ABSTRACT
How the obligatory intracellular bacterium Ehrlichia chaffeensis begins to replicate upon entry into human monocytes is poorly understood. Here, we examined the potential role of amino acids in initiating intracellular replication. PutA converts proline to glutamate, and GlnA converts glutamate to glutamine. E. chaffeensis PutA and GlnA complemented Escherichia coli putA and glnA mutants. Methionine sulfoximine, a glutamine synthetase inhibitor, inhibited E. chaffeensis GlnA activity and E. chaffeensis infection of human cells. Incubation of E. chaffeensis with human cells rapidly induced putA and glnA expression that peaked at 24 h postincubation. E. chaffeensis took up proline and glutamine but not glutamate. Pretreatment of E. chaffeensis with a proline transporter inhibitor (protamine), a glutamine transporter inhibitor (histidine), or proline analogs inhibited E. chaffeensis infection, whereas pretreatment with proline or glutamine enhanced infection and upregulated putA and glnA faster than no treatment or glutamate pretreatment. The temporal response of putA and glnA expression was similar to that of NtrY and NtrX, a two-component system, and electrophoretic mobility shift assays showed specific binding of recombinant E. chaffeensis NtrX (rNtrX) to the promoter regions of E. chaffeensis putA and glnA. Furthermore, rNtrX transactivated E. chaffeensis putA and glnA promoter-lacZ fusions in E. coli. Growth-promoting activities of proline and glutamine were also accompanied by rapid degradation of the DNA-binding protein CtrA. Our results suggest that proline and glutamine uptake regulates putA and glnA expression through NtrY/NtrX and facilitates degradation of CtrA to initiate a new cycle of E. chaffeensis growth.
IMPORTANCE
Human monocytic ehrlichiosis (HME) is one of the most prevalent, life-threatening emerging infectious zoonoses in the United States. HME is caused by infection with E. chaffeensis, an obligatory intracellular bacterium in the order Rickettsiales, which includes several category B/C pathogens, such as those causing Rocky Mountain spotted fever and epidemic typhus. The limited understanding of the mechanisms that control bacterial growth within eukaryotic cells continues to impede the identification of new therapeutic targets against rickettsial diseases. Extracellular rickettsia cannot replicate, but rickettsial replication ensues upon entry into eukaryotic host cells. Our findings will provide insights into a novel mechanism of the two-component system that regulates E. chaffeensis growth initiation in human monocytes. The result is also important because little is known about the NtrY/NtrX two-component system in any bacteria, let alone obligatory intracellular bacteria. Our findings will advance the field’s current conceptual paradigm on regulation of obligatory intracellular nutrition, metabolism, and growth.
INTRODUCTION
Ehrlichia chaffeensis, an obligatory intracellular bacterium, replicates within human white blood cells and causes human monocytic ehrlichiosis (HME). Discovered in 1986 and designated a nationally notifiable disease in 1998 by the Centers for Disease Control and Prevention, HME is one of the most prevalent, life-threatening emerging tick-borne zoonoses in the United States (1, 2). The symptoms of HME include severe flu-like fever and lethargy accompanied by hematologic abnormality and hepatitis. A broad-spectrum antibiotic, doxycycline, remains the drug of choice for treating patients. However, delayed initiation of therapy, the presence of underlying illness, or immunosuppression often leads to severe complications or death (3).
E. chaffeensis belongs to the order Rickettsiales, which includes Rickettsia prowazekii, a category B select agent, in the class Alphaproteobacteria. Uncovering a mechanism to limit pathogen nutrition and growth in animals has been a key research target to improve therapy against infectious diseases (4). E. chaffeensis has a small genome of 1.18 Mb, with a limited number of genes for biosynthesis and metabolism, which obliges E. chaffeensis to acquire host nutrients for growth (5). E. chaffeensis has a tricarboxylic acid (TCA) cycle and electron transport chain; however, it cannot synthesize most amino acids and cannot obtain carbon or energy from fatty acids or carry out glycolysis (5). Consequently, it is likely that E. chaffeensis takes up and uses amino acids as carbon, nitrogen, and energy sources. However, amino acid uptake or utilization by E. chaffeensis has not been studied.
Biosynthesis of nitrogenous compounds such as DNA, RNA, and proteins is dependent on maintaining intracellular pools of glutamate and glutamine (6). In Escherichia coli, these 2 amino acids are synthesized by a bifunctional glutamate synthetase proline utilization A (PutA), which converts proline to glutamate with its dual enzyme activities: proline dehydrogenase (PRODH) and proline/pyrroline-5-carboxylate dehydrogenase (P5CDH) (7), GlnA (glutamine synthetase [GS]) that converts glutamate to glutamine (8), and glutamate synthetase (glutamine-2-oxoglutarate-amidotransferase [GOGAT]) that produces glutamate from glutamine and α-ketoglutarate (9).
Our bioinformatic analysis indicates that the E. chaffeensis genome encodes PutA (ECH_0667) and GlnA (ECH_0089) (Table 1 and Fig. 1). E. chaffeensis can also convert glutamate to proline by a two-step reaction involving l-glutamate γ-semialdehyde dehydrogenase activity of PutA and pyrroline-5-carboxylate reductase (ECH_0013). However, E. chaffeensis lacks GOGAT for glutamate biosynthesis (Table 1) (5). In addition, members in the family Anaplasmataceae encode both carbamoyl phosphate synthase (ECH_0503/ECH_0378) and bifunctional glutamate synthase subunit beta/2-polyprenylphenol hydroxylase (ECH_0778), both of which can convert glutamine to ammonia and glutamate (Table 1). Glutamate can be further converted by glutamate dehydrogenase (ECH_0771) to 2-ketoglutarate, which enters the TCA cycle for energy production (Table 1).
TABLE 1 .
Distribution of genes involved in glutamate/glutamine biosynthesis/degradation in Ehrlichia chaffeensis and Chlamydia trachomatis
| Species | Presence of gene encodinga: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| NtrY/X | PutA | P5CR | GlnA | GlnA-C | CarA/B | GS/PH | GOGAT | GDH | |
|
Ehrlichia
chaffeensis Arkansas |
+ (ECH_0299/ ECH_0339) |
+ (ECH_0667) |
+ (ECH_0013) |
+ (ECH_0089) |
+ (ECH_0983) |
+ (ECH_0503/ ECH_0378) |
+ (ECH_0778) |
− | + (ECH_0771) |
|
Chlamydia
trachomatis IU824 |
− | − | − | − | − | − | − | − | − |
PutA, bifunctional proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase; P5CR, pyrroline-5-carboxylate reductase; GlnA, glutamine synthetase class I; GlnA-C, GlnA with a partial catalytic domain (Gln-Synt_C) (~270 aa); CarA/B, carbamoyl phosphate synthase small/large subunits; GS/PH, bifunctional glutamate synthase subunit beta/2-polyprenylphenol hydroxylase; GOGAT, glutamine-α-oxoglutarate aminotransferase (glutamate synthase); GDH, NAD-dependent glutamate dehydrogenase. +, present; −, absent. GenBank accession numbers: E. chaffeensis Arkansas, NC_007799.1; C. trachomatis IU824, NC_020511.1.
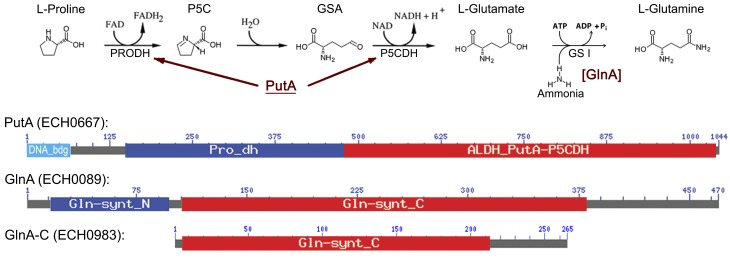
FIG 1 .
