Abstract
Ecological developmental biology (eco-devo) explores the mechanistic relationships between the processes of individual development and environmental factors. Recent studies imply that some of these relationships have deep evolutionary origins, and may even predate the divergences of the simplest extant animals, including cnidarians and sponges. Development of these early diverging metazoans is often sensitive to environmental factors, and these interactions occur in the context of conserved signaling pathways and mechanisms of tissue homeostasis whose detailed molecular logic remain elusive. Efficient methods for transgenesis in cnidarians together with the ease of experimental manipulation in cnidarians and sponges make them ideal models for understanding causal relationships between environmental factors and developmental mechanisms. Here, we identify major questions at the interface between animal evolution and development and outline a road map for research aimed at identifying the mechanisms that link environmental factors to developmental mechanisms in early diverging metazoans.
Keywords: Cnidaria, corals, environmental genomics, holobiont, hydra, sponges, symbiosis
Introduction
“Organisms determine what aspect of the part of the physical world is relevant to them and they construct out of these relevant bits and pieces a world of interaction” [1]. By analyzing the ecological factors that shape development, reproduction and survival, life history theory seeks to explain the evolution of the major features of life cycles [2, 3]. While significant progress has been made in explaining the diversity of life history strategies among species, the underlying mechanisms are still largely unknown [4-6]. In part, this is due to failure to incorporate ecological inputs into models describing phenotypic evolution, confounded by the complexity and our incomplete understanding of processes such as development and gene-regulation. However, evolutionary biologists are increasingly able to integrate information across organisms and levels of organization, providing insights not only into information processing and decision making in individual organisms but also the interaction networks of entire systems. Recently, integrative approaches in which ecology, molecular genetics, and physiology directly inform evolutionary biology and vice versa have proven highly fruitful and have given rise to entirely novel disciplines such as ecological developmental biology (eco-devo) [7-9].
Eco-devo seeks to understand developmental responses to environmental factors by focusing on three major questions: (i) How do environmental variables affect developmental processes? (ii) How do environmental interactions influence phenotypic evolution? (iii) How does developmental evolution impact the environment? It is now clear that developmental gene expression cannot be explained just in terms of interactions within the growing embryo [10-12]. Although the idea of animal development as an autonomous process directed by the genome has been replaced by models that accommodate internal “environ-mental” (cell-cell) signaling, we are unaccustomed to the notion of also integrating interactions with the external environment. Although underpinned by complex genetic programs, development must be reimagined as an orchestration of both animal-encoded ontogeny and environmental interactions.
Discussions about environmental impacts on development must take into account three different levels: (i) an ecological dimension because development responds to constantly changing environments; (ii) an evolutionary dimension because of the interplay between developmental plasticity and Darwinian selection; and (iii) a genomic dimension because epigenetic regulation allows environmental signals to be integrated at the genome level and because of the multiple responses in terms of gene expression to similar environmental cues, sometimes even within the same taxon. To date, eco-devo has focussed on a few specific cases, and general principles are not yet clear. Here, we provide examples of environmental effects on development in cnidarians and sponges, and highlight gaps in our understanding of the mechanisms involved. Understanding the mechanisms linking environment and development in these morphologically simple animals will provide new perspectives on the evolution of these processes, and may ultimately enable recognition of common principles.
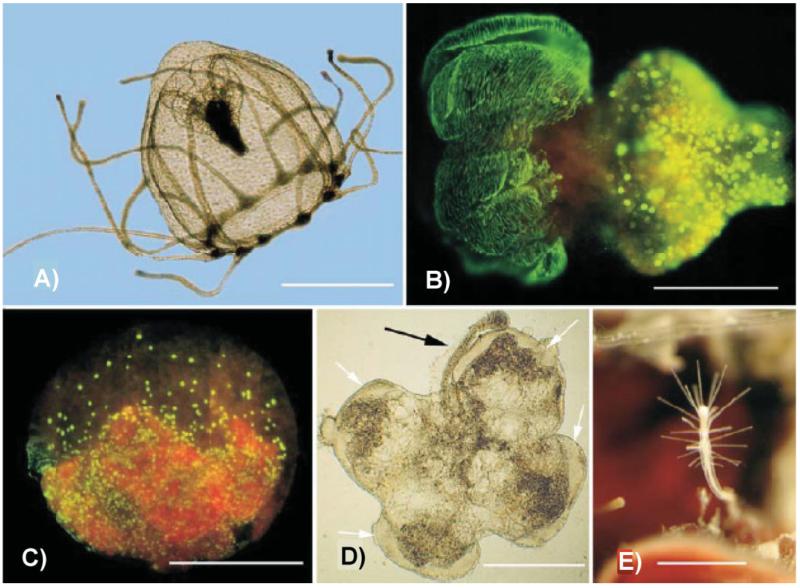
Under the definition of “development” adopted here we include not only changes occurring early in life history but also plasticity, regeneration and related processes occurring in mature stages. This broad interpretation is justified given the remarkable plasticity of development shown by cnidarians and sponges – a striking example being the fully reversible life-cycle of Turritopsis nutricula, a jellyfish whose “adult” stage can de-differentiate under environmental stress [13, 14] (Fig. 1). In Turritopsis, there is no point at which “development” ends, and hence distinguishing between “early” and “late” life history stages is not justified.
Figure 1.
Environmental stress has substantial impact on cnidarian life cycles. In Turritopsis nutricula, stress can reverse the development and make a medusa transforming into a polyp. A: Free-living healthy medusa. B: Stress-triggered transforming medusa. C: Balllike stage of transforming medusa. BrdU staining of replicating nuclei. D: Butterfly-shape remnant of adult medusa producing a hydrorhizal stolon characteristic of the polyp stage (black arrow). E: A newly formed polyp from reverse development of a medusa. Scale bars: A: 1 mm; B: 500 μm; C: 300 μm; D and E: 500 μm. Taken from Piraino et al. [14].
Advantages of early emerging animals for understanding environmental impacts on development
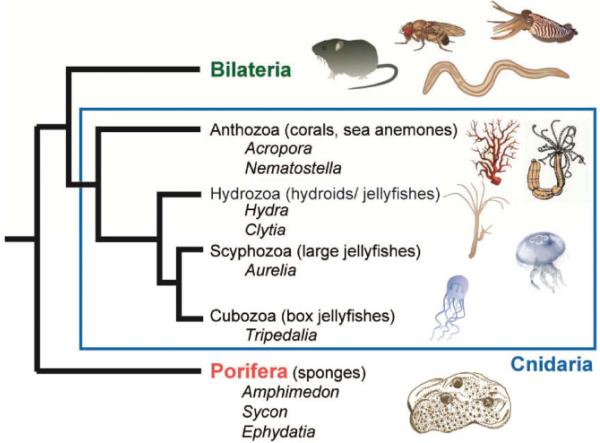
Extant cnidarians and sponges are representatives of phyla that diverged prior to the Bilateria (Fig. 2) [15, 16] and thus represent evolutionary success stories – these lineages have survived and prospered in a constantly changing world and are now near ubiquitous in aquatic habitats. What factors have contributed to the evolutionary success of sponges and cnidarians? One contributing factor may have been their high regenerative capacity, but so too has been their remarkable ability to form symbioses with microorganisms, both prokaryotic and eukaryotic.
