ABSTRACT
The prokaryotic pangenome partitions genes into core and dispensable genes. The order of core genes, albeit assumed to be stable under selection in general, is frequently interrupted by horizontal gene transfer and rearrangement, but how a core-gene-defined genome maintains its stability or flexibility remains to be investigated. Based on data from 30 species, including 425 genomes from six phyla, we grouped core genes into syntenic blocks in the context of a pangenome according to their stability across multiple isolates. A subset of the core genes, often species specific and lineage associated, formed a core-gene-defined genome organizational framework (cGOF). Such cGOFs are either single segmental (one-third of the species analyzed) or multisegmental (the rest). Multisegment cGOFs were further classified into symmetric or asymmetric according to segment orientations toward the origin-terminus axis. The cGOFs in Gram-positive species are exclusively symmetric and often reversible in orientation, as opposed to those of the Gram-negative bacteria, which are all asymmetric and irreversible. Meanwhile, all species showing strong strand-biased gene distribution contain symmetric cGOFs and often specific DnaE (α subunit of DNA polymerase III) isoforms. Furthermore, functional evaluations revealed that cGOF genes are hub associated with regard to cellular activities, and the stability of cGOF provides efficient indexes for scaffold orientation as demonstrated by assembling virtual and empirical genome drafts. cGOFs show species specificity, and the symmetry of multisegmental cGOFs is conserved among taxa and constrained by DNA polymerase-centric strand-biased gene distribution. The definition of species-specific cGOFs provides powerful guidance for genome assembly and other structure-based analysis.
IMPORTANCE
Prokaryotic genomes are frequently interrupted by horizontal gene transfer (HGT) and rearrangement. To know whether there is a set of genes not only conserved in position among isolates but also functionally essential for a given species and to further evaluate the stability or flexibility of such genome structures across lineages are of importance. Based on a large number of multi-isolate pangenomic data, our analysis reveals that a subset of core genes is organized into a core-gene-defined genome organizational framework, or cGOF. Furthermore, the lineage-associated cGOFs among Gram-positive and Gram-negative bacteria behave differently: the former, composed of 2 to 4 segments, have their fragments symmetrically rearranged around the origin-terminus axis, whereas the latter show more complex segmentation and are partitioned asymmetrically into chromosomal structures. The definition of cGOFs provides new insights into prokaryotic genome organization and efficient guidance for genome assembly and analysis.
INTRODUCTION
Prokaryotic genomes and their genes, albeit much smaller than those of eukaryotes, are proposed to be well organized in lineage-specific ways that are important for understanding genome structures and deciphering the genotype-phenotype relationship (1, 2). In the context of pangenome, genes of a given species are algorithmically divided into core and dispensable genes across a dozen or so genomes (3, 4). Such core genes often include those with essential functions and are considered coadapted over evolutionary time scales (5, 6). In addition, relative orders of core genes are often assumed to be stable for two reasons. First, core genes are assumed to be under strong selection in location and orientation to minimize interruption of their expression regulation (1, 7). Second, core genes are mostly vertically inherited, and their genome organization should have some degrees of robustness, being able to resist massive horizontal gene transfer (HGT) (8). Therefore, prokaryotic genome organization is expected not only to favor a set of conserved core genes but also to limit their order and orientation; such organizational frameworks are usually species specific or lineage associated, where horizontally acquired dispensable genes are allowed to squeeze into certain chromosome positions (9–11).
Although genome organization appears to be preserved under strong selection (12, 13), genome rearrangement is ubiquitous even among closely related isolates and has been assumed to be one of the mutational forces that drive genome evolution (14, 15). Sometimes genome rearrangement can be very intensive, involving up to half of the total genome length (16). However, previous studies on rearrangement have reported a symmetric pattern in a few species (17) and correlated rearrangement with variable ecological conditions (18, 19). Available algorithms on gene mobility focus on HGT or do not discriminate core genes from others (20, 21). Thus, the flexibility of core genome structures remains elusive.
In this study, we start with pangenomic analysis of 30 prokaryotic species from six phyla and hope to address four basic questions. First, within a pangenome, is there a set of framework-forming core genes (or a subset of the core genes) that are relatively stable in chromosome order and orientation? Second, if such a core-gene-defined genome organizational framework (cGOF) does exist for a given species or remain characteristic within lineages, how flexible and stable is it when HGT and genome rearrangement occur at different frequencies? Third, are there unique functional features specific to cGOF genes compared to non-cGOF genes? Fourth, how useful are cGOFs in assisting genome sequence assembly and finishing, as well as annotation and data mining?
RESULTS AND DISCUSSION
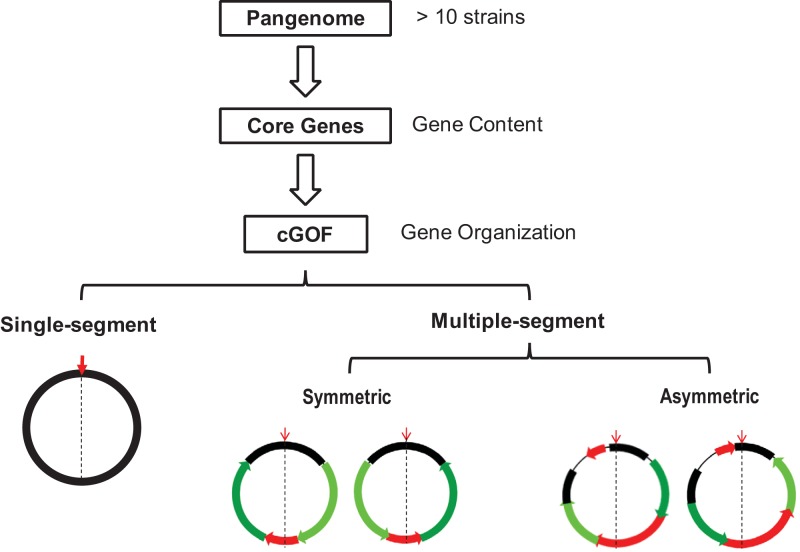
A subset of core genes in a pangenome defines cGOF.
The events altering gene order in prokaryotic chromosomes are mostly HGT and gene rearrangement; we thus classify all genes in a given species or a pangenome into three essential groups according to their relative mobility: (i) immobile genes, which maintain their order and orientation in all isolates, (ii) stable genes, which have conserved order and orientation in large segments but flip order or trade position with other chromosomal segments, and (iii) mobile genes, which relocate separately or together with neighbors in small blocks through HGT. To apply these definitions in pangenome analysis, we started with 425 strains from 30 species (Table 1), and each species has more than 10 complete genome sequences. First, we identified core genes common to all strains for a given species and their syntenic gene blocks as well (for conserved order and relative orientation toward the origin and terminus). Second, we used iteration (see Materials and Methods for detail of the algorithm) to find a series of gene blocks among core genes, which form the longest subsequence shared by all strains. In other words, we found a subset of core genes that serve as a backbone or framework (immobile genes, the 1st group) at the chromosomal level. As a result, the simplest cGOF has a single segment; among species with intensive rearrangement, we divided cGOFs into syntenic fragments at multiple recombination sites (i.e., identified multisegmental cGOFs). We used a cutoff of 20 genes (empirically defined [see Materials and Methods]) to exclude smaller gene blocks (containing mostly mobile genes, the 3rd group), and the rest (containing stable genes, the 2nd group) still form a stable structure.