Diagram of the glutamine biosynthesis pathway from proline and domain structures of related enzymes. E. chaffeensis encodes bifunctional proline dehydrogenase (PRODH; Pro_dh)/pyrroline-5-carboxylate dehydrogenase (P5CDH; ALDH_PutA-P5CDH) (116.3 kDa, GenBank no. YP_507475) (PutA), which has an N-terminal DNA-binding domain, and class I glutamine synthetase (GSI) GlnA (53.4 kDa, GenBank no. YP_506919). E. chaffeensis also encodes a glutamine synthetase domain-containing protein (ECH_0983; 30.1 kDa, GenBank no. YP_507770), which lacks the beta-Grasp domain (Gln-synt_N) found in GlnA and contains a partial glutamine synthetase catalytic domain (Gln-synt_C). P5C, pyrroline-5-carboxylate; GSA, glutamate semialdehyde; DNA_bdg, DNA-binding domain of proline dehydrogenase. Protein lengths of PutA and GlnA are not drawn to scale.
E. coli PutA is autoregulated through its DNA-binding domain, which functions as a transcriptional repressor (10). Expression of GS and GOGAT in E. coli is regulated by the two-component system NtrB/NtrC (11). NtrB/NtrC is not found in E. chaffeensis; however, NtrY/NtrX, the homologous pair to E. coli NtrB/NtrC, is identified in E. chaffeensis (12). Although NtrX/NtrY is present in most alphaproteobacteria, NtrY/NtrX function has been inferred in only a few species (13–16). In fact, E. chaffeensis NtrY/NtrX is the first and only pair for which biochemical evidence of NtrY autokinase activity and specific amino acid-dependent phosphotransfer from NtrY to NtrX has been documented (12, 17).
E. chaffeensis, a blood-borne bacterium, cannot replicate in the plasma or cell culture medium, but once intracellular it begins to replicate. Among all amino acids, glutamine, glutamate, and proline concentrations in the cells are particularly higher than in the plasma (18). Here, we investigated the functions of PutA and GlnA, the roles of their substrate and product amino acids (proline, glutamate, and glutamine), and the role of NtrX in the regulation of glnA and putA expression in E. chaffeensis. We also examined effects of these amino acids on another E. chaffeensis response regulator, CtrA (19), which is known to regulate chromosome replication in Caulobacter crescentus (20). The present study provides new information about the roles of PutA and GlnA and the amino acids critical for regulating replication of obligatory intracellular bacteria through the two-component system, which may assist in the discovery of the next generation of chemotherapeutic approaches for HME.
RESULTS
E. chaffeensis has functional PutA and GlnA enzymes.
PutA is conserved in many Gram-negative bacteria, including E. coli, Salmonella enterica serovar Typhimurium, Rhodobacter capsulatus, Agrobacterium tumefaciens, and Sinorhizobium meliloti (7, 21–24). Regulation mechanisms of putA expression are divergent among bacteria (10, 22–25). In the absence of proline, E. coli PutA remains in the cytoplasm and PutA binds to the putA regulatory region to repress putA transcription; in the presence of proline, E. coli PutA associates with the membrane to convert proline to glutamate (26). E. chaffeensis PutA (GenBank no. YP_507475, 116.3 kDa) and E. coli PutA (GenBank no. AAB59985, 143.8 kDa) share 47.8% amino acid identity. Residues 1 to 60 of E. chaffeensis PutA are predicted to serve as the DNA-binding domain (DNAbinder, http://www.imtech.res.in/raghava/dnabinder); however, this domain has only 11.5% identity with the E. coli PutA DNA-binding domain (residues 1 to 52) (Fig. 1).
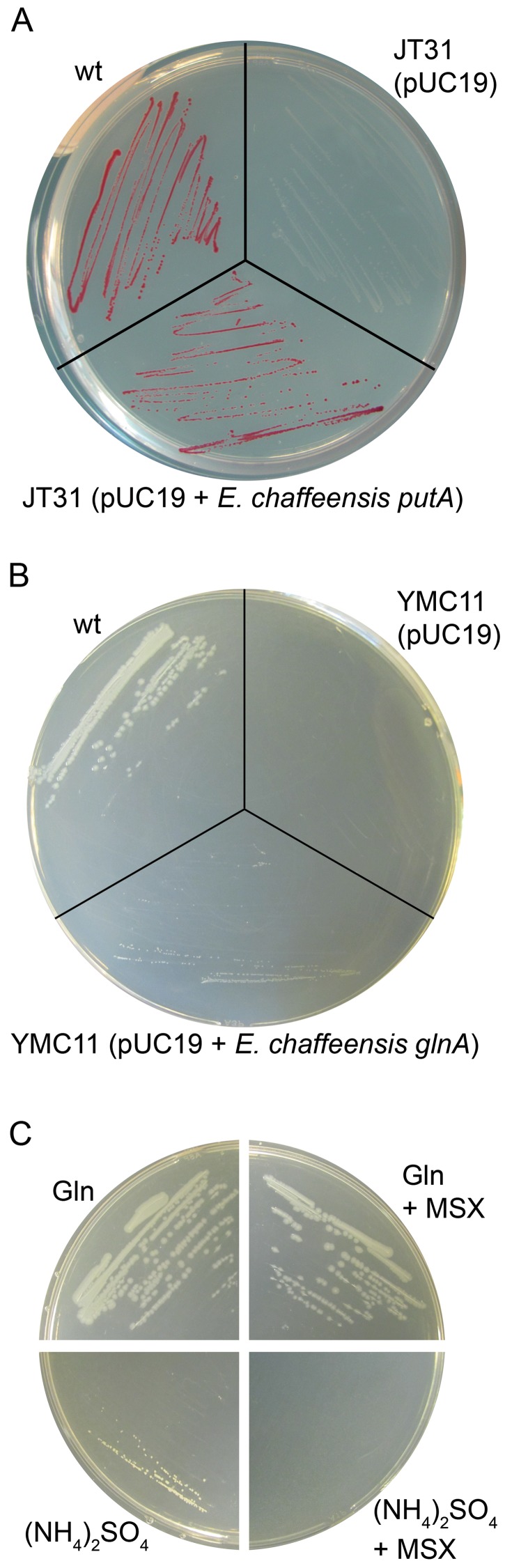
The E. coli putA mutant JT31, which cannot oxidize proline by reducing flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide (NAD), forms white colonies on plates containing triphenyltetrazolium chloride (TTC; redox indicator) and proline, whereas the wild-type (wt) strain CSH4 forms red colonies (27). We examined whether E. chaffeensis PutA complements JT31. After growth on TTC-proline indicator plates at 37°C for 24 h, E. coli JT31 transformed with plasmid expressing E. chaffeensis putA under control of the E. coli putA promoter formed red colonies, but JT31 transformed with pUC19 (empty vector, negative control) formed white colonies (Fig. 2A), demonstrating that E. chaffeensis PutA has flavoenzyme activity and complements E. coli mutants to utilize proline.
FIG 2 .
E. chaffeensis PutA and GlnA are functional enzymes, and E. chaffeensis GlnA is sensitive to MSX. (A) E. chaffeensis PutA complements an E. coli putA mutant (JT31). wt (CSH4) and JT31 transformed with the E. chaffeensis putA construct or pUC19 (negative control) were cultured on redox indicator TTC-proline plates. (B) E. chaffeensis GlnA complements an E. coli glnA mutant (YMC11). wt (XL1-Blue) and YMC11 transformed with the E. chaffeensis glnA construct or pUC19 (negative control) were cultured on M9 plates containing 20 mM (NH4)2SO4 as the sole nitrogen source. (C) MSX inhibits E. chaffeensis GlnA activity. YMC11 transformed with the E. chaffeensis glnA construct was cultured on M9 plates containing 2 mM glutamine (Gln) or 20 mM (NH4)2SO4 with or without 200 µM MSX.
GS, an essential enzyme in ammonia assimilation and glutamine biosynthesis from glutamate, has four distinctive types: GSI (GlnA), GSII, GlnT, and GSIII (28). Genes for GSI have been found only in bacteria (eubacteria) and archaea (archaebacteria), whereas GSII genes occur only in eukaryotes and a few soil-dwelling bacteria. GlnT is found in Rhizobium and Agrobacterium spp., and GSIII genes have been found only in a few bacterial species (29). E. chaffeensis glnA encodes GlnA (GenBank no. YP_506919, 53.4 kDa). E. chaffeensis GlnA has an N-terminal β-Grasp domain and a C-terminal GS catalytic domain, which condenses ammonia and glutamate (Fig. 1). Amino acid identity of E. chaffeensis GlnA and E. coli GlnA (GenBank no. YP_003056319, 51.9 kDa) is 53.9%. In addition, E. chaffeensis encodes a truncated GlnA (GlnA-C) (ECH_0983), but it unlikely has GS activity because of several internal deletions in the GS catalytic domain and the absence of an N-terminal β-Grasp domain (30, 31) (Fig. 1).