Figure 2.
Phylogeny of basal metazoan animals. Two lineages, the phylogenetic position of which remains contentious (placozoans and ctenophores), have been omitted.
Given recent technical advances, which include transcriptome and whole genome sequencing of representative sponges, cnidarians, and some of their symbionts [17-28], there are now also practical advantages in using early emerging animals to investigate complex genome/environment interactions. Representative cnidarians – Hydra, Hydractinia, Nematostella – are now amenable to knock-down and transgenic techniques, permitting functional analyses. Many species reproduce asexually by budding or colony fragmentation, and the apparent ability of early embryos to recover after fragmentation [29] enables clonal propagation. The ability to develop isogenic lines without a requirement for extensive back crossing facilitates the dissection of genome by environment interactions. These many advantages of animals, which are morphologically remarkably simple but have surprisingly complex gene repertoires encoding much of the signaling and sensory capacity of “higher” animals [26, 27, 30-33] lead us to propose that representatives of these early diverging phyla offer novel opportunities for studying environment/genotype interactions in development.
Five examples of environmental influences on developmental programs in cnidarians and sponges
Metamorphosis is environmentally triggered in cnidarians and sponges
Cnidarian life cycles often use environmental cues, both biotic (microbes and nutrients) and abiotic (temperature, light, and chemicals), to control key life-cycle transitions. Although their larvae generally are motile, most adult forms are sessile and therefore directly and constantly exposed to changing environments. Transition between these morphologically and ecologically distinct phases typically occurs when the developmentally competent larva encounters specific environmental cues, which act as a morphogenetic signal that induces metamorphosis. The selective value of the settlement behavior is likely to be strong, since in many cases the choice of settlement site determines the environmental conditions that the sedentary polyp or colony will be exposed to for its entire life. Although settlement cues are highly diverse and often specific, the larvae of many corals prefer only a few species of crustose coralline algae (CCA) [34-36], which have been described as serving as “chemical fly papers” for coral settlement [37] (Fig. 3A). Although cues for coral settlement and metamorphosis also are commonly associated with marine biofilms and their bacterial component in the absence of CCA [38-41], continued repopulation of coral reef ecosystems certainly is governed by the activities of specific bacteria [42]. How these cues are detected and the physiological mechanisms underlying settlement and metamorphosis are largely unknown. G-protein-coupled receptors (GPCRs) and their associated (AC/cAMP or PI/DAG/PKC) pathways have been implicated in perception of settlement cues in a number of cnidarian species ([41], and references therein).
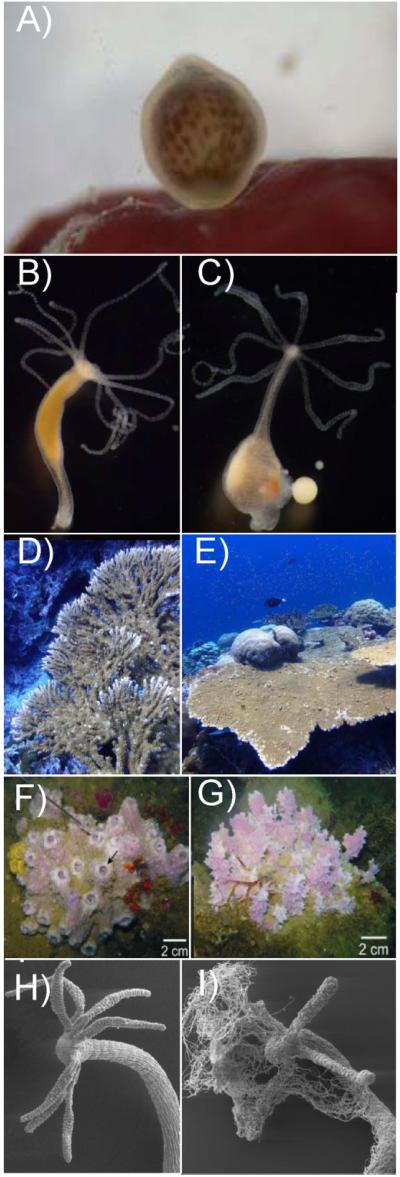
Figure 3.
A: A single larva of Stylophora pistillata, about 5 h after release, right at the moment it first contacts a possible settlement surface. Larvae from Stylophora pistillata are typically pear-shaped and have a mean length of 1 mm (photo credit: Peter J Edmunds; http://www.eoearth.org/view/article/150659/). B: Pelmatohydra oligactis polyp. Polyps are up to 10 mm long. C: Female Pelmatohydra oligactis carrying several eggs. Gametogenesis is triggered by a temperature shift. D and E: Acropora clathrata at 5 m in a sheltered location (D), at 20 m in a high current location (E). Reproduced with permission from Zoe Richards. F and G: Typical morphotypes of Dysidea avara sponges from depth sites of 8.8 m (F) and 14.3 m (G); pictures from Mendola et al. [58]. H: Hydra vulgaris under normal culture conditions. I: Hydra vulgaris cultured in the absence of bacteria and infected by fungi.
In the case of Hydractinia, a colonial hydroid frequently found in the North Sea covering shells inhabited by hermit crabs, in less than three days the fertilized egg develops into a mature planula larva that is competent to undergo metamorphosis. However, under sterile laboratory conditions Hydractinia planulae fail to undergo metamorphosis, are unable to take up food, and eventually die. The normal process of metamorphosis requires an external trigger that is thought to normally be provided by specific strains of Alteromonas. Although a lipophilic substance produced by these bacteria may act as the trigger [43], its identity is unknown. Neurosensory cells in the aboral pole of Hydractinia echinata larvae [44] may directly sense such external cues [45].
As pointed out by Maldonado et al. [46], very little is known about ecology of the sponge larvae, and multiple stimuli (including light, gravity, chemical cues, and substratum texture) are hypothesized to play roles in triggering settlement and metamorphosis. A recent study conducted on two demosponges demonstrated that, as in the case of coral larvae, exposure to CCA, or their extracts increased rates of larval metamorphosis. Surprisingly, similar effects were achieved when sponge larvae were exposed to the cnidarian neuropeptide GLW-amide, suggesting that the same signal transduction pathways might be involved in metamorphosis of these two phyla. Although mechanisms or cells responsible for signal perception in sponges remain unidentified [47], the broadly similar settlement cue requirements to corals are intriguing, not least because they occupy similar ecological niches.
