activated in distal compartments to assist tail opening (and consequent RDEL exposure) in the absence of clients. Thus, whilst the C29-based tail regulation is intrinsic to ERp44 and can be revealed in vitro, the histidine-dependent mechanisms are evident only in vivo as they require additional regulatory elements.
Figure 7. Schematic model of ERp44 regulation.
The lower pH encountered by ERp44 as it proceeds along the early secretory pathway, favors opening of the C-tail and KDELR binding regardless of the presence or absence of high-affinity client proteins. Accordingly, ERp44 mutants that bind few if any substrate in cells (e.g ERp44C29S, (Anelli et al., 2003) are not secreted. On the other hand, mutants lacking key conserved histidines or a loop at the border between the domain b′ and the tail (star) bind poorly to KDELRs, proceed towards the extracellular space and are O-glycosylated. Client binding induces KDELR-dependent retrieval of histidine mutants before (Ero1, Prx4) or also after (Sumf1) O-glycosylation takes place. Since histidine mutants remain pH sensitive in vitro, we hypothesize that histidine-region binding factor(s) (HRBF) favor tail movements in vivo, allowing efficient RDEL exposure.
Histidine mutants are more efficient than wt ERp44 in preventing Ero1α secretion. This feature, rather unexpected since they are partly secreted, may reflect their increased specificity towards Ero1α. Some clients would be more effective than others in competing with the C-tail for the substrate-binding site in the histidine mutants, which likely have a higher kinetic barrier for tail opening. In this context, Ero1 and Prx4 bind to ERp44 mainly before it undergoes O-glycosylation, whilst Sumf1 can interact also afterwards (Fig. 6A-B). These observations suggest that different proteins could be deposited sequentially in the exocytic pathway, depending on their affinity for ERp44 or on the pH dependency of the chaperone-client interaction. Along these lines, it is interesting to note that most ERp44 substrates/partners which are ER resident enzymes (Ero1α, Prx4, and Sumf1) lack a C-terminal KDEL sequence and rely on protein-protein interactions for their intracellular localization. Thus, the relative abundance of ERp44 is likely to regulate their localization and activity in and between different cells (Pagani et al., 2001; Swiatkowska et al., 2010; Zito et al., 2007).
Consistent with its many intracellular functions (Cortini and Sitia, 2010), ERp44 is efficiently retained by most cell types analysed. Notably, however, endometrial cells secrete O-glycosylated ERp44 and do so more efficiently in the secretory phase of the menstrual cycle (Fig. 6C-D). These findings suggest possible role(s) for ERp44 also in intercellular dialogues. Accordingly, ERp44 can be released by platelets following activation (Holbrook et al., 2010). It is thus possible that other mechanisms, including pH regulation, regulate the localization of ERp44 and in turn of many of its clients and interactors. Controlling the release of ERp44 might have relevant pathophysiological implications.
Material and methods
Reagents and cell cultures
Chemicals were from Sigma, unless otherwise indicated. Monoclonal and polyclonal anti-ERp44 antibodies (36C9 and JDA1 respectively), anti-myc (9E10) and anti-GM130 were previously described (Anelli et al., 2002; Anelli et al., 2007). Polyclonal anti-PDI was a kind gift of I. Braakman (Utrecht, NL). Unconjugated goat anti-mouse Ig were from Southern Biotechnology Associates, Inc. (Birmingham, AL), while horse-radish peroxidase (HRP) bound goat anti-mouse Ig and anti-rabbit Ig were from Jackson ImmunoResearch Laboratories, Inc. Fluorescent goat anti-mouse and anti-rabbit IgG (H+L) conjugated to Alexa Fluor 700, 647, 546 and 488 were from Invitrogen Molecular Probes (Eugene, Oregon, USA).
HeLa and HepG2 cell lines were purchased from ATCC. Tissue culture, transfection, and silencing were performed as described previously (Anelli et al., 2007). HepG2 cells were transfected by FugeneHD (Promega Corporation, Madison, USA) following the manufacturer instructions. Primary endometrial cells were collected from healthy donors and cultured as described by Di Blasio at al. (1995) (Di Blasio et al., 1995).