To determine the function of E. chaffeensis GlnA, E. coli glnA mutant complementation was performed. The E. coli glnA mutant YMC11 lacks GlnA and cannot grow on M9 minimal, defined growth plates containing NH4+ as the sole nitrogen source (32). After being cultured at 37°C for 48 h, the E. coli wt strain XL1-Blue and YMC11 transformed with the plasmid expressing E. chaffeensis glnA under control of the E. coli glnA promoter showed colonies on M9 plates containing (NH4)2SO4 as the sole nitrogen source but not YMC11 transformed with pUC19 (empty vector, negative control) (Fig. 2B), indicating that E. chaffeensis GlnA assimilates NH4+ and generates glutamine for YMC11 growth.
l-Methionine-SR-sulfoximine (MSX) is a membrane-permeable GS inhibitor (33); it inhibits the enzyme by serving as an analog of the tetrahedral intermediate or transient state formed by reaction of ammonia with γ-glutamyl phosphate (34). When MSX was added to M9 plates containing (NH4)2SO4, YMC11 transformed with the E. chaffeensis glnA construct did not grow, but the addition of 2 mM glutamine instead of (NH4)2SO4 in M9 plates abrogated the MSX-mediated inhibition in transformed YMC11 (Fig. 2C). These results demonstrated that the GS activity of E. chaffeensis GlnA is inhibited by MSX and that exogenous glutamine reverses MSX-mediated inhibition of bacterial growth.
Inhibition of glutamine synthesis prevents E. chaffeensis growth.
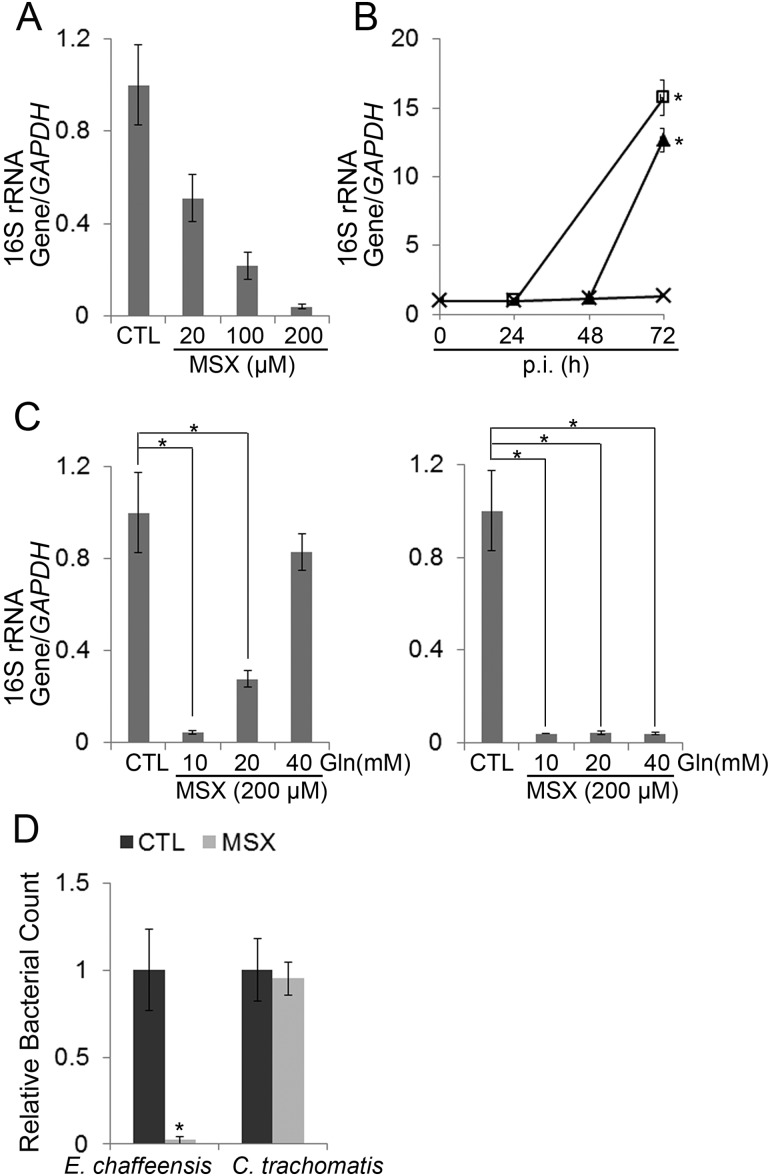
Because GS activity of E. chaffeensis GlnA expressed in E. coli was inhibited by MSX, we examined whether this inhibition affects E. chaffeensis growth. At 20 to 200 µM, MSX does not inhibit proliferation of mammalian cells (35). Pretreatment of the THP-1 human monocytic cell line with MSX (200 µM) for 30 min and removal of MSX immediately prior to the addition of E. chaffeensis had no effects on subsequent E. chaffeensis infection as assessed by quantitative PCR (qPCR) (data not shown). When added at 0 h, 20 to 200 µM MSX inhibited E. chaffeensis growth in THP-1 cells in a dose-dependent manner (Fig. 3A). Withdrawal of MSX at 24 or 48 h postinfection (p.i.) allowed growth of E. chaffeensis, indicating that MSX does not kill E. chaffeensis, even after 48 h of treatment, but rather suppresses its growth (Fig. 3B).
FIG 3 .
Inhibition of glutamine synthesis prevents E. chaffeensis growth. (A) MSX inhibits E. chaffeensis growth in THP-1 cells in a dose-dependent manner. DNA samples prepared from synchronously cultured E. chaffeensis in THP-1 cells treated at 0 h p.i. with 20, 100, or 200 µM MSX for 72 h were subjected to qPCR analysis. The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. (B) Inhibitory effect of MSX is reversible. MSX (200 µM) was added to E. chaffeensis-infected THP-1 cells at 0 h p.i. and removed after 24 or 48 h p.i. DNA samples were subjected to qPCR analysis. The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined at 0 h p.i. Symbols: ×, MSX was not removed; □, MSX was removed at 24 h p.i.; ▲, MSX was removed at 48 h p.i. Data indicate the means ± standard deviations from three independent experiments performed in triplicate. *, significantly different (P < 0.05; analysis of variance) compared with the value of the sample with MSX not removed. (C) Glutamine reverses the inhibitory effect of MSX. DNA samples prepared from synchronously cultured E. chaffeensis in THP-1 cells treated at 0 h p.i. with 200 µM MSX and 10, 20, or 40 mM glutamine (left) or glutamate (right) for 72 h were subjected to qPCR analysis. The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. *, significantly different (P < 0.05; analysis of variance) compared with the value of the control. (D) MSX has no effect on C. trachomatis growth in L929 cells. C. trachomatis- or E. chaffeensis-infected L929 cells were treated with 200 µM MSX for 72 h. Bacterial counts were scored from 100 cells and are relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. *, significantly different (P < 0.05; Student’s t test) compared with the value of the control.
Because glutamine supplementation reversed MSX-mediated inhibition of E. chaffeensis GlnA activity in E. coli, we examined whether glutamine supplementation could reverse MSX-mediated inhibition of E. chaffeensis growth. Supplementation of the culture medium with 10 to 40 mM glutamine reversed E. chaffeensis growth inhibition by MSX in a dose-dependent manner (Fig. 3C), indicating that glutamine is indeed the growth factor limited by MSX and that glutamine can be taken up by E. chaffeensis. In contrast, supplementation with 10 to 40 mM glutamate had no effect on the MSX-mediated inhibition (Fig. 3C).
As a control, growth of another obligatory intracellular bacterium, Chlamydia trachomatis, which lacks GlnA and nearly all genes involved in glutamate/glutamine biosynthesis/degradation (Table 1), was examined. Results showed that 200 µM MSX did not inhibit C. trachomatis growth (Fig. 3D), indicating that the inhibitory effect of MSX is specific and is not due to nonspecific toxicity to intracellular bacteria. Taken together, these results indicated that GS activity is important for E. chaffeensis growth and that E. chaffeensis obtains essential glutamine from endogenous glutamine synthesis and through uptake of exogenous glutamine.