Developmental programs in cnidarians and sponges are activated by temperature shifts
In both cnidarians and sponges, temperature frequently plays a critical role in mediating life cycle transitions. It is well established that gametogenesis can be induced in hydrozoans such as Pelmatohydra oligactis (Fig. 3B and C) by lowering the temperature by about 10° [48]. Increases in water temperature, in combination with light, also induce spawning in the anthozoan cnidarian Nematostella [49] and decreases in temperature induce strobilation (the production of juvenile medusae known as ephyrae) in polyps of the moon jellyfish Aurelia [50, 51]. This temperature dependence ensures that juvenile jellyfish develop during the more favorable conditions that follow the winter season. Since polyps must be able to distinguish between short-time temperature fluctuations and longer-term seasonal temperature decreases in the winter period, Aurelia polyps must also possess a timing mechanism that can sense the duration of the low-temperature period. The level of transcripts encoding peptide CL390 could constitute such a timing mechanism for measuring the duration of exposure to cold conditions [51].
Likewise, stem cells in the dormant gemmules of sponges can be activated by temperature shifts. Gemmules are a resistant stage formed in response to environmental stresses that may result in the death of the parent sponge, and are composed of thesocytes – an arrested form of the archaeocyte stem cells that enable regeneration in adult demosponges [52]. Gemmule hatching, while inducible by an increase in temperature in most species, can also occur at low temperatures [53]. During gemmule hatching, quiescent thesocytes are activated to become archeocytes. How temperature shifts are sensed by sponges and then transduced into changes in gene expression and cell behavior is not yet known. However, in the freshwater sponge Eunapius flagilis, high concentrations of sorbitol maintain high osmotic pressure, which appears to be required for holding gemmules in the resting state; when the temperature rises and gemmule hatching is initiated, the metabolic pathway that converts polyols to glycogen is activated, probably by up-regulation of sorbitol dehydrogenase (SDH) expression [54].
Environmental effects on morphology and tissue architecture in cnidarians and sponges
The adult morphologies of many cnidarians and sponges are quite plastic; these effects are most clear in the case of organisms that produce skeletal support, but probably occur more generally. Many species of hard corals (scleractinians) have radically different gross morphologies depending on the wave action to which they are exposed [55]. Acropora colonies from many West Australian reefs or from Lord Howe Island, where wave action is strong, are virtually unrecognizable to those of us accustomed to their “normal” morphologies when they are not subject to high flow rates (Fig. 3D and E). Similarly, sponges have to modify their skeleton to adapt to a constantly changing environment. Reciprocal transplantation of intertidal sponges between high and low wave action environments indicate that sponges rapidly initiate production of stiffer and stronger tissues in high wave energy environments but delay formation of new, less robust, tissues in calm habitats [56]. Moreover, as in corals, the gross morphology (body shape) of many sponges is affected by the environment, individuals exposed to higher flow rates being more compact than those growing in more sheltered locations [57, 58] (Fig. 3F and G). In addition to “global” changes of morphology, some sponges have been shown to rearrange their canal systems in response to changes of the water flow, either by differential growth or “replumbing” of existing canals to change their polarity [59, 60]. How do these animals adapt their morphology to a temporally variable, unpredictable environment? How do they sense the environmental signals that required modification of tissue rigidity or reorganization of canal systems? It has been recently suggested that short, non-motile cilia lining the inside of oscula (sponge exhalant canals) might be responsible for sensing of the water flow, thus the entire osculum might be viewed as a sensory organ [61]. But without a nervous system, how is the signal integrated to elicit a response of the entire sponge? What is the underlying acclimatory control mecha-nism? Although there is accumulating evidence that micro-environment controls cellular phenotype and thereby the architecture of the emerging multicellular structure or tissue, the microenvironmental factors whose signaling must be integrated in order to effect an organized, functional tissue morphology are still largely unknown.
Recent observations clearly demonstrate that the environment interacts with developmental processes. In turn, does developmental evolution affect the environment? It is quite obvious that “organisms change the world in which they are interacting. They both create it and destroy it” [1]. Reef-building corals are formidable structural engineers; the calcium carbonate skeletons that they secrete can accumulate over time into vast contiguous systems that modify ocean circulation, protect coastlines, and can even modify local climate. One role of the skeleton is to position the living coral tissue so as to optimize light availability to the photosynthetic symbionts. Coral species with tabular growth forms (Fig. 3D and E) are particularly important ecosystem engineers and key contributors to reef structural complexity [62]. They harbor distinctive understory communities [63] and provide shelter from predation and high flow for many mobile species [64] including juveniles of parrotfish species, which as adults play a critical role in controlling growth of macroalgae [65]. Some tabular species support corallivorous butterflyfish, which decline markedly in abundance without them [66]. Thus, corals are not only changing the world in which they are interacting on a massive scale, but have also enabled the diversification of associated animals such as reef fish. Although the Scleractinia have been major reef builders only since the Triassic period, in the deeper geological past the stromatoporids and rugose corals – extinct lineages distantly related to extant taxa – have played similar ecological roles. Thus, in all likelihood, cnidarians and sponges have modified the environment to a greater extent than have many other extant lineages.
Host-microbe interactions influence developmental processes in cnidarians and sponges
Bacteria are important components of cnidarian and sponge holobionts ([67], and references therein), and shifts in the composition of the microbiota can compromise the health of the whole animal [68]. The specificity and stability of interactions between host and microbiota point to the evolutionary significance of these associations. The maintenance of a characteristic microbial community appears to be a complex trait under genetic control, suggesting that hosts deprived of their normal microbiota should be at a disadvantage. From the earliest stages of development, Hydra employ sophisticated mechanisms to manage their microbial environment. Eggs in Hydra contain maternally derived antimicrobial peptides (AMPs) that impose chemical barriers and shape the composition of the associated microbiota [69, 70]. Conversely, adult polyps recruit specific bacteria to their epithelial surfaces to provide protection against fungal pathogens (Fraune, Schröder and Bosch, personal observation) (Fig. 3H and I). Further, a body of evidence supports the idea that acquired microbes are essential for a range of developmental functions in Hydra. These functions include induction of spatially and temporally restricted genes (Fraune and Bosch, personal observation), promotion of growth rate and activation of the innate immune system. Intriguingly, the regulatory pathways that control tissue homeostasis and stem cell behavior in Hydra appear also to have central roles in controlling the interactions with the associated microbiota, underscoring the intimate relationships between development and host-microbe interactions [71]. FoxO is strongly expressed in all three Hydra stem cell lineages, and its down-regulation leads not only to reduced stem cell numbers, but also results in dramatic changes in levels of expression of AMPs and thereby most probably the composition of the associated microbes [71].
Microbial communities, including protists, bacteria, archaea, and viruses, are also important components of the coral holobiont. The coral microbiome contributes to holobiont function due to its role in coral nutrition [72] and host defense [73]. Recent observations also indicate an impact of bacteria on the health of corals and coral reef ecosys-tems [74, 75]. Similarly, marine sponges frequently harbor dense and diverse microbial communities, at least some components of which are symbionts. Although direct evidence for functions of bacteria in sponge homeostasis is limited, several reports imply that cyanobacteria contribute significantly to the health and/or growth of the holobiont ([76] and references therein).
Perhaps the most pervasive example of microbial signaling in cnidarian and sponge development is in the above-mentioned induction of settlement and metamor-phosis of many marine larvae [77]. Together, this recognition has led to a new understanding of biology; one that reflects the strong interdependencies that exist between these complex multicellular organisms and their associated microbes.