TABLE 1 .
cGOF characteristics of representative species
| cGOF class and species | Gram stain | Phylum | Habitata | DnaE groupb | No. of segments | No. of cGOF genes | No. of core genes | % of cGOF/core genes | Genome size (Mb) | No. of coding genes | GC % | LeGPf % | No. of samples |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single segment | |||||||||||||
| Bifidobacterium animalis | + | Actinobacteria | H | 1 | 1 | 1,305 | 1,305 | 100.0 | 1.94 | 1,570 | 60.50 | 0.7 | 11 |
| Corynebacterium diphtheriae | + | Actinobacteria | H | 2 | 1 | 1,570 | 1,573 | 99.8 | 2.47 | 2,276 | 53.55 | 0.62 | 13 |
| Corynebacterium pseudotuberculosis | + | Actinobacteria | H | 2c | 1 | 1,369 | 1,370 | 99.9 | 2.32 | 2,078 | 52.18 | 0.57 | 15 |
| Bacillus cereus | + | Firmicutes | S | 3 | 1 | 3,102 | 3,109 | 99.8 | 5.52 | 5,608 | 35.35 | 0.75 | 13 |
| Bacillus subtilis | + | Firmicutes | S | 3 | 1 | 2,778 | 2,780 | 99.9 | 4.40 | 4,363 | 43.93 | 0.74 | 11 |
| Clostridium botulinum | + | Firmicutes | H, S, A | 3 | 1 | 2,283 | 2,303 | 99.1 | 3.92 | 3,604 | 28.08 | 0.83 | 10 |
| Staphylococcus aureus | + | Firmicutes | H | 3 | 1 | 1,455 | 1,455 | 100.0 | 2.84 | 2,525 | 32.85 | 0.76 | 12 |
| Alteromonas macleodii | − | Gamma-proteobacteria | A | 2 | 1 | 1,385 | 1,550 | 89.4 | 4.59 | 3,939 | 44.77 | 0.54 | 12 |
| Escherichia coli | − | Gamma-proteobacteria | H | 1 | 1 | 1,486 | 2,542 | 58.5 | 5.10 | 4,766 | 50.67 | 0.55 | 19 |
| Treponema pallidum | − | Spirochaetes | H | 1 | 1 | 814 | 814 | 100.0 | 1.14 | 1,026 | 52.80 | 0.66 | 10 |
| Symmetric | |||||||||||||
| Bifidobacterium longum | + | Actinobacteria | H | 1 | 2 | 865 | 885 | 97.7 | 2.46 | 2,005 | 59.98 | 0.66 | 10 |
| Mycobacterium tuberculosis | + | Actinobacteria | H | 2 | 2 | 2,666 | 2,666 | 100.0 | 4.40 | 3,941 | 65.57 | 0.59 | 22 |
| Propionibacterium acnes | + | Actinobacteria | H | 2 | 2 | 1,648 | 1,649 | 99.9 | 2.52 | 2,295 | 60.05 | 0.6 | 10 |
| Bacillus amyloliquefaciens | + | Firmicutes | S | 3 | 2 | 2,808 | 2,812 | 99.8 | 4.00 | 3,929 | 46.23 | 0.75 | 11 |
| Bacillus thuringiensis | + | Firmicutes | S | 3 | 2 | 2,264 | 2,982 | 75.9 | 6.03 | 6,055 | 35.15 | 0.75 | 11 |
| Listeria monocytogenes | + | Firmicutes | S, A | 3 | 2 | 797 | 798 | 99.9 | 2.94 | 2,906 | 38.04 | 0.79 | 29 |
| Streptococcus suis | + | Firmicutes | H | 3 | 4d | 948 | 962 | 98.5 | 2.09 | 1,999 | 41.22 | 0.79 | 16 |
| Streptococcus pneumoniae | + | Firmicutes | H | 3 | 4 | 794 | 1,197 | 66.3 | 2.11 | 2,035 | 39.70 | 0.79 | 26 |
| Streptococcus pyogenes | + | Firmicutes | H | 3 | 4 | 1,173 | 1,173 | 100.0 | 1.86 | 1,808 | 38.53 | 0.78 | 9 |
| Asymmetric | |||||||||||||
| Sulfolobus islandicus | NAe | Crenarchaeota (Archaea) | A | NAe | 7 | 1,677 | 1,827 | 91.8 | 2.65 | 2,745 | 35.17 | 0.49 | 10 |
| Prochlorococcus marinus | NA | Cyanobacteria | A | 1 | 9 | 472 | 754 | 62.6 | 1.86 | 2,050 | 35.98 | 0.49 | 12 |
| Neisseria meningitidis | − | Betaproteobacteria | H | 1 | 7 | 1,046 | 1,204 | 86.9 | 2.22 | 1,953 | 51.58 | 0.53 | 14 |
| Campylobacter jejuni | − | Epsilon-proteobacteria | H | 1 | 6 | 916 | 953 | 96.1 | 1.69 | 1,674 | 30.51 | 0.62 | 11 |
| Helicobacter pylori | − | Epsilon-proteobacteria | H | 1 | 6 | 638 | 923 | 69.1 | 1.63 | 1,508 | 38.91 | 0.59 | 17 |
| Acinetobacter baumannii | − | Gamma-proteobacteria | A | 1 | 5 | 1,065 | 1,271 | 83.8 | 3.97 | 3,689 | 39.02 | 0.59 | 15 |
| Francisella tularensis | − | Gamma-proteobacteria | A | 1 | 13 | 498 | 995 | 49.1 | 1.90 | 1,619 | 32.25 | 0.61 | 12 |
| Legionella pneumophila | − | Gamma-proteobacteria | A | 1 | 2 | 2,148 | 2,200 | 97.6 | 3.50 | 3,097 | 38.35 | 0.57 | 12 |
| Pseudomonas putida | − | Gamma-proteobacteria | H, S, A | 2 | 32 | 497 | 1,234 | 40.3 | 6.10 | 5,516 | 61.84 | 0.55 | 11 |
| Salmonella enterica | − | Gamma-proteobacteria | Host | 1 | 12 | 2,204 | 2,268 | 97.2 | 4.87 | 4,595 | 52.13 | 0.59 | 29 |
| Yersinia pestis | − | Gamma-proteobacteria | Host | 1 | 30 | 1,283 | 2,290 | 56.1 | 4.75 | 4,072 | 47.65 | 0.59 | 12 |
H, host; S, soil; A, aquatic.