Uptake of proline and glutamine is critical for subsequent E. chaffeensis infection.
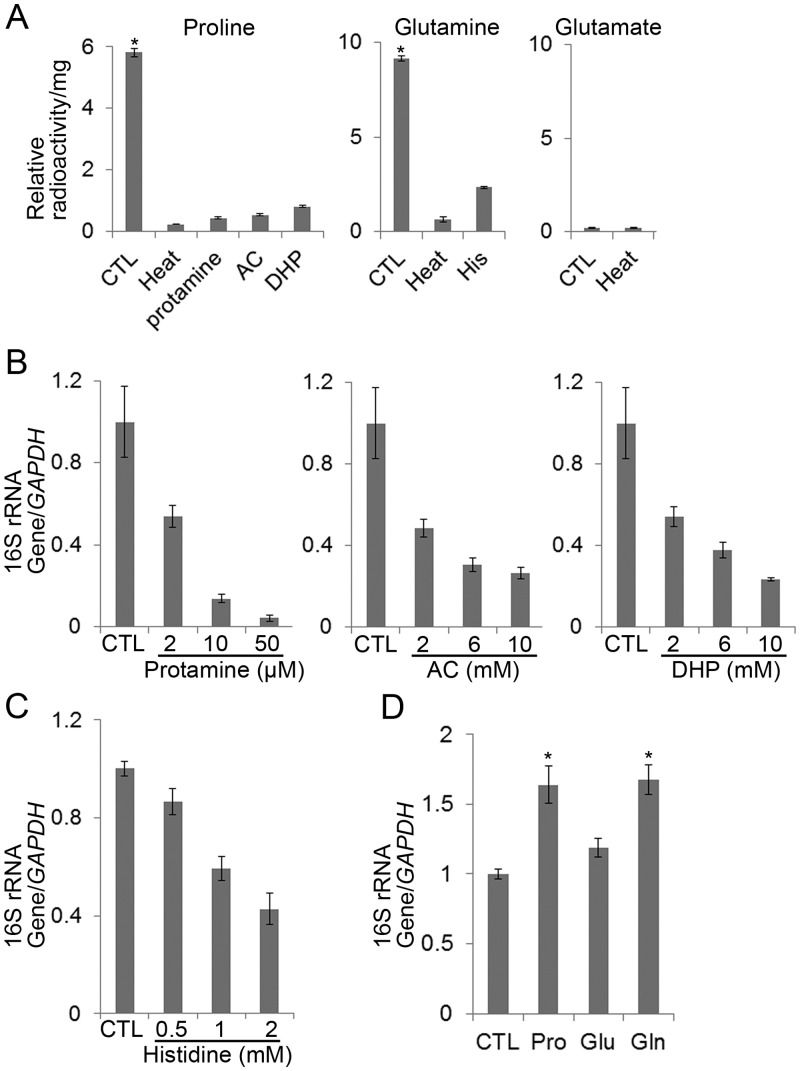
Because E. chaffeensis PutA and GlnA could complement the respective E. coli mutants, and because exogenous glutamine reversed MSX-mediated inhibition of bacterial growth, we examined whether E. chaffeensis can import proline, glutamate, and glutamine. Incubation of host cell-free E. chaffeensis with 3H-labeled proline, glutamate, or glutamine showed that E. chaffeensis can take up proline and glutamine but not glutamate (Fig. 4A). Protamine inhibits proline uptake, causes rapid efflux of proline from preloaded cells, and reduces ATP content in S. Typhimurium (36). The proline analogs, l-azetidine-2-carboxylic acid (AC) and 3,4-dehydro-dl-proline (DHP), are toxic to Staphylococcus saprophyticus, a proline auxotroph (37). Pretreatment of host cell-free E. chaffeensis with protamine, AC, or DHP significantly inhibited E. chaffeensis proline uptake (Fig. 4A) and inhibited subsequent bacterial infection in THP-1 cells in a dose-dependent manner as assessed by qPCR (Fig. 4B), indicating that proline uptake by E. chaffeensis is critical for bacterial infection. Histidine is a competitive inhibitor of glutamine uptake by mitochondria (38). Pretreatment of host cell-free E. chaffeensis with histidine reduced E. chaffeensis glutamine uptake (Fig. 4A) and growth in THP-1 cells (Fig. 4C), indicating that glutamine uptake by E. chaffeensis is critical for bacterial infection.
FIG 4 .
E. chaffeensis can import proline and glutamine, but not glutamate, and requires proline and glutamine uptake for infection. (A) E. chaffeensis was incubated at 65°C for 15 min (Heat) or pretreated with 50 µM protamine, 10 mM AC or DHP, or 2 mM histidine (His) on ice for 30 min and incubated with 3H-labeled proline, glutamate, or glutamine at 37°C for 2 h to determine amino acid uptake. Relative radioactivity per milligram E. chaffeensis protein was measured. Data indicate the means ± standard deviations from three independent experiments. CTL, nontreated control. *, significantly different (P < 0.05; analysis of variance) among all groups. (B) Dose-dependent inhibitory effect of protamine, AC, or DHP on E. chaffeensis growth in THP-1 cells. DNA samples for qPCR were prepared at 72 h p.i. from THP-1 cells infected with host cell-free E. chaffeensis pretreated with protamine (2, 10, or 50 µM), AC (2, 6, or 10 mM), or DHP (2, 6, or 10 mM). The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. (C) Dose-dependent inhibitory effect of histidine on E. chaffeensis growth in THP-1 cells. DNA samples for qPCR were prepared at 72 h p.i. from THP-1 cells infected with host cell-free E. chaffeensis pretreated with histidine (0.5, 1, or 2 mM). The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. (D) Proline and glutamine, but not glutamate, enhance E. chaffeensis infection. DNA samples for qPCR were prepared at 48 h p.i. from THP-1 cells infected with host cell-free E. chaffeensis pretreated with 20 mM proline, glutamate, or glutamine. CTL, control pretreatment; Pro, proline pretreatment; Glu, glutamate pretreatment; Gln, glutamine pretreatment. The values reflect bacterial 16S rRNA gene normalized against human GAPDH DNA, relative to the amount determined for the sham-treated control (CTL). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. *, significantly different (P < 0.05; analysis of variance) compared with the value of the control.
Given that uptake of both proline and glutamine is critical for E. chaffeensis infection, we examined the effects of excess proline and glutamine on E. chaffeensis growth. We found that pretreatment of E. chaffeensis with proline or glutamine enhanced its growth compared with pretreatment with glutamate or no treatment (Fig. 4D).