In addition to abiotic factors and microbes, photosynthetic symbionts can shape the host phenotype
In many cases, the phenotype of the sponge or cnidarian “holobiont” is determined in large part by its associated symbionts. Some of these are photosynthetic – the dinofla-gellate Symbiodinium in reef-building corals, the green alga Chorella in the case of Hydra viridis, or the photosynthetic cyanobacteria associated with some sponges [78–80]. Many corals in high-temperature habitats host Symbiodinium symbionts that appear to confer greater heat-tolerance [81, 82], but this may be at the cost of decreased output of photosynthates [83]. Thus, for at least some coral-algal symbioses, a degree of acclimation to particular temperature environments may occur through changes in the identity of the algal symbiont. Elegant work by the Chen group at Academia Sinica suggests that the flexibility of the coral Platygyra verweyi with respect to its Symbiodinium partners enables the association to thrive in the warmer water of a nuclear power plant outflow as well as surrounding waters [84]. Near the Kenting (Taiwan) site of the nuclear power plant, Platygyra hosts Symbiodinium C3 (heat sensitive), D (heat-tolerant), or both; D completely dominates close to the outflow, while the frequency of the C3 association increases with distance from the outflow. The ability of some corals to “shuffle” their dinoflagellate symbionts has been interpreted as an adaptive mechanism [83], but the significance of this process is not clear. At least under acute stress, some corals that appear inflexible with respect to symbiont strain (e.g. Porites spp.) are significantly more stress tolerant than other corals that can “shuffle” (e.g. Acropora spp.). Moreover, corals hosting clade C3 Symbiodinium occur and appear to thrive in the Persian Gulf under temperature regimes that would normally be associated with bleaching [85]. Thus, both host and photosynthetic symbiont genotype influence thermal tolerance, and there is no simple relationship between the ability to “shuffle” and survival on biological time scales. In Symbiodinium, many photosynthetic regulatory processes have been investigated, including oxygen production, PSII quantum yield [86–88], carbon assimilation [89], and pigment content [88]. However, it is still unclear how the photosynthetic ability of the algae affects coral molecular regulation and behavior [90].
Evidence for evolutionarily conserved mechanisms linking environment to development
As discussed above, much of the genetic complexity of higher animals, including many of the receptors and signaling pathways, is also present in sponges and cnidarians. Given that in some cases physical and biological factors affect the development of these morphologically simple animals and bilaterians in similar ways, it is reasonable to hypothesize that some of the mechanisms involved may be conserved. Of course, some mechanisms will be taxon-specific – environmental influences on sex determination, for example – but others may have deeper evolutionary origins. Three examples of possible mechanisms involved are given below:
Nutrient sensing
The mammalian protein target of rapamycin (mTOR) lies at the heart of a nutrient-sensing signaling network that controls cellular metabolism. mTOR is an evolutionarily conserved kinase complex integrating inputs arising from energy status, amino acid levels, cellular stresses, and growth factors. Acting as master integrator, mTOR signaling not only adjusts protein synthesis, lipid synthesis, gene expression, and autophagy but is also a key mediator of insulin, insulin-like growth factor 1, and other growth-factor signals to the cell growth machinery. In mammals, mTOR acts downstream to the PI3K/Akt signaling pathway to activate protein synthesis and cell growth and, together with the modulators raptor and LST8, forms the TOR complex 1 (TORC1) [91]. Forkhead transcription factors of the FoxO subfamily are involved in the regulation of the cell cycle, apoptosis, and metabolism [92]. In Hydra, expression of the single FOXO gene has been shown to induce stemness [71]. Given the impact of environmental factors on development [9] and aging [93-95], proteins of the FoxO subfamily are tightly regulated to ensure that transcription of specific target genes is responsive to environmental conditions. A major form of regulation is Akt-mediated phos-phorylation of FOXO and nuclear exclusion in response to insulin or other growth factors. As anticipated, the PI3K/Akt pathway is well conserved in Hydra [96, 97]. In addition to the metabolic sensor TOR, other components of TORC1, Raptor and LST8, the TOR regulators RHEB and TSC2, and the TOR target Atg1 are present in Hydra ([97], Klimovich and Bosch, unpubl.), as are three genes encoding insulin-like peptides [98]; these latter could possibly act through a known insulin receptor gene HTK7 [99]. Thus, although functions are yet to be established, many of the components of the nutrient sensing systems of bilaterians are present in cnidarians.
Bacterial sensing
Toll-like receptors (TLRs) are conserved throughout animal evolution and are involved in eliminating pathogens and controlling commensal colonization by recognizing conserved microbe-associated molecular patterns (MAMPs) including lipopolysaccharides, flagellin, and peptidoglycans [100, 101]. Cnidarians including Nematostella, Acropora, and Hydra possess a bona fide TLR-signaling cascade. In vivo experiments in Hydra using MyD88 loss-of-function approaches via down-regulating the level of MyD88 transcripts implicate TLR signaling in bacterial recognition [102]. TLR signaling in Hydra is also linked to JNK/p38 MAP kinases [102] and affects expression of AMPs; although the primary role of AMPs is to act as effector molecules of innate immunity, these molecules also regulate the composition of commensal microbiota [67, 103]. The discovery that AMPs are downstream targets of the stem cell transcription factor FoxO in Hydra [95] points to a mutual dependency and interaction between the stem cell regulatory machinery of the host and the composition of the resident microbiota, such that disturbances in one trigger a restructuring and resetting of the other [104, 105].
Light sensing
Despite extensive research in the last few years, many gaps remain in our understanding of the perception of light in organisms at the base of animal evolution. Gorbunov and Falkowski [106] suggested that detection of the blue region of the spectrum of moonlight might act as a cue that determines the specific night of spawning in corals, because several species were found to be sensitive to light in this region of the spectrum. Levy et al. [107] showed that in corals, there is a night time preference for DNA replication. Furthermore, orthologs of various photoreceptors and many core circadian genes common to mammals and Drosophila are present in the coral Acropora millepora and in the sea anemone Nematostella vectensis [25, 28, 108, 109]. The expression of several opsins (acropsins 1–3) in planulae of Acropora palmata and the demonstration that specific Acropora G proteins can be activated by acropsins in a light-dependent manner in vitro [110] indicates that functional photoreceptors can be formed that may play a role in color preference during settlement, vertical positioning, and other light-guided behaviors observed in coral larvae. Molecular research on the coral A. millepora suggests that cnidarian cryptochromes may act as photoreceptors that mediate environmental signals, such as moonlight, in synchronizing the central pacemaker [111]. Sponges lack a bona fide nervous system [112], but nevertheless show the competence to react to light [113, 114] and possess cryptochrome/photolyase genes responsive to wavelengths of light that also mediate larval phototactic behavior [115].