The three DnaE groups are classified based on the presence of different DNA polymerase III gene isoforms and other related mutator genes: 1, dnaE1-dnaE1; 2, dnaE1-dnaE1-dnaE2; 3, polC-dnaE3-polV.
This species is proposed to be one of the DnaE2 group members since the dnaE2 gene has been found in almost all species in this genus and sometime is carried in plasmids that are not included in the chromosomal sequences.
This species is proposed to have a four-segment symmetric cGOF, but the arm segment is very short and transfers between the opposite positions along the origin-terminus axis.
NA, not available.
LeGP, leading-strand gene proportion.
Among the 30 species in the data set, 10 species, or a third of the total, have single-segment cGOFs, and the rest contain multiple segments (Fig. 1). Notably, single-segment cGOFs are not confined to a specific phylum or lineage but are found in all three major phyla investigated in this study—Proteobacteria, Firmicutes, and Actinobacteria—as well as a single representative from Spirochaetes (see Fig. S1A in the supplemental material). In addition, even closely related species appeared to have different cGOFs in terms of their segmental patterns. For instance, although both E. coli and S. enterica belong to the family Enterobacteriaceae, the former has a single-segment cGOF and that of the latter is multisegmental. The scenario is similar among the four species of the genus Bacillus (Table 1).
FIG 1 .
The workflow of cGOF definition. cGOF is a subset of the order-stable core genes of a given pangenome and is divided into multiple segments when genome rearrangement occurs. All multiple-segment cGOFs are grouped as symmetric or asymmetric according to their symmetry in segmentation and rearrangement. The downward red arrows indicate the origin of replication, and black dashed lines indicate the origin-terminus axis. Segments are colored according to their movement patterns: black bars indicate those immobile with respect to the origin, dark and light green bars indicate arm segments that exchange their locations and orientations, and red bars indicate segments that are either locally inverted or moved to other locations. Chromosomal regions without cGOF genes are indicated with thin lines.
Aside from taxonomy, species with single-segment and multisegment cGOFs share many genome-wide characteristics, including Gram staining, strand-biased gene distribution (based on strand-specific gene counts), genome size, and GC content. We also evaluated ecological parameters, such as soil, water, and host, and found no significant association with GOF segmentation (see Table S1 in the supplemental material). Hence, a single-segment cGOF may be a transient definition, since the definition of whether a cGOF is single segmental or multisegmental is often limited by both sample size (the number of isolates sequenced) and the degree of genome diversity (or the frequency of gene rearrangement events). For example, S. aureus is generally considered to have a conserved chromosome without apparent rearrangement; i.e., it has a single-segment cGOF, but a recent study has reported a rare isolate that has an inverted chromosome (16). If this isolate is included, S. aureus becomes a species possessing a two-segment cGOF.
Multisegment cGOFs are either symmetric or asymmetric.
We classified the rest of the 20 species with multisegment cGOFs as symmetric or asymmetric according to their rearrangement patterns toward the origin-terminus axis (Table 1). The symmetric cGOF is observed exclusively among the Gram-positive species (six Firmicutes and three Actinobacteria species); these cGOFs are generally divided into two or four segments and rearranged basically symmetrically around the origin-terminus axis (see Fig. S1B in the supplemental material). The asymmetric cGOF was observed among 9 Gram-negative Proteobacteria species, one Cyanobacteria species, and one Archaea species, comprised of 2 to 30 segments and rearranged asymmetrically (see Fig. S1C). We also searched for genome parameters related to the symmetry of cGOFs and found significant association with strand-biased gene distribution but not with genome size and GC content. We further looked into the DnaE (α subunit of DNA polymerase III) grouping scheme, whose isoforms are known to associate with strand-biased gene distribution (22) (Table 2). According to the DnaE isoforms contained, bacterial genomes are classified into three groups: (i) DnaE1, whose isoform is a dnaE1-dnaE1 homodimer; (ii) DnaE2, whose isoform is a dnaE1-dnaE1 homodimer plus a mutator gene, dnaE2; and (iii) DnaE3, whose isoform is a polC-dnaE3 heterodimer plus polV, another mutator gene (23, 24). In this study, we found that the species with symmetric cGOFs have strong strand-biased gene distribution (indicated by leading-strand gene proportion) and are classified into the DnaE2 and DnaE3 groups, except Bifidobacterium longum (which belongs to the DnaE1 group), whereas the species with asymmetric cGOFs are generally balanced in gene distribution between the two strands and are all classified into the DnaE1 group, except one species in DnaE2 group, Pseudomonas putida (Table 1).
TABLE 2 .
Additional characteristics of symmetric and asymmetric cGOFs
| Parameter | cGOFs |
P valuea | |
|---|---|---|---|
| Symmetric | Asymmetric | ||
| Gram staining (no./total) | 0.045 × 10−3 | ||
| Positive | 9/9 | 0/11 | |
| Negative | 0/9 | 9/11 | |
| NAb | 0/9 | 2/11 | |
| Phylum (no.) | 0.17 × 10−3 | ||
| Firmicutes | 6/9 | 0/11 | |
| Actinobacteria | 3/9 | 0/11 | |
| Proteobacteria | 0/9 | 9/11 | |
| Others | 0/9 | 2/11 | |
| DnaE (no./total) | 3.4 × 10−3 | ||
| DnaE1 | 1/9 | 9/11 | |
| DnaE2 | 2/9 | 1/11 | |
| DnaE3 | 6/9 | 0/11 | |
| NA | 0/9 | 1/11 | |
| Habitats (no./total) | 70 × 10−3 | ||
| Host | 6/9 | 5/11 | |
| Soil | 2/9 | 0/11 | |
| Aquatic | 0/9 | 5/11 | |
| Positive | 1/9 | 1/11 | |
| Leading-strand gene proportion (%) | 72.2 ± 8.3 | 57.3 ± 3.9 | 0.090 × 10−3 |
| Genome size (Mb) | 3.16 ± 1.39 | 3.19 ± 1.54 | 0.96 |
| GC content (%) | 47.2 ± 11.5 | 42.1 ± 9.8 | 0.31 |
The P value was calculated using the chi-square test for count data and Student’s t test for measurement data.
NA, not available.
Since the replication of prokaryotic genomes is symmetrical around the origin-terminus axis, the leading and lagging strands are different in gene expression priorities (22). Furthermore, strand-biased gene distribution appears to be species specific or lineage associated and has been considered under selection (13, 25). In the case of symmetric cGOF, an inversion of the terminus segment or an exchange of the two arm segments after inversion does not change the gene distribution on the two strands as reported previously (17). Therefore, the limited segmentation and rearrangement in symmetric cGOFs appear to be dependent on selection force that maintains their strand-biased gene distribution. In the case of species with asymmetric cGOF, selections on cross-strand rearrangement are more relaxed so that cGOFs may have better tolerance of the complicated GOF segmentation and rearrangement. The underlying reason appears to be the function of specific DnaE isoforms (the principle components of prokaryotic DNA replication and repair apparatuses), which are proposed to govern strand-biased gene distribution and consequently the limitation to the rearrangement symmetry.