E. chaffeensis putA and glnA are upregulated at an early stage of growth, which is accelerated by proline and glutamine.
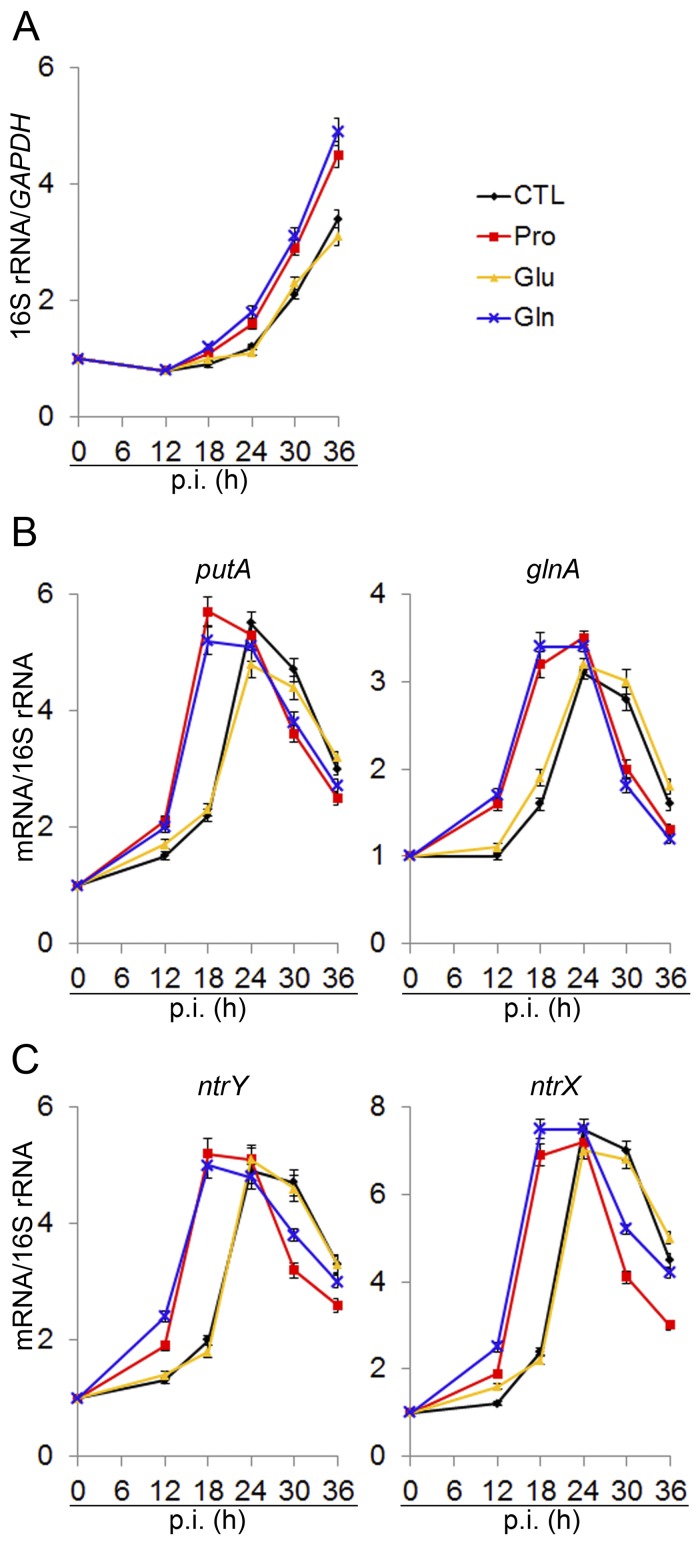
Under our synchronized infection conditions in THP-1 cells, the amount of bacterial 16S rRNA normalized against the amount of host cell GAPDH mRNA indicated that intracellular bacterial growth begins after a lag phase of ~24 h, followed by exponential replication that ends ~72 h p.i. upon the rupture of host cells owing to overwhelming infection (19). As shown in Fig. 5A, pretreatment of E. chaffeensis with proline or glutamine, compared with pretreatment with glutamate or no treatment, shortened the lag phase and enhanced E. chaffeensis growth. Because glutamine and proline uptake is critical for initiating bacterial proliferation (Fig. 4A to D and 5A), we examined when putA and glnA are expressed during the E. chaffeensis intracellular infection cycle. Our results showed that the mRNA levels of glnA and putA normalized by bacterial 16S rRNA, although not encoded in a single operon, were almost synchronously upregulated and peaked at 24 h p.i. (Fig. 5B). Pretreatment of E. chaffeensis with proline or glutamine accelerated the induction of putA and glnA expression (Fig. 5B). In contrast, pretreatment with glutamate had no effect on putA or glnA expression (Fig. 5B). Collectively, these results showed that putA and glnA expression was low at 0 h p.i. but was upregulated at an early phase prior to exponential growth; moreover, induction of putA and glnA was accelerated by pretreatment with proline and glutamine compared with pretreatment with glutamate or no treatment.
FIG 5 .
putA and glnA, and ntrY and ntrX are synchronously upregulated prior to exponential growth, which is accelerated by proline and glutamine. RNA samples for quantitative RT-PCR were prepared at different time points p.i. from THP-1 cells infected with host cell-free E. chaffeensis for E. chaffeensis 16S rRNA normalized against human GAPDH mRNA (A) and for putA and glnA (B) or ntrY and ntrX (C) normalized against bacterial 16S rRNA. Values relative to the amount at 0 h p.i. are shown. Data indicate the means ± standard deviations from three independent experiments performed in triplicate.
E. chaffeensis ntrY and ntrX are upregulated prior to exponential growth, which is accelerated by proline and glutamine.
The two-component system NtrY/NtrX has been suggested to regulate nitrogen assimilation in some nitrogen-fixing bacteria (16, 39). We previously showed histidine autokinase activity of E. chaffeensis NtrY and specific amino acid-dependent phosphotransfer from E. chaffeensis NtrY to E. chaffeensis NtrX and demonstrated that NtrY and NtrX are highly expressed by E. chaffeensis in human leukocytes (12, 17, 40). We found that mRNA levels of ntrY and ntrX, although they are not part of the same operon, were synchronously upregulated and peaked at 24 h p.i., similar to the synchronous expression of putA and glnA (Fig. 5C). Pretreatment of E. chaffeensis with proline or glutamine accelerated the induction of ntrY and ntrX expression (Fig. 5C). The similarity in temporal transcriptional patterns and the effects of proline and glutamine suggested that the expression of glnA and putA might be regulated by NtrY/NtrX in E. chaffeensis.
NtrY/NtrX binds to the promoter regions of glnA and putA and transactivates the expression of these genes.
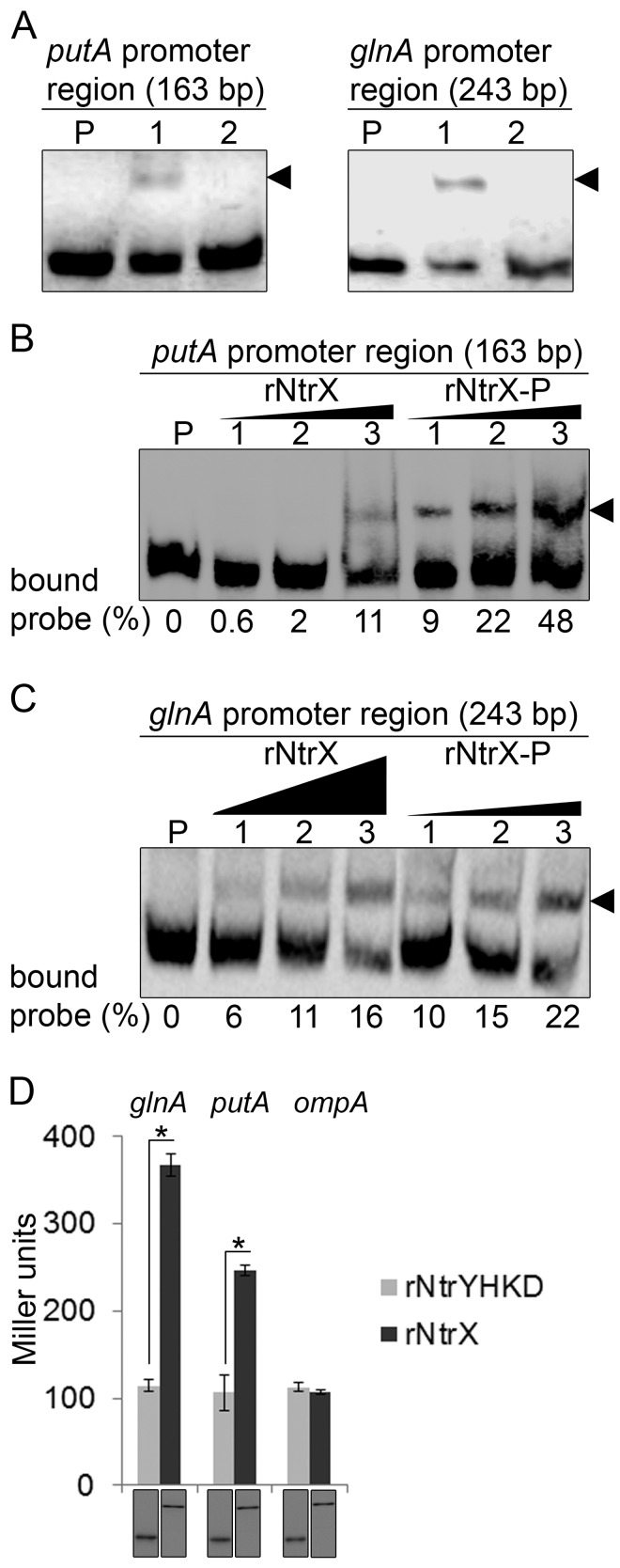
To determine whether NtrX regulates glnA and putA expression, we used the electrophoretic mobility shift assay (EMSA) to examine whether NtrX binds to the promoter regions of glnA and putA. Upon incubation of purified recombinant E. chaffeensis NtrX (rNtrX) with biotinylated probes derived from the promoter regions of glnA and putA, shifted bands were detected (Fig. 6A). The binding specificity was confirmed by the result that no band shift occurred upon addition of a 50-fold excess of the corresponding unlabeled competitor (Fig. 6A). In R. capsulatus, the sensor kinase NtrB phosphorylates NtrC, and the affinity of phosphorylated NtrC for DNA-binding sites is 4-fold greater than that of unphosphorylated NtrC (41). Because there is no information regarding the DNA-binding affinity of phosphorylated NtrX, we examined whether phosphorylation would increase the affinity of rNtrX for the promoter regions of glnA and putA. Acetyl-phosphate is a small-molecule phosphodonor that can phosphorylate response regulators in vivo and in vitro (42). After in vitro phosphorylation of rNtrX with acetyl-phosphate, binding curves were generated based on intensity measurements of bound and unbound bands in each reaction, which allowed comparison of the strength of rNtrX and rNtrX-P to glnA and putA DNA probes. The affinity of rNtrX-P for the promoter regions of glnA and putA was greater than rNtrX by severalfold (Fig. 6B and C).