A roadmap toward answering outstanding questions
To better understand how the environment influences developmental pathways in sponges and cnidarians, we first need to select species with appropriate ecological niches and link morphological traits in different environments with gene expression landscapes. This is not a trivial undertaking, because the selected species need to be amenable to laboratory work in addition to providing insight into fundamental evolutionary processes. While genome projects on single representative species have often provided new perspectives on animal evolution, broader scale genome projects such as that underway to generate large molecular datasets from 20 to 30 other Drosophila species, promise a much clearer view of the evolutionary process itself and to revolutionize animal eco-devo studies. A similar approach applied to an appropriate range of cnidarians and sponges would add another dimension to eco-devo, but what criteria should be applied in selecting candidates? Eco-devo requires well-characterized genomes, and a robust phylogeny. Of the presently available cnidarian (Nematostella vectensis, Hydra magnipapillata, two Acropora species) and sponge (Amphimedon queenslandica) genome sequences, none approach the level of completeness of the D. melanogaster assembly. When targets have been identified, the first priority is to work toward high-quality genome assemblies, backed up by large-scale transcriptomics. Many cnidarians have relatively tractable genomes (moderate (A + T)-content, 300–500 Mbp size range, etc), so this is clearly achievable. Ideally, lineages of organisms that have undergone extensive genomic rearrangements with respect to the metazoan ancestor (i.e. large amounts of gene losses) should be avoided.
Perhaps the most challenging issue in selecting target species is the desirable range of phenotypic and genetic variation – what is the “Goldilocks” range of variation for an eco-devo model? While data to enable this kind of choice do not yet exist, and such choices may be somewhat arbitrary, extremes of variation should be avoided if possible. Too little variation indicates strong canalization (or robustness), whereas very high phenotypic variability suggests very limited adaptation to a wide range of conditions.
Onto these biological constraints, in choosing appropriate species all practical considerations will include the possibility of experimental manipulation. Ideally, target species would have short generation times and be genetically tractable. However, while these are desirable characteristics, the primary criteria in making choices should be biological rather than strictly practical. Species of particular evolutionary significance should not be overlooked just because they are not “easy” to work with. For instance, the practical difficulties of working on amphioxus have not overridden recognition of its significance as an important target for analysis [116-118]. Experience suggests that practical tools can be developed for “difficult” organisms – witness recent successes in closing the life cycle of amphioxus in vitro [119].
Moreover, it will be important to develop monitoring methods to dissect the gene by environment interactions in cnidarians and/or sponges. Although there have been major advances in genomics technologies and their application has become ever more affordable, our understanding of how environmental conditions influence genome structures and gene functions, and, in turn, how individuals and populations cope with changing environments, remains limited. We have to understand the basis and extent of variation of gene expression that manifests in diverse environments. As observed in yeast [120], many genes demonstrate variable expression across environments, across genetic backgrounds, or both. Are there steps in biochemical pathways that are more robust than others? It will be important to examine in greater depth the correlation between genotype, gene expression, and epigenetic modification. The term “epigenetics” was coined in 1940 by biologist Conrad Waddington, who defined it as “the interactions of genes with their environment, which bring the phenotype into being” [121]. The fact that the epigenome differs not only over an individual’s lifetime but is also affected by environmental factors, certainly adds complexity to all efforts of understanding the interactions of genes with their environment. Carefully selected cnidarian model species may contribute to developing “eco-epigenomics”, which requires the identification of changes not only among different individuals, but also among different tissues, developmental stages, and in different environments. As a first step, the major histone modifications and their relationships to neighboring genes have now been mapped in the sea anemone N. vectensis [122].
Last but not least, both mathematical and experimental modeling approaches are likely to inform our understanding of this issue. Therefore, collaborative approaches that bring together scientists using high-throughput molecular biology and ecology, zoologists, geneticists, physiologists, and mathematicians are critical to the investigation and under-standing of genome-environment interactions in cnidarians and sponges.
Conclusions and outlook
In summary, we are only beginning to comprehend the diversity of genome-environment interactions. Understanding genome structure and gene expression under different environmental conditions requires integrative, multidisciplinary, and modeling-based approaches. Representative cnidarians and sponges provide novel opportunities to investigate complex biological questions such as the mechanisms by which developmental programs respond to environmental stimuli. Advantages of these “simple” animals include the ease with which clonal material can be generated and propagated and the availability of simple genetic manipulation technologies. Moreover, the complex and vertebrate-like gene repertoires of at least some of these animals (Nem-atostella, Acropora, and Hydra) are much less derived than are those of the usual model animals (Drosophila, Caenorhabditis, Ciona, etc.). Quite conceivably, ancestral mechanisms may be more clearly seen in these early diverging animals, providing insights into how the environment interacts with developmental processes and, in turn, how developmental evolution affects the environment.
Acknowledgment
We thank the three anonymous reviewers for their valuable comments on our manuscript.
This paper is the product of an international workshop, “The case of the missing mechanism: how does the environment influence developmental pathways in sponges and cnidarians”, which took place in Aidling (Germany) in September 2013. As the manuscript is based on ideas that arose in open discussions to which all of the participants contributed, all are listed as co-authors on the basis that all were essential in enabling this manuscript. Thomas Bosch and David Miller were together responsible for synthesizing ideas from the individual contributions, generating the overall structure and writing a substantial proportion of the paper, and on this basis are listed as first and last authors, respectively.