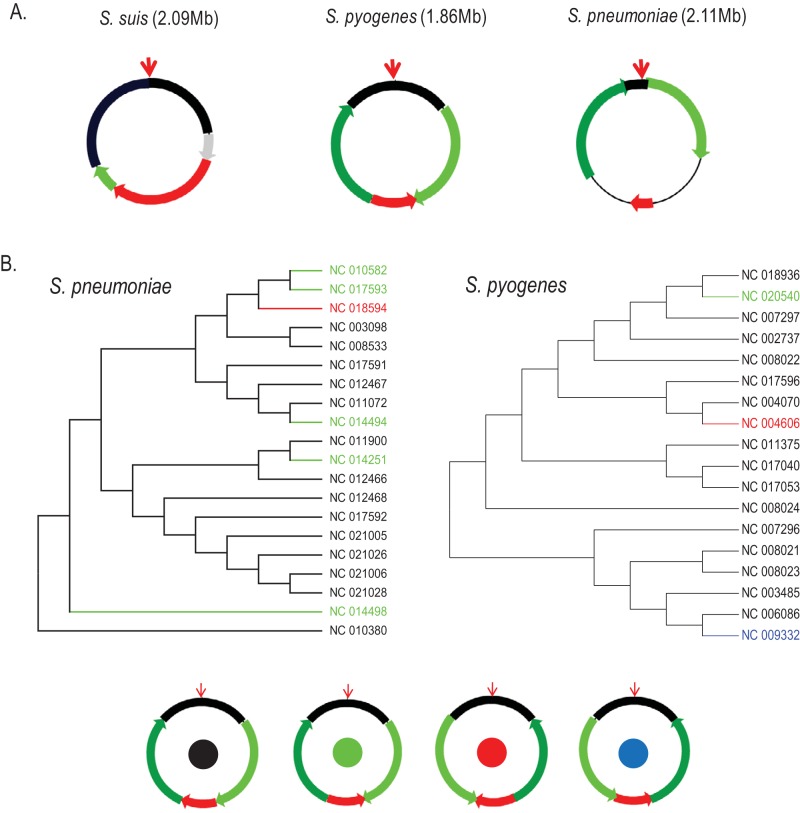
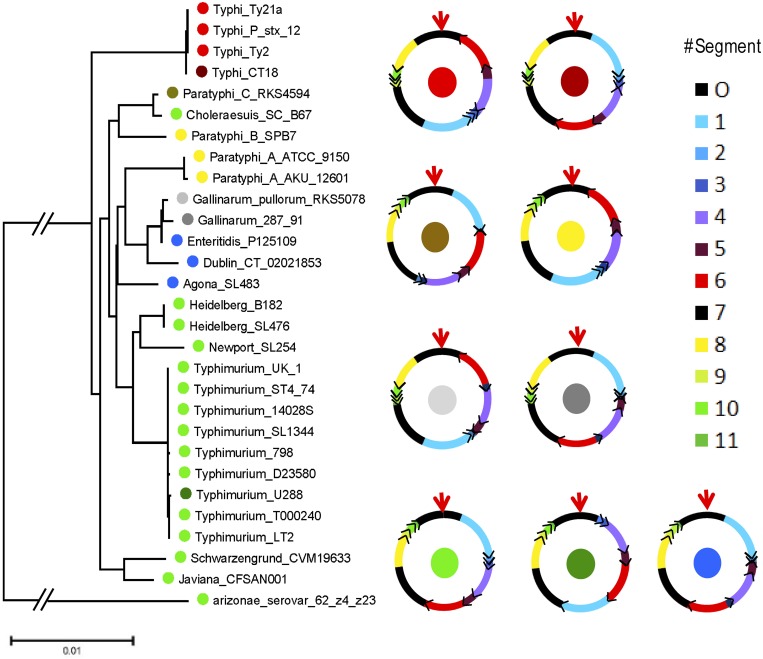
In the symmetric cGOF group, we investigated three species from the genus Streptococcus (S. suis, S. pneumoniae, and S. pyogenes) in detail. First, they all share a four-segment symmetric cGOF but differ from one another in terms of segment size. An exception is S. suis, which bears a variant of the four-segment symmetric cGOF with only one arm segment. Interestingly, this arm segment is very short and resides at either the left or right half of the chromosome between the origin segment and the terminus segment (Fig. 2A). Further investigation of S. pneumoniae and S. pyogenes revealed that their cGOF rearrangements are not congruent with the phylogenic relationship in multilocus sequence typing (MLST) trees (Fig. 2B); i.e., isolates with the same cGOF configurations (permutation of each cGOF segment) can be found in different lineages. The complex distribution of cGOF configurations indicates that gene rearrangement can be independent of phylogenetic relationship: they must occur at the same recombination sites multiple times. Such a reversible rearrangement in symmetric GOF was experimentally confirmed in a recent report where an S. aureus strain periodically inverts half of its chromosome back and forth (16). In contrast, the segment rearrangement scenarios of the asymmetric group are largely different. For example, in Salmonella enterica, the organization of its 12 cGOF segments in each subspecies (such as S. Typhi and S. Paratyphi) are distinct (Fig. 3), indicating that the genome was rearranged in the ancestor strain and such an event has not been reversed since. Our overall impression is that, as opposed to reversible rearrangement found in symmetric cGOFs, the rearrangement of asymmetric cGOFs may behave as a driving force of evolution because rearrangement events are often accompanied by the emergence of new lineages or subspecies, as previously illustrated in Yersinia pestis, another Proteobacteria species with an asymmetric cGOF (15). Among Gram-positive bacteria, reversible events of cGOFs homogenize lineage specificity in their cGOF permutation and do not increase species diversity (at most four cGOF configurations) and so are unlikely to be taken into account as an evolutionary force. Such different effects of Gram-positive and Gram-negative bacteria in contributing to genome evolution need to be noted, and the different replication/recombination machineries are thought to be responsible.
FIG 2 .
Segmentation and rearrangement of symmetric cGOFs. (A) cGOF segmentation and rearrangement in three Streptococcus spp. Segments are colored as follows: black, segment with origin site; red, segment with terminus site; light and dark green, left and right arm segments on both sides; gray, a potential location of the arm segment of Streptococcus suis to distinguish it from the standard four-segment cGOF of the other species. (B) Multilocus sequence typing (MLST) trees of Streptococcus pneumoniae (left) and Streptococcus pyogenes (right). The four types of four-segment symmetric cGOF are indicated by the color of the solid circles at the center of each ring; isolates in the MLST trees are colored accordingly.
FIG 3 .
Asymmetric cGOF segmentation and permutation. (A) MLST tree for 29 Salmonella enterica isolates. (B) cGOF segment orders in S. enterica. The colors of the solid circles represent the leading isolate names, and solid circles in the center of the rings indicate the segment orders.