FIG 6 .
NtrX regulates putA and glnA expression. (A) EMSA for rNtrX binding to the promoter regions of putA (left) and glnA (right). The length (bp) of the probe is shown above each panel. For each panel, biotinylated DNA probe (2 nM) was incubated alone (P), with rNtrX (10 nM, lane 1), or with rNtrX in the presence of 50-fold excess of the corresponding unlabeled DNA competitor (lane 2). Shifted bands are indicated by arrowheads. (B) EMSA for dose-dependent binding of rNtrX and rNtrX-P to the putA promoter. Biotinylated DNA probe (2 nM) was incubated alone (P) or with rNtrX or rNtrX-P at different concentrations (lanes 1 to 3: 2, 4, and 8 nM, respectively). Shifted bands are indicated by an arrowhead. Black triangles show proportions of protein amounts. The numbers below the panel indicate the percentage of bound probe with respect to the total of each lane. (C) EMSA for dose-dependent binding of rNtrX and rNtrX-P to the glnA promoter. Biotinylated DNA probe (2 nM) was incubated alone (P) or with rNtrX at different concentrations (lanes 1 to 3: 6, 20, and 60 nM, respectively) or with rNtrX-P at different concentrations (lanes 1 to 3: 1.5, 5, and 15 nM, respectively). Shifted bands are indicated by an arrowhead. Black triangles show proportions of protein amounts. The numbers below the panel indicate the percentage of bound probe with respect to the total of each lane. (D) NtrX transactivates glnA and putA promoter-lacZ fusions in E. coli. E. coli strains containing pET-33b(+) encoding rNtrX and rNtrYHKD were transformed with the glnA-, putA-, or ompA-lacZ fusions containing the promoter regions of glnA (243 bp), putA (163 bp), or ompA (368 bp), respectively. After induction of rNtrX or rNtrYHKD with IPTG, β-galactosidase activity was measured. Top, β-galactosidase activity (Miller units). Data indicate the means ± standard deviations from three independent experiments performed in triplicate. *, significantly different (P < 0.05) by Student’s t test. Bottom, Western blot analysis of samples from the β-galactosidase assays was performed using a monoclonal antibody against the His tag to verify the expression of rNtrX and rNtrYHKD. A representative of three independent experiments is shown. Each lane received 1 µg protein.
To examine whether rNtrX transactivates the expression of glnA and putA, E. chaffeensis glnA or putA promoter-lacZ fusion was transformed into E. coli BL21(DE3) expressing E. chaffeensis rNtrX or the E. chaffeensis NtrY histidine kinase domain (rNtrYHKD) as a negative control (12). rNtrX induction by isopropyl-thio-β-d-galactoside (IPTG) resulted in a significant increase in β-galactosidase activity compared with rNtrYHKD induction (Fig. 6D), demonstrating transactivation of putA and glnA by NtrX but not by NtrY. The expression of rNtrX and rNtrYHKD was confirmed by Western blotting (Fig. 6D). The lacZ fusion containing the E. chaffeensis ompA promoter (negative control) was not transactivated despite the expression of rNtrX (Fig. 6D).
CtrA is rapidly degraded in a proline and glutamine uptake-dependent manner upon E. chaffeensis entry.
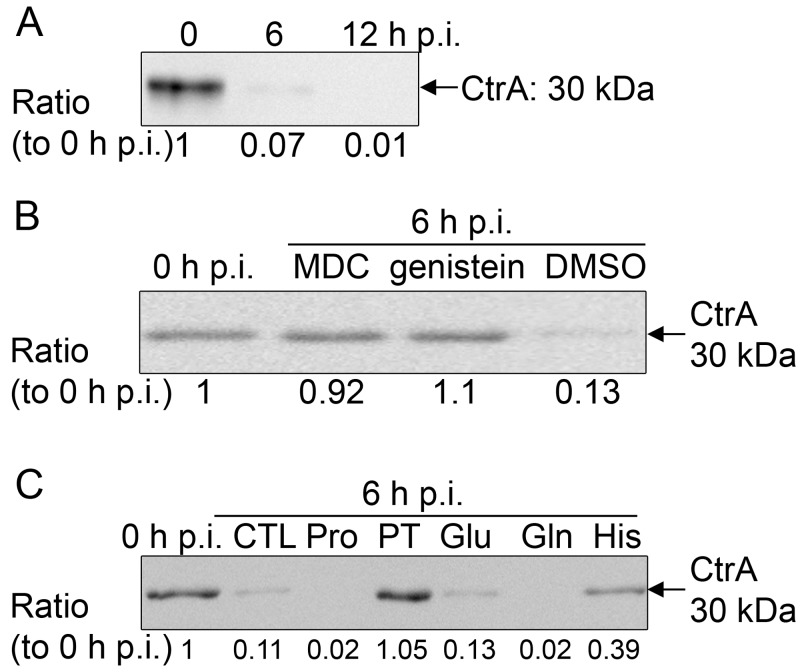
E. chaffeensis has three pairs of two-component systems, and only NtrX and CtrA response regulators have DNA-binding motifs (17). CtrA is highly upregulated at the stationary stage of E. chaffeensis growth and induces resistance and infectious traits of E. chaffeensis to prepare for the next infection cycle (19). In the free-living alphaproteobacterium C. crescentus, CtrA serves as a negative regulator of bacterial growth because it binds the origin of chromosome replication and blocks DNA replication, and CtrA needs to be degraded prior to chromosome replication (20). CtrA consensus binding sequences are present at the chromosome origin of E. chaffeensis (43). Therefore, we tested for CtrA degradation during early E. chaffeensis infection. Our result showed that at 0 h p.i., bacteria contained a high level of CtrA when they were prepared from the stationary stage of growth as previously described (19). Upon incubation with human leukocytes, CtrA was rapidly degraded as early as 6 h p.i. (Fig. 7A). Transglutaminase and protein-tyrosine kinase activities are required for E. chaffeensis entry into (but not for binding to) THP-1 cells to initiate a new infection cycle (44). To examine whether bacterial internalization is required for initiation of CtrA degradation, THP-1 cells were pretreated with monodansylcadaverine (transglutaminase inhibitor), genistein (generic protein-tyrosine kinase inhibitor), or dimethyl sulfoxide (DMSO; solvent control) at 37°C for 3 h and then incubated with host cell-free bacteria. At 6 h p.i., CtrA degradation in the monodansylcadaverine- and genistein-treated groups was blocked compared with that in the DMSO control group (Fig. 7B). The result suggested that bacterial internalization is required for the initiation of CtrA degradation.
FIG 7 .
CtrA is rapidly degraded upon bacterial entry, and proline and glutamine uptake is needed for CtrA degradation. (A) Western blot analysis of CtrA levels in E. chaffeensis-infected THP-1 cells at different time points p.i.; (B) Western blot analysis of CtrA levels in E. chaffeensis-infected THP-1 cells at 0 h p.i. and 6 h p.i. with different pretreatments for 3 h; (C) Western blot analysis of CtrA levels in E. chaffeensis-infected THP-1 cells at 6 h p.i. with different pretreatments (CTL, control; Pro, proline; PT, protamine; Glu, glutamine; Gln, glutamine; His, histidine) for 30 min. For panels A to C, all samples were normalized to the bacterial 16S rRNA gene level as determined by quantitative RT-PCR. The numbers below the panels indicate the relative intensity of each protein band.