Plasmids and vectors
The cDNA encoding human ERp44 without the signal sequence from pGEX-4T-1-ERp44 and the vector for sGFP-RDEL expression were previous described (Vavassori et al., 2013). Vectors driving the expression of human ERp44 were previously described (Anelli et al., 2002). Mutants were obtained by PCR o by site-directed mutagenesis (SDM) using the primers listed in Supplementary Table 2. All the primers were purchased from PRIMM srl (Milano, Italy). Plasmids encoding mutated secretory Ig-μ chains (μΔCH1) were previously described (Cenci et al., 2006; Mattioli et al., 2006).
Halo ERp44 vector construction
A Halo tag was inserted after the ERp44 leader sequence cleavage site as follows: ERp44 no tag was cleaved from the pcDNA3.1 (−) vector by XhoI and KpnI and inserted in pBlueScript II KS (+). A SgfI site was inserted after the leader sequence cleavage site by PCR (p44sgfI Fw: “CCTGTAACAACTGAAATAGCGATCGCTGAAATAACAAGT” and p44sgfI Rv: “ACTTGTTATTTCAGCGATCGCTATTTCAGTTGTTACAGG”). The PCR product was then re-inserted into pcDNA 3.1 (−) vector by XhoI and KpnI. In pHTN-HaloTagCMV-neoVector® (Promega Corporation), HaloTag was mutagenized inserting a Sgf I site at N terminal extremity by PCR (pHTN SgfI Fw: “GCCGCGATCGCTGAAGCAGAAATCGGTAACTGGCTTTCCATTC” and pHTN SgfI Rv: “GGAAGCGATCGCGTTATCGCTCTG”). Halo-ERp44 was obtained by ligating SgfI fragments over night at 16°C, screened by SmaI digestion and validated by sequencing.
Western Blotting, Densitometric Quantifications and Biochemical Techniques
Fluorograms or western blot images were acquired with the Chemidoc-it Imaging System (UVP, Upland, CA) or with FLA-900 Starion (Fujifilm Life Science, USA) and quantified with Image J as described (Anelli et al., 2007).
To assess O-glycosylation, aliquots from HeLa cell lysates were denatured at 95°C for 10 minutes in glycoprotein denaturing buffer (New England BioLabs Inc.), digested with neuraminidase and O-glycosidase over night at 37°C and analyzed by western blot with specific antibodies.
As loading controls for Western blotting assays we used Ponceau and/or anti-tubulin staining and, in some experiments, some aspecific bands present in blots decorated anti-HA (see for instance red asterisks in Figs. 2 and 4). Signal intensity was quantified with image J. The loading of the lanes rarely, if ever, differed more than 5%, with the exception for O-glycosidase treatment where the protein levels were about 20% lower with respect to BFA or untreated samples. Immunoprecipitation of endogenous ERp44 from primary endometrial cell lysates and spent medium was performed with Sepharose-immobilized 36C9 as previously described (Anelli et al., 2012).
Wt and mutant ERp44 were purified and analyzed as described (Masui et al., 2011; Vavassori et al., 2013). Briefly, ANS fluorescence spectra were recorded in 1 cm cuvettes on a Hitachi F-2500 spectrofluorometer. ERp44 and mutants (5 μM) were mixed with 100 μM ANS in 20mMTris-HCl (pH 7.5; 7.0) or MES (pH 6.5) containing 150mM NaCl and incubated at 293 K for 10 min before measurement.
For maleimidyl PEG-2K modification of ERp44 Cys29, each ERp44 derivative (5 μM) was incubated on ice for 30 min in various pH buffers containing 100 mM sodium phosphate and 150 mM NaCl, followed by incubation with maleimidyl PEG2K (300 μM) for 10 min at room temperature. The reaction was stopped by the addition of 5% trichloroacetic acid, and the protein pellet was washed with acetone and dissolved in buffer containing 50 mM Tris-HCl (pH 7.0) and 1% SDS before loading onto a reducing SDS gel (10%).
Radioactive pulse and chase assays
Cells were starved for 5 min in cysteine and methionine-free DMEM (GIBCO, Invitrogen), pulsed for 10 min with (35S) cysteine and methionine (Easy Tag, Perkin-Elmer), washed twice, and chased in complete medium. After the indicated chase times, cells were treated with 10 mM NEM and lysed in RIPA as described (Vavassori et al., 2013). Immunoprecipitates were resolved by SDS-PAGE under reducing conditions, transferred to nitrocellulose and filters visualized by autoradiography with FLA900 Starion (Fujifilm Life Science, Tokyo, Japan).