Since gene essentiality has been proposed to play roles in strand-biased gene distribution (26), we further investigated the essentiality of cGOF genes. Among the 30 species recruited for this study, only eight are included in the Database of Essential Genes (http://www.essentialgene.org). Essential genes in all of these species exhibit both enrichment in cGOF genes (ranging from 60.7% to 84.6%, except for H. pylori [49.5%]) and strand-biased distribution toward the leading strand (ranging from 60.7% to 90.4%) regardless of their cGOF types: single segment versus multisegment and symmetric versus asymmetric. It seems that the strand-biased distribution of essential genes does not depend on cGOFs in the context of genome organization and rearrangement. However, we are unable to show a clear picture of essential genes in all species with regard to their stability and flexibility in cGOFs and relevance to genome organization in general due to inadequate data and their greater variation within a pangenome.
cGOFs have functional implications.
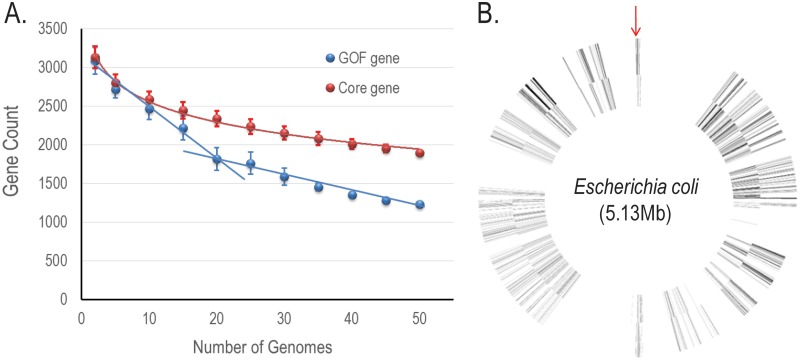
To further examine the biological significance of cGOF, we took E. coli and its isolates as an example. First, we investigated the proportion of cGOF genes in the pangenome based on a total of 53 complete genomes from the NCBI databases (see Fig. S2 in the supplemental material). This Gram-negative species (Gammaproteobacteria) has a single-segment cGOF, even when all 53 isolates are included in the analysis. As the sample size increases, the number of cGOF genes exhibits two linear decreases: the fast decrease is followed by a slow decrease when the sample size exceeds 20 genomes (Fig. 4A). From the phylogenetic tree including all isolates, we selected 19 from most taxonomic groups for maximal sequence diversity in the pangenomic analysis (see Fig. S2). The cGOF genes from the 19 selected isolates form a single-segment framework (Fig. 4B). There are also four large cGOF gene-free spaces that contain mobile (mostly dispensable) genes; in these spaces, small gene blocks are found rearranged asymmetrically around the origin-terminus axis, similar to its close relative, S. enterica, which has a multisegment asymmetric cGOF with rearranged small segments. The emerging rearrangement caused by these small gene blocks in E. coli is most likely the reason for the continuing decrease in the cGOF gene count (Fig. 4A).
FIG 4 .
cGOF of E. coli. (A) The plot illustrates the average number of E. coli core genes (blue) and cGOF genes (red) for n = 2, 5, 10, 15, 20 … 50 genomes, based on a maximum of 500 random combinations of genomes for each n. (B) cGOF gene distribution in a virtual E. coli genome. cGOF genes are depicted by thin lines. The spaces between neighboring cGOF genes are scaled to the average distance between genes in 19 E. coli genomes. The outer and inner layers represent positive and negative strands, respectively. The downward arrow points to the replication origin.
Next, we investigated if cGOF comprises naturally immobile genes as opposed to the null hypothesis that all genes in genome share the same mobility (i.e., genes are transferred or rearranged randomly within the genome). We proposed a pangenome model in which all genes move around in a random fashion at a rate comparable to that of the actual E. coli pangenome (see the supplemental methods in Text S1 in the supplemental material). In this model, each simulation randomly generates a list of genes that do not change their locations; however the number of such genes is nowhere near the actual number of cGOF genes in E. coli, even after the transfer rate is gradually reduced to 40% of the actual rate (see Fig. S3 in the supplemental material). Therefore, the null hypothesis has to be rejected, and a set of orientation-stable core genes does exist. The existence of the set of order-conserved core genes in each species supports the core genome hypothesis as previously described (27, 28).
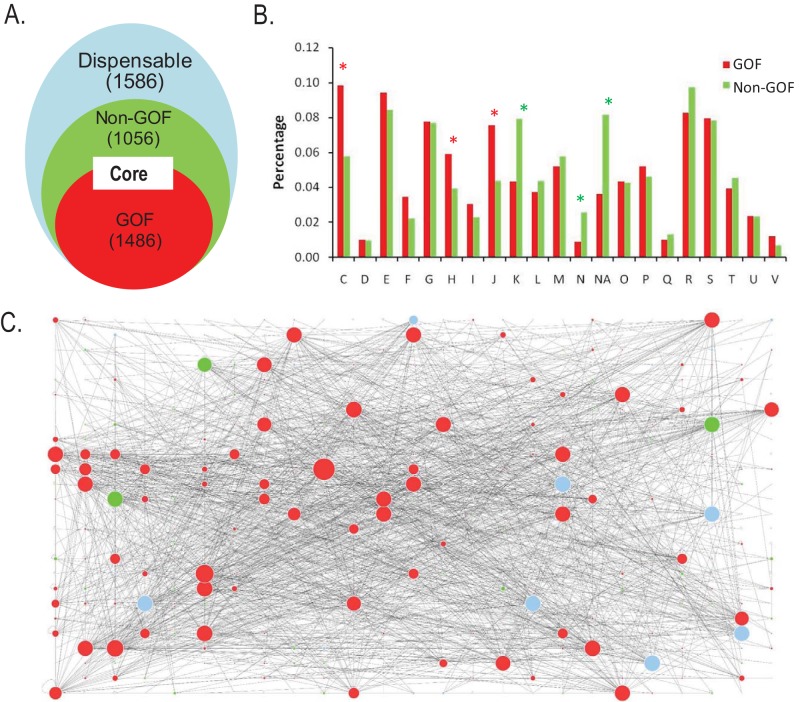
Finally, we evaluated functional implications of cGOF genes in a representative E. coli strain (DH10B). In this strain, the numbers of cGOF, non-cGOF core, and dispensable genes (defined based on the 19-isolate pangenome) are 1,486, 1,056, and 1,586, respectively (Fig. 5A). When comparing the cGOF and non-cGOF genes, we observed several unique features. First, the essential genes of E. coli are enriched in the cGOF genes (see Table S2A in the supplemental material). Second, the cGOF genes in COG categories are significantly enriched in categories C (energy production and conversion), J (translation, ribosomal structure and biogenesis), and H (coenzyme transport and metabolism), all of which represent the most ancient biological functions (Fig. 5B; see Table S2B) (29, 30). The categories in which non-cGOF genes are enriched, K (transcription), N (cell mobility), and NA (“not assigned”), stood out. Third, the cGOF genes show a significant bias in codon usage (see Fig. S4 in the supplemental material), which is also characteristic for genes with key functions (31, 32). Fourth, the cGOF genes have increased connectivity in gene interaction network, i.e., they often serve as hub genes in the network (Fig. 5C; see Table S2C). According to studies on the gene interaction network (33), there is a core set of genes, usually ancestral in evolution and critical for functions, involved in a large number of interactions with other genes (e.g., functioning as hub genes), whereas genes with auxiliary functions, usually horizontally transferred or dispensable, are less connected. Fifth, the genes with the top 50 connections in the cGOF, non-cGOF, and dispensable gene sets are involved in three major functional categories: ribosomal, energy production/conversion, and transporter activities, respectively (see Table S2D). The ribosomal protein genes that carry out the most fundamental functions in cellular activities are almost all cGOF genes. Therefore, although the non-cGOF core genes are present in all isolates and recognized as core genes in classic pangenome analyses, they do not encode proteins that function as hubs of cellular activities and instead exhibit properties more characteristic of dispensable genes.