References
- 1.Lewontin R. The Charles M. and Martha Hitchcock lectures. Univ California Television (UCTV); UC Berkeley: 2008. Gene, organism and environment: bad metaphors and good biology. Cited from. https://www.youtube.com/watch?v=we4ZzjKxFHM. [Google Scholar]
- 2.Stearns SC. The Evolution of Life Histories. Oxford University Press; London: 1992. [Google Scholar]
- 3.Flatt T, Heyland A. Mechanisms of Life History Evolution: The Genetics and Physiology of Life History Traits and Trade-Offs. 2011;13 Oxford Scholarship Online Print ISBN-9780199568765. [Google Scholar]
- 4.Roff DA. The Evolution of Life Histories: Theory and Analysis. Chapman and Hall; New York: 1992. [Google Scholar]
- 5.Stearns SC. Life history evolution: successes, limitations, and prospects. Naturwissenschaften. 2000;87:476–86. doi: 10.1007/s001140050763. [DOI] [PubMed] [Google Scholar]
- 6.Roff DA, Fairbairn DJ. The evolution of trade-offs: where are we now? J Evol Biol. 2007;20:433–47. doi: 10.1111/j.1420-9101.2006.01255.x. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert SF. Ecological developmental biology: developmental biology meets the real world. Dev Biol. 2001;233:1–12. doi: 10.1006/dbio.2001.0210. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert SF, Epel D. Ecological Developmental Biology. Sinauer; Sunderland, MA: 2008. [Google Scholar]
- 9.Gilbert SF. Ecological developmental biology: environmental signals for normal animal development. Evol Dev. 2012;14:20–8. doi: 10.1111/j.1525-142X.2011.00519.x. [DOI] [PubMed] [Google Scholar]
- 10.Nijhout HF. Control mechanisms of polyphonic development in insects. BioScience. 1999;49:181–92. [Google Scholar]
- 11.Mead KS, Epel D. Beakers versus breakers: how fertilisation in the laboratory differs from fertilisation in nature. Zygote. 1995;3:95–9. doi: 10.1017/s096719940000246x. [DOI] [PubMed] [Google Scholar]
- 12.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carla’ EC, Pagliara P, Piraino S, Boero F, et al. Morphological and ultrastructural analysis of Turritopsis nutricula during life cycle reversal. Tissue Cell. 2003;35:213–22. doi: 10.1016/s0040-8166(03)00028-4. [DOI] [PubMed] [Google Scholar]
- 14.Piraino S, De Vito D, Schmich J, Bouillon J, et al. Reverse development in Cnidaria. Can J Zool. 2004;82:1748–54. [Google Scholar]
- 15.Young GA, Hagadorn JW. The fossil record of cnidarian medusa. Palaeoworld. 2010;19:212–21. [Google Scholar]
- 16.Erwin DH, Valentine JW. The Construction of Animal Biodiversity. Roberts and Company; Greenwood Village: 2013. The Cambrian Explosion. [Google Scholar]
- 17.Bayer T, Aranda M, Sunagawa S, Yum LK, et al. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One. 2012;7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conaco C, Neveu P, Zhou H, Arcila ML, et al. Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics. 2012;13:209. doi: 10.1186/1471-2164-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riesgo A, Farrar N, Windsor PJ, Giribet G, et al. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 2014;31:1102–20. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- 20.Howells EJ, Willis BL, Bay LK, van Oppen MJ. Spatial and temporal genetic structure of Symbiodinium populations within a common reef-building coral on the Great Barrier Reef. Mol Ecol. 2013;22:3693–708. doi: 10.1111/mec.12342. [DOI] [PubMed] [Google Scholar]
- 21.Barshis DJ, Ladner JT, Oliver TA, Seneca FO, et al. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci USA. 2013;110:1387–92. doi: 10.1073/pnas.1210224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10:641–54. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 23.Meyer E, Weis VM. Study of cnidarian-algal symbiosis in the “omics” age. Biol Bull. 2012;223:44–65. doi: 10.1086/BBLv223n1p44. [DOI] [PubMed] [Google Scholar]
- 24.Radax R, Rattei T, Lanzen A, Bayer C, et al. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ Microbiol. 2012;14:1308–24. doi: 10.1111/j.1462-2920.2012.02714.x. [DOI] [PubMed] [Google Scholar]
- 25.Shinzato C, Shoguchi E, Kawashima T, Hamada M, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–3. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava M, Simakov O, Chapman J, Fahey B, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–6. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman JA, Kirkness EF, Simakov O, Hampson SE, et al. The dynamic genome of Hydra. Nature. 2010;464:592–6. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoguchi E, Shinzato C, Kawashima T, Gyoja F, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23:1399–408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 29.Heyward AJ, Negri AP. Turbulence, cleavage, and the naked embryo: a case for coral clones. Science. 2012;335:1064. doi: 10.1126/science.1216055. [DOI] [PubMed] [Google Scholar]
- 30.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13:2190–5. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 31.Technau U, Rudd S, Maxwell P, Gordon PM, et al. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–9. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Putnam NH, Srivastava M, Hellsten U, Dirks B, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 33.Hemmrich G, Khalturin K, Boehm AM, Puchert M, et al. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol. 2012;29:3267–80. doi: 10.1093/molbev/mss134. [DOI] [PubMed] [Google Scholar]
- 34.Arnold SN, Steneck RS, Mumby PJ. Running the gauntlet: inhibitory effects of algal turfs on the processes of coral recruitment. Mar Ecol Prog Ser. 2010;414:91–105. [Google Scholar]
- 35.Price N. Habitat selection, facilitation, and biotic settlement cues affect distribution and performance of coral recruits in French Polynesia. Oecologia. 2010;163:747–58. doi: 10.1007/s00442-010-1578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritson-Williams R, Paul VJ, Arnold S, Steneck R. Larval settlement preferences and post-settlement survival of the threatened Caribbean corals Acropora palmata and A. cervicornis. Coral Reefs. 2010;29:71–81. [Google Scholar]
- 37.Morse DE, Morse AN, Raimondi PT, Hooker N. Morphogen-based chemical flypaper for Agaricia humilis coral larvae. Biol Bull. 1994;186:172–81. doi: 10.2307/1542051. [DOI] [PubMed] [Google Scholar]
- 38.Negri AP, Webster NS, Hill RT, Heyward AJ. Metamorphosis of broadcast spawning corals in response to bacteria from crustose algae. Mar Ecol Prog Ser. 2001;223:121–31. [Google Scholar]
- 39.Webster NS, Smith LD, Heyward AJ, Watts JEM, et al. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl Environ Microbiol. 2004;70:1213–21. doi: 10.1128/AEM.70.2.1213-1221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tebben J, Tapiolas DM, Motti CA, Abrego D, et al. Induction of larval metamorphosis of the coral Acrepora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One. 2011;6:e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran C, Hadfield MG. Are G-protein-coupled receptors involved in mediating larval settlement and metamorphosis of coral planulae? Biol Bull. 2012;222:128–36. doi: 10.1086/BBLv222n2p128. [DOI] [PubMed] [Google Scholar]
- 42.Sharp KH, Ritchie KB. Multi-partner interactions in corals in the face of climate change. Biol Bull. 2012;223:66–77. doi: 10.1086/BBLv223n1p66. [DOI] [PubMed] [Google Scholar]
- 43.Leitz T, Wagner T. The marine bacterium Alteromonas espejiana induces metamorphosis of the hydroid Hydractina echinata. Mar Biol. 1993;115:173–8. [Google Scholar]