Immunofluorescence
HepG2 cells were plated on 15 mm glasses and transfected. After 48 hours of transfection cells were fixed in 4% PFA, permeabilized with 0.2% Triton X-100 and stained with different antibodies. For Halo tagged proteins, cells were incubated overnight with 10 nM TMR Halo ligand (Promega Corporation). Samples were analyzed on an Olympus inverted fluorescence microscope (model IX70) with DeltaVision RT Deconvolution System (Alembic, HSR, Milano). After deconvolution, images were processed with Adobe Photoshop CS4 (Adobe Systems Inc.).
GPHR silencing
Total RNA from untreated and GPHR silenced (GPHRi) HeLa transfectants was extracted with Tryzol from Thermo Fisher Scientific (Waltham, USA) following the manufacturer instructions and its quality was checked by OD 260/280 readings and electrophoresis. An aliquot of each sample was retro-transcribed by Super-Script II kit (Invitrogen, USA). Real time polymerase chain reaction was performed with Syber green Master Mix in 25ul of volume by ABI7900 (Applied Biosystem, Foster City, CA). Finally data were analyzed by SDS 2.1 software (Applied Biosystem).
Supplementary Material
Acknowledgements
We thank Stefano Vavassori for his contribution in designing and producing the ΔHis mutant, and for many stimulating discussions. We are indebted to Maria Francesca Mossuto, Anna Rubartelli, Eelco van Anken, Edgar Yoboue and other members of our laboratories for suggestions, criticisms and reagents, the Alembic facility for help in imaging and Roberta Colzani for secretarial assistance. We specially thank Federica Quattrone and Paola Panina for kindly providing primary endometrial cells and for helpful discussions. This work was supported by grants from Telethon (GGP11077), AIRC (IG 10721 and 5x1000 Special Project 9965) to RS, and the Next Generation World-Leading Researchers program (MEXT) to KI.
Abbreviations
- ER
endoplasmic reticulum
- ESC
early secretory pathway
References
- Anelli T, Alessio M, Bachi A, Bergamelli L, Bertoli G, Camerini S, Mezghrani A, Ruffato E, Simmen T, Sitia R. Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 2003;22:5015–5022. doi: 10.1093/emboj/cdg491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Alessio M, Mezghrani A, Simmen T, Talamo F, Bachi A, Sitia R. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002;21:835–844. doi: 10.1093/emboj/21.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T, Bergamelli L, Margittai E, Rimessi A, Fagioli C, Malgaroli A, Pinton P, Ripamonti M, Rizzuto R, Sitia R. Ero1a regulates Ca 2+ fluxes at the endoplasmic reticulum-mitochondria interface (MAM) Antioxidants and Redox Signaling. 2012;16:1077–1087. doi: 10.1089/ars.2011.4004. [DOI] [PubMed] [Google Scholar]
- Anelli T, Ceppi S, Bergamelli L, Cortini M, Masciarelli S, Valetti C, Sitia R. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26:4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci S, Mezghrani A, Cascio P, Bianchi G, Cerruti F, Fra A, Lelouard H, Masciarelli S, Mattioli L, Oliva L, et al. Progressively impaired proteasomal capacity during terminal plasma cell differentiation. EMBO J. 2006;25:1104–1113. doi: 10.1038/sj.emboj.7601009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortini M, Sitia R. ERp44 and ERGIC-53 synergize in coupling efficiency and fidelity of IgM polymerization and secretion. Traffic. 2010;11:651–659. doi: 10.1111/j.1600-0854.2010.01043.x. [DOI] [PubMed] [Google Scholar]
- Di Blasio AM, Centinaio G, Carniti C, Somigliana E, Vigano P, Vignali M. Basic fibroblast growth factor messenger ribonucleic acid levels in eutopic and ectopic human endometrial stromal cells as assessed by competitive polymerase chain reaction amplification. Mol. Cell. Endocrinol. 1995;115:169–175. doi: 10.1016/0303-7207(95)03687-3. [DOI] [PubMed] [Google Scholar]
- Fraldi A, Zito E, Annunziata F, Lombardi A, Cozzolino M, Monti M, Spampanato C, Ballabio A, Pucci P, Sitia R, et al. Multistep, sequential control of the trafficking and function of the multiple sulfatase deficiency gene product, SUMF1 by PDI, ERGIC-53 and ERp44. Hum. Mol. Genet. 2008;17:2610–2621. doi: 10.1093/hmg/ddn161. [DOI] [PubMed] [Google Scholar]