FIG 5 .
Function of cGOF genes in E. coli DH10B. (A) Partition of cGOF, non-cGOF, and dispensable genes in DH10B, where cGOF and non-cGOF genes are both core genes. (B) Distribution of cGOF and non-cGOF genes in COG categories. Red asterisks indicate categories where cGOF genes are significantly enriched, and green asterisks indicate those of non-cGOF genes (P < 0.05). (C) Genes and their actions with other genes in the gene-gene interaction network. Red, green, and blue solid circles denote cGOF, non-cGOF core, and dispensable genes, respectively. The radius of each solid circle is scaled to the number of the corresponding gene actions.
cGOF provides a powerful guide for genome assembly and finishing.
The order conservation of cGOF genes provides a discrete sequence anchorage for scaffold orientation and finishing: both are critical and difficult steps in genome sequence assembly. To test the efficiency of cGOF-assisted assembly, we selected 91 genomes as a testing set (see Table S3A in the supplemental material) from four species with adequate numbers of complete genomes for the exercise: E. coli (single-segment GOF, Gram negative), S. aureus (single-segment GOF, Gram positive), S. pyogenes (symmetric GOF, Gram positive), and H. pylori (asymmetric GOF, Gram negative). The cGOFs of these species are highly conserved in that only 3 genomes exhibit inversions inside segments (see Table S3B). To simulate genome assembly, the test genomes were further broken into contigs (see Fig. S4A and the supplemental methods in Text S1 in the supplemental material) and reoriented according to the order of its cGOF. In all four species, the average accuracy of contig orientation reached 97% of the total contig length, and only contigs without cGOF genes (up to 3%) were missed (see Table S3B). In the test assembly of the 34 virtual E. coli drafts, the cGOF-based method was further compared to the routine assembly directed by a single reference genome, and was found to yield a much lower (approximately one-third) error rate (see Fig. S4B). Moreover, the cGOF-based strategy also exhibited high efficiency with respect to the empirical draft. In an assembly of 10 self-sequenced wild-type E. coli isolates (30 to 80 scaffolds per draft), up to 97.18% of the total scaffold length is oriented after some of the cGOF-free scaffolds are linked to cGOF scaffolds according to paired-end information. All of the neighboring relationships between the scaffolds determined based on cGOF, including those without supporting reads, were experimentally verified to be correct with polymerase chain reactions (see Table S3C). In addition, for species with multiple-segment cGOF, we can connect neighboring segments according to the rules of scaffolding (see Fig. S4C) as we achieved one scaffold in 32% of 260 simulated assemblies of H. pylori genomes (a six-segment cGOF). However, the practicality of this method in species with too many segments, such as Yersinia pestis, remains to be challenged.
Although the next-generation sequencing (NGS) platforms provide sequencing reads with high coverage (such as data from Illumina’s Hiseq 2500), long repetitive sequences (such as the ribosome RNA gene clusters) are still problematic in sequence assembly as they tend to break assemblies into multiple scaffolds. The cGOF-assisted assembly certainly provides a strategy to overcome this problem by predicting permutations of scaffolds with maximal probabilities based on pangenomic data. Although Pacific Biosciences’ SMRT system provides enough read contiguity to overcome the problem of long repeats (34), the high cost and low throughput still limit its application, especially for a large amount of samples. Our cGOF-based method provides an efficient and economical solution when pangenomic data are available.
Conclusions.
A cGOF, a stable set of core genes in linear orders and representing the majority of core genes, provides species-specific information on genome organization. The two basic cGOF types, according to the symmetry of syntenic gene blocks distributed around the origin-terminus axis, symmetric and asymmetric cGOFs, appear to have been adopted by Gram-positive and Gram-negative bacteria, respectively. Species with symmetric cGOFs exhibit strong strand-biased gene distribution that works against asymmetric rearrangement, and its relationship with specific DnaE isoforms implies that the DNA replication and repair apparatus may control the choice of cGOF types. Interestingly, symmetric cGOFs are rearranged reversibly while maintaining strand-specific orientation, whereas asymmetric GOFs are irreversible, serving as benchmarks for genome diversity and evolution studies. Functional analyses demonstrate that cGOF genes are hub related in gene-gene interaction networks and functionally unique. Moreover, cGOF construction provides a useful tool for guiding scaffold orientation in genome assembly. Therefore, cGOF is a basic structural organization of prokaryotic genes and essential for pangenomic analysis as well as the understanding of structure-function relationship among prokaryotic genomes.
MATERIALS AND METHODS
Data collection.
We used two data sets for this analysis: one contains 425 complete genome sequences from 30 species for cGOF identification, and the other includes sequences of 91 complete genomes (including the species Escherichia coli, Staphylococcus aureus, Streptococcus pyogenes, and Helicobacter pylori, as listed in Table S3A in the supplemental material) and 10 drafts of wild E. coli isolates from various vertebrates for testing the cGOF-based assembling. All complete genomes were downloaded from the public databases (ftp://ftp.ncbi.nih.gov) on 14 April 2013, and all drafts were generated with average depth of 200× in our own laboratory (Hiseq 2000; Illumina, Inc., San Diego, CA) and de novo assembled by using SOAPdenovo into 30 to 80 scaffolds per draft.
We limited pangenome definition to 10 to 29 strains for each species, and classified a pangenome into core and dispensable genes using the Pan-Genome Analysis Pipeline (PGAP; http://pgap.sf.net/) (35). Among the 30 species investigated, there are nine species whose core gene numbers do not clearly converge, implying a higher possibility of dispensable genes to be falsely identified as core genes. The inaccuracy of core genes may impair the consequent cGOF identification, whereas in the process of identifying cGOF, we apply an iteration algorithm and segment size cutoff (described below), which filter out most of the mobile genes (always dispensable genes as well), and thus moderate the problem. To evaluate the effects of core genome misconvergence on cGOF identification, we used a subset of Salmonella enterica strains (with asymmetric cGOF) to define its core gene and cGOF and found that if only all cGOF configurations were represented in the sub-pangenome, the cGOF structure (segmentation and rearrangement) is almost the same as that deduced from the whole pangenome, except for including a few more cGOF genes. The situation in Listeria monocytogenes (with symmetric cGOF) is almost the same. This means that the cGOF structure depends on the convergence of the cGOF configuration but not that of the core gene. Therefore, pangenome size and core genome convergence are not critical problems for cGOF identification, and we retained the nine species without a converged core gene number in the following analysis.
cGOF identification.