- 44.Müller WA, Leitz T. Metamorphosis in the Cnidaria. Can J Zool. 2002;80:1755–71. [Google Scholar]
- 45.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51:167–83. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 46.Maldonado M, Durfort M, McCarthy DA, Young CM. The cellular basis of photobehavior in the tufted parenchymella larva of demosponges. Mar Biol. 2003;143:427–41. [Google Scholar]
- 47.Whalan S, Webster NS, Negri AP. Crustose coralline algae and a cnidarian neuropeptide trigger larval settlement in two coral reef sponges. PLoS One. 2012;7:e30386. doi: 10.1371/journal.pone.0030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hyman LH. Miscellaneous observations on hydra, with special reference to reproduction. Biol Bull. 1928;54:65–108. [Google Scholar]
- 49.Fritzenwanker JH, Technau U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa) Dev Genes Evol. 2002;212:99–103. doi: 10.1007/s00427-002-0214-7. [DOI] [PubMed] [Google Scholar]
- 50.Purcell JE, Hoover RA, Schwarc NT. Interannual variation of strobilation by the scyphozoan Aurelia labiata in relation to polyp density, temperature, salinity, and light conditions in situ. Mar Ecol Prog Ser. 2009;375:139–49. [Google Scholar]
- 51.Fuchs B, Wang W, Graspeuntner S, Li Y, et al. Regulation of polyp to jellyfish transition in Aurelia aurita. Curr Biol. 2014;24:263–73. doi: 10.1016/j.cub.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Funayama N. The stem cell system in demosponges: suggested involvement of two types of cells: archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells) Dev Genes Evol. 2013;223:23–38. doi: 10.1007/s00427-012-0417-5. [DOI] [PubMed] [Google Scholar]
- 53.Simpson TL, Fell PE. Dormancy among the Porifera: gemmule formation and germination in freshwater and marine sponges. Trans Am Microsc Soc. 1974;93:544–77. [Google Scholar]
- 54.Loomis SH, Bettridge A, Branchini BR. The effects of elevated osmotic concentration on control of germination in the gemmules of freshwater sponges Eunapius fragilis and Anheteromeyania ryderi. Physiol Biochem Zool. 2009;82:388–95. doi: 10.1086/589901. [DOI] [PubMed] [Google Scholar]
- 55.Veron JEN. Corals in Space and Time: The Biogeography and Evolution of the Scleractinia. Cornell University Press; Ithaka, London: 1995. [Google Scholar]
- 56.Palumbi SR. Tactics of acclimation: morphological changes of sponges in an unpredictable environment. Science. 1984;225:1478–80. doi: 10.1126/science.225.4669.1478. [DOI] [PubMed] [Google Scholar]
- 57.Kaandorp JA. Morphological analysis of growth forms of branching marine sessile organisms along environmental gradients. Mar Biol. 1999;134:295–306. [Google Scholar]
- 58.Mendola D, de Caralt S, Uriz MJ, van den End F, et al. Environmental flow regimes for Dysidea avara sponges. Mar Biotechnol. 2008;10:622–30. doi: 10.1007/s10126-008-9102-0. [DOI] [PubMed] [Google Scholar]
- 59.McDonald JI, McGuinness KA, Hooper JNA. Influence of re-orientation on alignment to flow and tissue production in a Spongia sp (Porifera: Demospongiae: Dictyoceratida) J Exp Mar Biol Ecol. 2003;296:13–22. [Google Scholar]
- 60.Mendola D, van den Boogaart JG, van Leeuwen JL, Wijffels RH. Re-plumbing in a Mediterranean sponge. Biol Lett. 2007;3:595–8. doi: 10.1098/rsbl.2007.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ludeman DA, Farrar N, Riesgo A, Paps J, et al. Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. BMC Evol Biol. 2014;14:3. doi: 10.1186/1471-2148-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madin JS, Hughes TP, Connolly SR. Calcification, storm damage and population resilience of tabular corals under climate change. PLoS One. 2012;7:e46637. doi: 10.1371/journal.pone.0046637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baird AH, Hughes TP. Competitive dominance by tabular corals: an experimental analysis of recruitment and survival of understorey assemblages. J Exp Mar Biol Ecol. 2000;251:117–32. doi: 10.1016/s0022-0981(00)00209-4. [DOI] [PubMed] [Google Scholar]
- 64.Pratchett MS, Marnane MJ, Berumen ML, Eagle JE, et al. Habitat associations of juvenile versus adult butterflyfishes. Coral Reefs. 2008;27:541–51. [Google Scholar]
- 65.DR Bellwood, TP Hughes, C Folke, Nyström M. Confronting the coral reef crisis. Nature. 2004;429:827–33. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- 66.Berumen ML, Pratchett MS. Trade-offs associated with dietary specialization in corallivorous butterflyfishes (Chaetodontidae: Chaetodon) Behav Ecol Sociobiol. 2008;62:989–94. [Google Scholar]
- 67.Franzenburg S, Walter J, Künzel S, Baines JF, et al. Distinct antimicrobial tissue activity shapes host species-specific bacterial associations. Proc Natl Acad Sci USA. 2013;110:E3730–8. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fraune S, Bosch TCG. Why bacteria matter in animal development and evolution. BioEssays. 2010;32:571–80. doi: 10.1002/bies.200900192. [DOI] [PubMed] [Google Scholar]
- 69.Fraune S, Augustin R, Bosch TCG. Embryo protection in contemporary immunology: why bacteria matter. Commun Integr Biol. 2011;4:369–72. doi: 10.4161/cib.4.4.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107:18067–72. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boehm AM, Hemmrich G, Khalturin K, Puchert M, et al. FoxO is a critical regulator of stem cell maintenance and immortality in Hydra. Proc Natl Acad Sci USA. 2012;109:19697–702. doi: 10.1073/pnas.1209714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 73.Kelman D, Kashman Y, Rosenberg E, Kushmaro A, et al. Antimicrobial activity of Red Sea corals. Mar Biol. 2006;149:357–63. [Google Scholar]
- 74.Kimes NE, Johnson WR, Torralba M, Nelson KE, et al. The Montastraea faveolata microbiome: ecological and temporal influences on a Caribbean reef-building coral in decline. Environ Microbiol. 2013;15:2082–94. doi: 10.1111/1462-2920.12130. [DOI] [PubMed] [Google Scholar]
- 75.Roder C, Arif C, Daniels C, Weil E, et al. Bacterial profiling of White Plague Disease across corals and oceans indicates a conserved and distinct disease microbiome. Mol Ecol. 2014;23:965–74. doi: 10.1111/mec.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2012;14:335–46. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- 77.Hadfield MG. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu Rev Mar Sci. 2011;3:453. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 78.Muscatine L, Lenhoff HM. Symbiosis: on the role of algae symbiotic with Hydra. Science. 1963;142:956–8. doi: 10.1126/science.142.3594.956. [DOI] [PubMed] [Google Scholar]
- 79.Bosch TCG. What hydra has to say about the role and origin of symbiotic interactions. Biol Bull. 2012;223:78–84. doi: 10.1086/BBLv223n1p78. [DOI] [PubMed] [Google Scholar]
- 80.Keesing JK, Usher KM, Fromont J. First record of photosynthetic cyanobacterial symbionts from mesophotic temperate sponges. Mar Freshwater Res. 2012;63:403–8. [Google Scholar]
- 81.Baker AC, Starger CJ, McClanahan TR, Glynn PW. Corals’ adaptive response to climate change. Nature. 2004;430:741. doi: 10.1038/430741a. [DOI] [PubMed] [Google Scholar]
- 82.Oliver TA, Palumbi SR. Many corals host thermally resistant symbionts in high-temperature habitat. Coral Reefs. 2011;30:241–50. [Google Scholar]
- 83.Little AF, van Oppen MJH, Willis BL. Flexibility in algal endosymbiosis shapes growth in reef corals. Science. 2004;304:1492–4. doi: 10.1126/science.1095733. [DOI] [PubMed] [Google Scholar]