- Hamby SE, Hirst JD. Prediction of glycosylation sites using random forests. BMC Bioinformatics. 2008;9:500. doi: 10.1186/1471-2105-9-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook L-, Watkins NA, Simmonds AD, Jones CI, Ouwehand WH, Gibbins JM. Platelets release novel thiol isomerase enzymes which are recruited to the cell surface following activation. Br. J. Haematol. 2010;148:627–637. doi: 10.1111/j.1365-2141.2009.07994.x. [DOI] [PubMed] [Google Scholar]
- Kakihana T, Araki K, Vavassori S, Iemura S-, Cortini M, Fagioli C, Natsume T, Sitia R, Nagata K. Dynamic regulation of Ero1a and peroxiredoxin 4 localization in the secretory pathway. J. Biol. Chem. 2013;288:29586–29594. doi: 10.1074/jbc.M113.467845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Ide T, Koike M, Uchiyama Y, Kinoshita T. GPHR is a novel anion channel critical for acidification and functions of the golgi apparatus. Nat. Cell Biol. 2008;10:1135–1145. doi: 10.1038/ncb1773. [DOI] [PubMed] [Google Scholar]
- Masui S, Vavassori S, Fagioli C, Sitia R, Inaba K. Molecular bases of cyclic and specific disulfide interchange between human ERO1a protein and protein-disulfide isomerase (PDI) J. Biol. Chem. 2011;286:16261–16271. doi: 10.1074/jbc.M111.231357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli L, Anelli T, Fagioli C, Tacchetti C, Sitia R, Valetti C. ER storage diseases: A role for ERGIC-53 in controlling the formation and shape of russell bodies. J. Cell. Sci. 2006;119:2532–2541. doi: 10.1242/jcs.02977. [DOI] [PubMed] [Google Scholar]
- Otsu M, Bertoli G, Fagioli C, Guerini-Rocco E, Nerini-Molteni S, Ruffato E, Sitia R. Dynamic retention of Ero1a and Ero1ß in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxidants and Redox Signaling. 2006;8:274–282. doi: 10.1089/ars.2006.8.274. [DOI] [PubMed] [Google Scholar]
- Pagani M, Pilati S, Bertoli G, Valsasina B, Sitia R. The C-terminal domain of yeast Ero1p mediates membrane localization and is essential for function. FEBS Lett. 2001;508:117–120. doi: 10.1016/s0014-5793(01)03034-4. [DOI] [PubMed] [Google Scholar]
- Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-la. Mol. Cell. Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzoni R, Anelli T, Brunati M, Cortini M, Fagioli C, Sitia R. Pathogenesis of ER storage disorders: Modulating russell body biogenesis by altering proximal and distal quality control. Traffic. 2010;11:947–957. doi: 10.1111/j.1600-0854.2010.01071.x. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-G, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatkowska M, Padula G, Michalec L, Stasiak M, Skurzynski S, Cierniewski CS. Ero1a is expressed on blood platelets in association with protein-disulfide isomerase and contributes to redox-controlled remodeling of aIIbß3. J. Biol. Chem. 2010;285:29874–29883. doi: 10.1074/jbc.M109.092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavassori S, Cortini M, Masui S, Sannino S, Anelli T, Caserta IR, Fagioli C, Mossuto MF, Fornili A, van Anken E, et al. A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol. Cell. 2013;50:783–792. doi: 10.1016/j.molcel.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang L, Vavassori S, Li S, Ke H, Anelli T, Degano M, Ronzoni R, Sitia R, Sun F, et al. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 2008;9:642–647. doi: 10.1038/embor.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DW, Lewis MJ, Pelham HRB. pH-dependent binding of KDEL to its receptor in vitro. J. Biol. Chem. 1993;268:7465–7468. [PubMed] [Google Scholar]
- Zito E, Buono M, Pepe S, Settembre C, Annunziata I, Surace EM, Dierks T, Monti M, Cozzolino M, Pucci P, et al. Sulfatase modifying factor 1 trafficking through the cells: From endoplasmic reticulum to the endoplasmic reticulum. EMBO J. 2007;26:2443–2453. doi: 10.1038/sj.emboj.7601695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.