For each species, we ordered the single-copy core genes according to their original positions in each genome. We also developed an iteration algorithm (described below) to obtain the longest common subsequence of the single-copy core genes shared by all strains (i.e., the maximal set of order-conserved genes). These genes and their order formed a cGOF of this species. When a cGOF was interrupted (i.e., when rearrangement breaks the contiguity of the framework), we identified all recombination sites and divided the putative cGOF into segments. Short segments containing less than the cutoff of the gene number were removed from the cGOF assembly, and the flanking segments were joined if their orientation was maintained. After optimizing cutoffs ranging from 10 to 60 genes, we found that a cutoff of 20 genes gave the best result—it not only effectively eradicated HGT fragments but also left enough cGOF segments to cover the chromosomes for all species investigated.
Iteration algorithm.
For a set of gene permutations of 1, 2, 3 … n (allowing missing some numbers), we need to find the longest common subsequences of them. First, we construct a directed graph as follows. Vertices are numbers from 0 to n. There exists a directed edge from vertex i to j, if and only if in all sequences i appears prior to j. If there exist edges i→j, j→k … l→m, then path i→j→k→ … →l→m corresponds to a common subsequence (i, j, k … l, m). Thus, the longest common subsequence corresponds to the longest path in this graph. Since all of the genes we used are single-copy genes and all of the sequences start with the first gene after oriC (usually dnaA), there is no loop in our sequences. Next, we use an iteration algorithm to achieve the longest common path. To a vertex i, define F(i) to be the vertex next to i in the longest path from i to n and G(i) to be the length of this path. Since there exists edge i→n, F can be defined for all vertices excluding n. Define G(n) = 0. The longest path (where Fi means the ith iteration of F) is 0→F(0)→F2(0)→F3(0)→ … →Fk − 1 (0)→Fk(0) = n. The iteration equations are shown as follows and were carried out with the programming language perl: (i) F(i) = j, where j ∈(k | there exists edge i→k) and maximizes G(k) and (ii) G(i) = G[F(i)] + 1.
SUPPLEMENTAL MATERIAL
The supplemental methods file describes the methods used in simulation of the equal-movement-rate E. coli pangenome, comparison of cGOF and non-cGOF genes in their connectivity and essentiality, generation of simulated drafts and assembly, and assembly of 10 empirical E. coli drafts. Download
cGOF maps of representative species. (A) Four examples of single-segment cGOFs come from Firmicutes, Actinobacteria, Proteobacteria, and Spirochaetes. (B) Two examples of symmetric cGOFs come from Actinobacteria and Firmicutes. (C) Helicobactor pylori (which belongs to Betaproteobacteria) as an example of the asymmetric cGOF. In each species, cGOFs are presented as circles or multiple segments with fine lines indicating cGOF genes, and distances between neighboring lines are scaled to the average distance between corresponding cGOF genes in all sample strains. In multisegment cGOFs (B and C), sketch maps present all observed arrangements of segments. In symmetric cGOFs (B), O and T represent segments with origin of replication and terminus sites, respectively; L and R are the left and right segments. In asymmetric cGOFs (C), all segments are sequentially numbered, except for the O segment. Download
Strains used in E. coli pangenome analysis. The phylogenetic tree includes the 53 strains with completed genomes. The 19 strains used for pangenome analysis are labeled with red rectangles, and the others were used for testing of cGOF-assisted assembly. Download
Distribution of the unmoved operon counts for the simulated and actual pangenome sets. The five curves from left to right are calculated with 100%, 85%, 70%, 55%, and 40% actual movement rate of the simulated 19-genome pangenome (500 simulations for each movement rate). The cGOF operon count in the actual pangenome is indicated with a vertical blue line at the position of the 876 operon count. Download
Codon usage bias in cGOF, non-cGOF, shared, and unique gene sets of E. coli. CDC represents the codon deviation coefficient, a measurement of codon usage bias as previously described (32). Download
cGOF-directed genome assembly. (A) Flowchart showing how to generate virtual drafts from completed genomes. The inset shows the distribution of contig sizes in drafts after de novo assembly. (B) Accuracy of cGOF- or reference-based finishing strategy. A box plot compares the total contig lengths that are misassembled based on either cGOF- or reference-based strategies (E. coli, 340 simulations). (C) Segment coordination for species of segmented cGOF. The upper panel shows the schematic diagram illustrating how to join multiple small segments into a limited number of supersegments according to sequence scaffolding principles. Red vertical bars, anchorage sequences; blue horizontal bar, anchorage segment; and green horizontal bars, joined segments. The lower panel is a bar diagram of the distribution of supersegment counts (H. pylori, 260 simulations). Download
Species distribution and characteristics in segmented and nonsegmented cGOF groups.
Comparison of cGOF and non-cGOF genes in functions.
cGOF-directed genome assembly.
ACKNOWLEDGMENTS
This study was supported by grants from the Ministry of Health of PR. China (China Mega-Project on Infectious Disease Prevention no. 2013ZX10004-605): NSFC 11271029, NSFC 31471248, and NSFC 11171024. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Yu Kang had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The study was conceived and designed by Y.K., Z.Z., and J.Y. Data acquisition was performed by Y.K., L.Y., C.G., Y.Z., CL, and A.A.K. Analysis and interpretation of data were performed by Y.K., L.Y., Z.Z., and J.Y. The manuscript was drafted by Y.K., Z.Z., and J.Y. Critical revision of the manuscript for important intellectual content was performed by J.X., F.C., and D.J. Statistical analysis was performed by Y.K. and C.G. Funding was obtained by J.Y. Administrative, technical, and material support was provided by Y.W., X.L., Q.L., and M.Q. Supervision was provided by J.Y.
Footnotes
Citation Kang Y, Gu C, Yuan L, Wang Y, Zhu Y, Li X, Luo Q, Xiao J, Jiang D, Qian M, Ahmed Khan A, Chen F, Zhang Z, Yu J. 2014. Flexibility and symmetry of prokaryotic genome rearrangement reveal lineage-associated core-gene-defined genome organizational frameworks. mBio 5(6):e01867-14. doi:10.1128/mBio.01867-14.