Because preloading E. chaffeensis with proline and glutamine accelerated E. chaffeensis growth and because inhibition of initial proline and glutamine uptake blocked E. chaffeensis infection, we next examined whether proline and glutamine induce CtrA degradation in E. chaffeensis prior to initiation of growth. At 6 h p.i., compared with no treatment or glutamate pretreatment for 30 min, pretreatment of bacteria with proline and glutamine accelerated CtrA degradation (Fig. 7C). Importantly, blocking of proline or glutamine uptake by protamine or histidine treatment for 30 min, respectively, inhibited CtrA degradation upon incubation with THP-1 cells (Fig. 7C). These results indicated that proline and glutamine uptake by E. chaffeensis induces CtrA degradation, which may be necessary to initiate E. chaffeensis replication. Taken together, these results suggested that E. chaffeensis entry and subsequent proline and glutamine uptake from host cells signal CtrA degradation.
DISCUSSION
The present study reveals the importance of the PutA/GlnA pathway in E. chaffeensis infection. Proline is utilized as an extracellular source of carbon and energy; upon import by E. chaffeensis, it is converted into glutamate by PutA (10). Glutamate can be further converted by glutamate dehydrogenase (present in all sequenced Rickettsiales species) to 2-ketoglutarate, which is subsequently channeled into the TCA cycle to generate ATP via oxidative phosphorylation. By assimilating ammonium, GlnA converts glutamate to glutamine, which is a key nitrogen donor in nitrogen metabolism (45, 46). The present study showed that E. chaffeensis can take up exogenous proline and glutamine but not glutamate. The related obligatory intracellular bacteria Neorickettsia (formerly Rickettsia or Ehrlichia) sennetsu and Neorickettsia (formerly Ehrlichia) risticii can utilize glutamine, but not glutamate, as an energy source (47, 48). In contrast, glutamate rather than glutamine is a critical energy source for Rickettsia typhi (49). Thus, the E. chaffeensis growth-accelerating effects of proline and glutamine uptake are due to rapid energy generation and subsequent metabolic stimulation through the PutA/GlnA pathway. This is distinct from Chlamydia psittaci, for which leucine, phenylalanine, and valine are the most critical amino acids for infection (50). Whether proline and glutamine are the most critical amino acids required for E. chaffeensis growth remains to be determined. In Ehrlichia ruminantium-infected BA886 cells, a bovine endothelial line, proline and glutamine are the most depleted amino acids after a 3-day culture (51), suggesting that these amino acids are required for Ehrlichia spp.
Among intracellular bacteria, Mycobacterium tuberculosis is sensitive to MSX in vitro and in vivo, because it makes bacteria fragile by blocking GlnA activity, which is required for extrabacterial polyglutamine capsule formation (35, 45, 46). E. chaffeensis is the second intracellular bacterium shown to be sensitive to MSX, but the mechanism for MSX growth inhibition of E. chaffeensis is distinct from that of Mycobacterium because E. chaffeensis does not produce a polyglutamine capsule. Because E. chaffeensis can import exogenous glutamine, its high sensitivity to MSX suggests that accumulation of NH4+ is detrimental to the proliferation of intracellular E. chaffeensis and/or that endogenous glutamine synthesis is needed owing to insufficient glutamine within inclusions, especially at the onset of growth. Although we cannot deny the possibility that MSX has other inhibitory targets in E. chaffeensis, the design of MSX analogs with an improved therapeutic-to-toxic ratio remains a promising strategy for the development of novel drugs to treat E. chaffeensis infection.
GlnA and PutA were identified as part of the NtrX regulon in the present work. In Azorhizobium caulinodans, NtrX is involved in regulating nif genes (39). The NtrX-binding DNA sequence has been determined between +42 and +80 from the puf transcriptional start site in R. capsulatus, but no consensus binding sequence has been determined (52). E. chaffeensis lacks nif and puf homologs, and the NtrX-binding DNA sequence remains to be studied, which may help identify the remaining NtrX regulon. Whether NtrX phosphorylation enhances its binding to DNA was unknown until this study, which demonstrated that after phosphorylation, the DNA-binding affinity of E. chaffeensis NtrX increased at levels similar to those of R. capsulatus NtrC (41).
In R. capsulatus, NtrY/NtrX is involved in nitrogen metabolism and photosynthetic gene expression (15, 52). NtrY is predicted to be a transmembrane protein and to sense the extrabacterial nitrogen level in Azorhizobium caulinodans (39) and Azospirillum brasilense (16). In contrast, NtrB in E. coli is a cytoplasmic protein that senses the intrabacterial nitrogen level (53). In A. brasilense, the genes encoding NtrY/NtrX and NtrB/NtrC occur as an operon and coregulate the fixation of atmospheric nitrogen (16). Interestingly, Brucella abortus NtrY contains a Per-ARNT-SIM (PAS) domain and acts as a redox sensor within bacteria (14), but E. chaffeensis NtrY lacks the PAS domain. Whether and what NtrY senses in E. chaffeensis are unknown. E. chaffeensis NtrY lacks a signal peptide and is predicted to be retained in the bacterial cytoplasm; indeed, it is found in the soluble fraction of E. chaffeensis rather than in the membrane fraction (12, 17), suggesting that NtrY detects an intrabacterial signal.
In E. coli, S. Typhimurium, and S. meliloti, PutA is autoregulated, but in R. capsulatus and A. tumefaciens, PutA expression is regulated by PutR (10, 22–25). E. chaffeensis lacks PutR, but E. chaffeensis PutA has the DNA-binding domain for autoregulation, albeit with a low amino acid sequence identity to that of E. coli PutA. Thus, E. chaffeensis PutA may also be autoregulated, and this autoregulation may have a synergistic effect with NtrX regulation.
CtrA is conserved among alphaproteobacteria, including all sequenced members of the family Anaplasmataceae. CtrA not only binds to the origin of chromosome replication but also acts as a molecular switch that coordinates cell cycle progression and morphogenesis by positively or negatively regulating at least 95 genes in C. crescentus (54). The 60 genes repressed by CtrA in C. crescentus include genes involved in cell cycle regulation, cell division, and cell wall synthesis (55). CtrA transcriptional regulation and CtrA degradation are quite complex and have been best studied in C. crescentus (55). CtrA is degraded prior to chromosome replication by ATP-dependent protease ClpXP in C. crescentus (20). E. chaffeensis has abundant ClpXP (40); it remains to be determined whether ClpXP is responsible for rapid degradation of CtrA upon E. chaffeensis entry into host cells. Although addition of proline and glutamine accelerated CtrA degradation in E. chaffeensis, this rapid degradation was dependent on bacterial entry, so it is likely that these amino acids are acquired from host cells upon entry. The same 2 amino acids also accelerated ntrY and ntrX expression and putA and glnA expression and subsequent bacterial growth. Because E. chaffeensis entry via its surface invasin EtpE and the EtpE host receptor DNase X does not require bacterial energy (56), the availability of higher concentrations of proline and glutamine upon bacterial entry may trigger the cascade of E. chaffeensis energy-requiring events that initiate the intracellular growth cycle. In summary, the present study uncovers a novel mechanism of regulation of obligatory intracellular growth by regulation of the two-component systems and a downstream regulon in response to critical amino acids. The identified pathway may provide a platform for developing new prophylactic and therapeutic strategies against E. chaffeensis.
MATERIALS AND METHODS
Bacterial strains and culture.
E. chaffeensis Arkansas (57) was propagated in THP-1 cells in RPMI 1640 (Corning Cellgro, Manassas, VA) medium supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 2 mM l-glutamine (Invitrogen, Grand Island, NY) (complete culture medium) at 37°C in 5% CO2 and 95% air. C. trachomatis was cultivated in L929 in Dulbecco’s modified Eagle medium (DMEM; Corning Cellgro) with 10% FBS, containing 2 µg/ml cycloheximide. E. coli strains NovaBlue (Novagen, San Diego, CA) and BL21(DE3) (Novagen) for DNA cloning and protein expression were cultured in Luria-Bertani (LB) broth (58). When required, the medium was supplemented with kanamycin (30 µg/ml) (Fisher Bioreagents, Pittsburgh, PA) or chloramphenicol (34 µg/ml) (Sigma, St. Louis, MO).