- 84.Keshavramurthy S, Hsu C-M, Kuo C-Y, Meng P-J, et al. Symbiont communities and host genetic structure of the brain coral Platygyra verweyi at the outlet of a nuclear power plant and adjacent areas. Mol Ecol. 2012;21:4393–407. doi: 10.1111/j.1365-294X.2012.05704.x. [DOI] [PubMed] [Google Scholar]
- 85.Hume B, D’Angelo C, Burt J, Baker AC, et al. Corals from the Persian/Arabian Gulf as models for thermotolerant reef-builders: prevalence of clade C3 Symbiodinium, host fluorescence and ex situ temperature tolerance. Mar Pollut Bull. 2013;72:313–22. doi: 10.1016/j.marpolbul.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 86.Sorek M, Levy O. Influence of the quantity and quality of light on photosynthetic periodicity in coral endosymbiotic algae. PLoS One. 2012;7:e43264. doi: 10.1371/journal.pone.0043264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sorek M, Levy O. The effect of temperature compensation on the circadian rhythmicity of photosynthesis in Symbiodinium, coral-symbiotic alga. Sci Rep. 2012;2:1–8. doi: 10.1038/srep00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorek M, Yacobi YZ, Roopin M, Berman-Frank I, et al. Photosynthetic circadian rhythmicity patterns of Symbiodinium, the coral endosymbiotic algae. Proc R Soc B. 2013;280:20122942. doi: 10.1098/rspb.2012.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chalker BE. In: Taylor DL, editor. Daily variation of in: Daily variation of in the calcification capacity of Acropora cervicornis; Proceedings: Third International Coral Reef Symposium; Miami. 1977; pp. 417–23. Rose-nstiel School of Marine and Atmospheric Science. [Google Scholar]
- 90.Sorek M, Díaz-Almeyda EM, Medina M, Levy O. Circadian clocks in symbiotic corals: the duet between Symbiodinium algae and their coral host. Mar Genomics. 2014;14:47–57. doi: 10.1016/j.margen.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 91.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 92.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 93.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 94.Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann N Y Acad Sci. 2010;1204:156–62. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- 95.Boehm AM, Rosenstiel P, Bosch TCG. Stem cells and aging from a quasi-immortal point of view. BioEssays. 2013;35:994–1003. doi: 10.1002/bies.201300075. [DOI] [PubMed] [Google Scholar]
- 96.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 97.Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in Hydra: a response to starvation and stress in early animal evolution. Biochim Biophys Acta. 2009;1793:1432–43. doi: 10.1016/j.bbamcr.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 98.Böttger A, Strasser D, Alexandrova O, Levin A, et al. Genetic screen for signal peptides in Hydra reveals novel secreted proteins and evidence for non-classical protein secretion. Eur J Cell Biol. 2006;85:1107–17. doi: 10.1016/j.ejcb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 99.Steele RE, Lieu P, Mai NH, Shenk MA, et al. Response to insulin and the expression pattern of a gene encoding an insulin receptor homologue suggest a role for an insulin-like molecule in regulating growth and patterning in Hydra. Dev Genes Evol. 1996;206:247–59. doi: 10.1007/s004270050050. [DOI] [PubMed] [Google Scholar]
- 100.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 101.Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Adv Exp Med Biol. 2005;560:11–8. doi: 10.1007/0-387-24180-9_2. [DOI] [PubMed] [Google Scholar]
- 102.Franzenburg S, Fraune S, Künzel S, Baines JF, et al. MyD88 deficient Hydra reveal ancient function of TLR-signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. 2012;109:19374–9. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev Comp Immunol. 2009;33:559–69. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 104.Bosch TCG. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Ann Rev Microbiol. 2013;67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 105.Bosch TCG. Rethinking the role of immunity: lessons from Hydra. Trends Immunol. 2014 doi: 10.1016/j.it.2014.07.008. in press, DOI 10.1016/j.it.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 106.Gorbunov MY, Falkowski PG. Photoreceptors in the cnidarian hosts allow symbiotic corals to sense blue moonlight. Limnol Oceanogr. 2002;47:309–15. [Google Scholar]
- 107.Levy O, Kaniewska P, Alon S, Eisenberg E, et al. Complex diel cycles of gene expression in coral-algal symbiosis. Science. 2011;331:175. doi: 10.1126/science.1196419. [DOI] [PubMed] [Google Scholar]
- 108.Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadianclock. PLoS One. 2010;5:e12805. doi: 10.1371/journal.pone.0012805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vize PD. Transcriptome analysis of the circadian regulatory network in the coral Acropora millepora. Biol Bull. 2009;216:131–7. doi: 10.1086/BBLv216n2p131. [DOI] [PubMed] [Google Scholar]
- 110.Mason B, Schmale M, Gibbs P, Miller MW, et al. Evidence for multiple phototransduction pathways in a reef-building coral. PLoS One. 2012;7:e50371. doi: 10.1371/journal.pone.0050371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Levy O, Appelbaum L, Leggat W, Gothlif Y, et al. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science. 2007;318:467–70. doi: 10.1126/science.1145432. [DOI] [PubMed] [Google Scholar]
- 112.Hyman LH. Invertebrates: Protozoa Through Ctenophora. McGraw-Hill; New York: 1940. [Google Scholar]
- 113.Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biol Bull. 2001;201:323–8. doi: 10.2307/1543611. [DOI] [PubMed] [Google Scholar]
- 114.Leys SP, Cronin TW, Degnan BM, Marshall JN. Spectral sensitivity in a sponge larva. J Comp Physiol A. 2002;188:199–202. doi: 10.1007/s00359-002-0293-y. [DOI] [PubMed] [Google Scholar]
- 115.Rivera AS, Ozturk N, Fahey B, Plachetzki DC, et al. Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J Exp Biol. 2012;215:1278–86. doi: 10.1242/jeb.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu JK. The evolutionary origin of the vertebrate neural crest and its developmental gene regulatory network – insights from amphioxus. Zoology. 2010;113:1–9. doi: 10.1016/j.zool.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 117.Bertrand S, Escriva H. Evolutionary crossroads in developmental biology: amphioxus. Development. 2011;138:4819–30. doi: 10.1242/dev.066720. [DOI] [PubMed] [Google Scholar]
- 118.Holland LZ. Genomics, evolution and development of amphioxus and tunicates: the goldilocks principle. J Exp Zool B Mol Dev Evol. 2014 doi: 10.1002/jez.b.22569. in press, DOI: 10.1002/jez.b.22569. [DOI] [PubMed] [Google Scholar]
- 119.Benito-Gutiérrez E, Weber H, Bryant DV, Arendt D. Methods for generating year-round access to amphioxus in the laboratory. PLoS One. 2013;8:e71599. doi: 10.1371/journal.pone.0071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hodgins-Davis A, Adomas AB, Warringer J, Townsend JP. Abundant gene-by-environment interactions in gene expression reac-tion norms to copper within Saccharomyces cerevisiae. Genome Biol Evol. 2012;4:1188. doi: 10.1093/gbe/evs084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Waddington CH. Organisers and Genes. Cambridge University Press; Cambridge: 1940. [Google Scholar]
- 122.Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, et al. Evolutionary conservation of the eumetazoan gene regulatory land-scape. Genome Res. 2014;24:639–50. doi: 10.1101/gr.162529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]