REFERENCES
- 1. Sobetzko P, Travers A, Muskhelishvili G. 2012. Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. U. S. A. 109:E42–E50. 10.1073/pnas.1108229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. 2010. Spatial organization of the flow of genetic information in bacteria. Nature 466:77–81. 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mira A, Martín-Cuadrado AB, D’Auria G, Rodríguez-Valera F. 2010. The bacterial pan-genome: a new paradigm in microbiology. Int. Microbiol. 13:45–57. [DOI] [PubMed] [Google Scholar]
- 4. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955. 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 6. Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36:760–766. 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 7. Kuhlman TE, Cox EC. 2012. Gene location and DNA density determine transcription factor distributions in Escherichia coli. Mol. Syst. Biol. 8:610. 10.1038/msb.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318:1449–1452. 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 9. Pál C, Papp B, Lercher MJ. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372–1375. 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 10. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreimer A, Borenstein E, Gophna U, Ruppin E. 2008. The evolution of modularity in bacterial metabolic networks. Proc. Natl. Acad. Sci. U. S. A. 105:6976–6981. 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campo N, Dias MJ, Daveran-Mingot ML, Ritzenthaler P, Le Bourgeois P. 2004. Chromosomal constraints in Gram-positive bacteria revealed by artificial inversions. Mol. Microbiol. 51:511–522. 10.1046/j.1365-2958.2003.03847.x. [DOI] [PubMed] [Google Scholar]
- 13. Esnault E, Valens M, Espéli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 3:e226. 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darmon E, Leach DR. 2014. Bacterial genome instability. Microbiol. Mol. Biol. Rev. 78:1–39. 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darling AE, Miklós I, Ragan MA. 2008. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 4:e1000128. 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui L, Neoh HM, Iwamoto A, Hiramatsu K. 2012. Coordinated phenotype switching with large-scale chromosome flip-flop inversion observed in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:E1647–E1656. 10.1073/pnas.1204307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eisen JA, Heidelberg JF, White O, Salzberg SL. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 1:Research0011. 10.1186/gb-2000-1-6-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol. Evol. 5:783–793. 10.1093/gbe/evt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andersen MT, Liefting LW, Havukkala I, Beever RE. 2013. Comparison of the complete genome sequence of two closely related isolates of “Candidatus Phytoplasma australiense” reveals genome plasticity. BMC Genomics 14:529. 10.1186/1471-2164-14-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shifman A, Ninyo N, Gophna U, Snir S. 2014. Phylo SI: a new genome-wide approach for prokaryotic phylogeny. Nucleic Acids Res. 42:2391–2404. 10.1093/nar/gkt1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brilli M, Liò P, Lacroix V, Sagot MF. 2013. Short and long-term genome stability analysis of prokaryotic genomes. BMC Genomics 14:309. 10.1186/1471-2164-14-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rocha E. 2002. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 10:393–395. 10.1016/S0966-842X(02)02420-4. [DOI] [PubMed] [Google Scholar]
- 23. Zhao XQ, Hu JF, Yu J. 2006. Comparative analysis of eubacterial DNA polymerase III alpha subunits. Genomics Proteomics Bioinformatics 4:203–211. 10.1016/S1672-0229(07)60001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu H, Zhang Z, Hu S, Yu J. 2012. On the molecular mechanism of GC content variation among eubacterial genomes. Biol. Direct 7:2. 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rocha EP. 2004. The replication-related organization of bacterial genomes. Microbiology 150:1609–1627. 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 26. Rocha EP, Danchin A. 2003. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 31:6570–6577. 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riley MA, Lizotte-Waniewski M. 2009. Population genomics and the bacterial species concept. Methods Mol. Biol. 532:367–377. 10.1007/978-1-60327-853-9_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feil EJ. 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2:483–495. 10.1038/nrmicro904. [DOI] [PubMed] [Google Scholar]
- 29. Ferré-D’Amaré AR. 2011. Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:2942–2948. 10.1098/rstb.2011.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2:a003483. 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karberg KA, Olsen GJ, Davis JJ. 2011. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc. Natl. Acad. Sci. U. S. A. 108:20154–20159. 10.1073/pnas.1109451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Z, Li J, Cui P, Ding F, Li A, Townsend JP, Yu J. 2012. Codon deviation coefficient: a novel measure for estimating codon usage bias and its statistical significance. BMC Bioinformatics 13:43. 10.1186/1471-2105-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pál C, Papp B, Lercher MJ. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372–1375. 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 34. Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10:563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 35. Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. 10.1093/bioinformatics/bts416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplemental methods file describes the methods used in simulation of the equal-movement-rate E. coli pangenome, comparison of cGOF and non-cGOF genes in their connectivity and essentiality, generation of simulated drafts and assembly, and assembly of 10 empirical E. coli drafts. Download
cGOF maps of representative species. (A) Four examples of single-segment cGOFs come from Firmicutes, Actinobacteria, Proteobacteria, and Spirochaetes. (B) Two examples of symmetric cGOFs come from Actinobacteria and Firmicutes. (C) Helicobactor pylori (which belongs to Betaproteobacteria) as an example of the asymmetric cGOF. In each species, cGOFs are presented as circles or multiple segments with fine lines indicating cGOF genes, and distances between neighboring lines are scaled to the average distance between corresponding cGOF genes in all sample strains. In multisegment cGOFs (B and C), sketch maps present all observed arrangements of segments. In symmetric cGOFs (B), O and T represent segments with origin of replication and terminus sites, respectively; L and R are the left and right segments. In asymmetric cGOFs (C), all segments are sequentially numbered, except for the O segment. Download
Strains used in E. coli pangenome analysis. The phylogenetic tree includes the 53 strains with completed genomes. The 19 strains used for pangenome analysis are labeled with red rectangles, and the others were used for testing of cGOF-assisted assembly. Download
Distribution of the unmoved operon counts for the simulated and actual pangenome sets. The five curves from left to right are calculated with 100%, 85%, 70%, 55%, and 40% actual movement rate of the simulated 19-genome pangenome (500 simulations for each movement rate). The cGOF operon count in the actual pangenome is indicated with a vertical blue line at the position of the 876 operon count. Download
Codon usage bias in cGOF, non-cGOF, shared, and unique gene sets of E. coli. CDC represents the codon deviation coefficient, a measurement of codon usage bias as previously described (32). Download
cGOF-directed genome assembly. (A) Flowchart showing how to generate virtual drafts from completed genomes. The inset shows the distribution of contig sizes in drafts after de novo assembly. (B) Accuracy of cGOF- or reference-based finishing strategy. A box plot compares the total contig lengths that are misassembled based on either cGOF- or reference-based strategies (E. coli, 340 simulations). (C) Segment coordination for species of segmented cGOF. The upper panel shows the schematic diagram illustrating how to join multiple small segments into a limited number of supersegments according to sequence scaffolding principles. Red vertical bars, anchorage sequences; blue horizontal bar, anchorage segment; and green horizontal bars, joined segments. The lower panel is a bar diagram of the distribution of supersegment counts (H. pylori, 260 simulations). Download
Species distribution and characteristics in segmented and nonsegmented cGOF groups.
Comparison of cGOF and non-cGOF genes in functions.
cGOF-directed genome assembly.