E. coli complementation.
The E. coli putA or glnA promoter region (401 or 383 bp, respectively) was inserted upstream of the promoterless E. chaffeensis glnA or putA, respectively, in pUC19 (New England Biolabs) by overlap extension PCR (59). The JT31 (ΔputA) (CGSC, New Haven, CT) or YMC11 (ΔglnA) (32) strain was transformed with the plasmid containing E. chaffeensis putA or glnA, respectively, or pUC19 as a negative control. All E. coli strains were cultured in LB broth at 37°C overnight and then inoculated on plates. For E. coli putA complementation, E. coli strains CSH4 (putA wt) (CGSC) and transformed JT31 (ΔputA) were cultured on TTC (Sigma)-proline (Sigma) indicator plates at 37°C for 24 h (27). For E. coli glnA complementation, E. coli strains XL1-Blue (glnA wt) (60) (Stratagene, La Jolla, CA) and transformed YMC11 (ΔglnA) were cultured on M9 medium plates containing 0.4% glucose (Sigma) supplemented with 20 mM (NH4)2SO4 (Sigma) at 37°C for 48 h (32). The expression of E. chaffeensis glnA and putA in E. coli was confirmed by reverse transcription (RT)-PCR.
Pretreatment of host cell-free E. chaffeensis.
E. chaffeensis-infected THP-1 cells (8 × 107 cells, >90% infected cells) were harvested by centrifugation at 400 × g for 5 min. The pellet was resuspended in 5 ml SPK buffer (200 mM sucrose, 50 mM potassium phosphate, pH 7.4) supplemented with 2 mM l-glutamine and sonicated on ice twice at setting 2 for 10 s using a W-380 Sonicator (Heat Systems Ultrasonics, Farmingdale, NY). Unbroken cells and cell debris were removed by centrifugation at 1,000 × g for 5 min. Host cell-free bacteria in SPK buffer were sonicated on ice twice at setting 4.5 for 30 s to disrupt the fragile bacteria (reticulate cells). The sonication-resistant bacterial dense-cored cells were harvested by centrifugation at 18,000 × g for 5 min at 4°C (61). Dense-cored cells were resuspended in 200 µl SPK buffer supplemented with 2 mM l-glutamine with 2, 10, or 50 µM protamine (Sigma), 2, 6, or 10 mM AC (Sigma) or DHP (Sigma), or 0.5, 1, or 2 mM histidine (Sigma) on ice for 30 min or 20 mM proline, glutamate (Sigma), or glutamine (Sigma) at 37°C for 30 min. Pretreated dense-cored cells were washed twice with 500 µl cold SPK buffer and then used to infect THP-1 cells.
MSX treatment.
Transformed E. coli strain YMC11 was cultured in LB broth at 37°C overnight and then was inoculated on M9 medium plates containing 0.4% glucose supplemented with 20 mM (NH4)2SO4 or 2 mM glutamine with or without 200 µM MSX (Sigma) and cultured at 37°C for 48 h (32). E. chaffeensis-infected THP-1 cells in complete culture medium were treated with 20, 100, or 200 µM MSX (46) at 0 h p.i., and infected cells were harvested at 72 h p.i. To evaluate the reversibility of inhibition, 200 µM MSX added at 0 h p.i. was removed at 24 or 48 h p.i., and infected cells were harvested at 0, 24, 48, and 72 h p.i. To evaluate the effects of glutamate and glutamine on MSX-mediated inhibition, 200 µM MSX was added to complete culture medium at 0 h p.i. with 10, 20, or 40 mM glutamate or glutamine, and infected cells were harvested at 72 h p.i. Bacteria were quantitated by qPCR using primers specific to E. chaffeensis 16S rRNA gene and normalized against the expression of human GAPDH (19). C. trachomatis- or E. chaffeensis-infected L929 cells were cultured in DMEM supplemented with 10% FBS containing 2 µg/ml cycloheximide, with or without 200 µM MSX, at 37°C for 72 h p.i. Bacterial count was scored in 100 host cells on Diff-Quik-stained slides.
Amino acid uptake.
E. chaffeensis (purified as described above) in 200 µl SPK buffer with 2 mM l-glutamine was incubated at 65°C for 15 min or pretreated with 50 µM protamine, 10 mM AC or DHP, or 2 mM histidine on ice for 30 min as described above. Each sample in SPK buffer with 2 mM l-glutamine received 0.2 µCi 3H-labeled l-proline (25 to 55 Ci/mmol; PerkinElmer, Waltham, MA), l-glutamate (40 to 80 Ci/mmol; PerkinElmer), or l-glutamine (30 to 60 Ci/mmol; PerkinElmer) along with 1 µM cold l-proline, l-glutamate, or l-glutamine, respectively, and then incubated at 37° C for 2 h, washed twice with cold SPK buffer, and resuspended in 200 µl SPK buffer (62). ScintiVerse E cocktail (1 ml; Fisher Scientific, Waltham, MA) was added to each sample. Data were acquired with a model 1450 liquid scintillation counting (LSC) luminescence counter (PerkinElmer).
Synchronous culture of E. chaffeensis.
E. chaffeensis was synchronously cultured as described (19). Samples were collected at various time points by centrifugation, and total RNA was isolated from each sample using the RNeasy kit (Qiagen, Valencia, CA). The amount of E. chaffeensis 16S rRNA as well as the levels of E. chaffeensis ntrX, ntrY, putA, and glnA mRNA were determined by quantitative RT-PCR using specific primers (12). The level of E. chaffeensis mRNA at each time point was normalized against bacterial 16S rRNA, given that this roughly correlates with live bacterial counts (19).
Cloning and purification of recombinant proteins and phosphorylation.
The recombinant full-length E. chaffeensis NtrX (rNtrX) was cloned into pET-33b(+), expressed by IPTG induction in E. coli BL21(DE3) and purified by nickel-affinity resin (Sigma) as described (17). The purified rNtrX was dialyzed against storage buffer (50 mM KCl, 50 mM Tris-HCl, pH 8.0) with 50% glycerol. Phosphorylation was carried out as described (63). Briefly, rNtrX was incubated with 25 mM acetyl phosphate in a buffer consisting of 50 mM Tris-HCl (pH 7.6), 50 mM KCl, and 20 mM MgCl2 at 37°C for 2 h.
EMSA.
EMSA was performed as described (19). Briefly, rNtrX or phosphorylated rNtrX (rNtrX-P) was incubated with biotinylated DNA probe (2 nM) in a 50-µl reaction mixture containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.05% (wt/vol) NP-40, and 50 ng/µl poly(dI·dC) on ice for 30 min. As a control for binding specificity, a separate reaction mixture was prepared containing the above-given components plus a 50-fold excess of the corresponding unlabeled DNA competitor. Samples were subjected to native polyacrylamide gel electrophoresis (5% acrylamide), transferred to a nylon membrane, and UV cross-linked. The biotinylated DNA fragments were detected using a LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL) and an LAS-3000 luminescent image analyzer (Fujifilm, Stamford, CT). The density of the band was analyzed using Multi-Gauge v 3.0 software (Fujifilm).
Construction of lacZ fusions.
The lacZ fusions were constructed as described (19). Briefly, the lacZ transcriptional fusion plasmid was constructed by cloning the amplified glnA, putA, or ompA (negative-control) promoter fragments upstream of the promoterless lacZ gene amplified from plasmid pQF50 into plasmid pACYC184 (New England Biolabs). BL21(DE3) strains containing pET-33b(+) encoding rNtrX, or rNtrYHKD as a negative control, were transformed with the lacZ fusions. After inducing the recombinant proteins with 0.05 mM IPTG, β-galactosidase activity was measured (19, 64). Recombinant protein expression was confirmed by Western blotting using horseradish peroxidase-conjugated mouse antibody against the His tag (Sigma) at room temperature for 3 h. The membrane was incubated with ECL Western blotting detection reagents (Amersham, Piscataway, NJ), and bands were visualized with an LAS-3000.
CtrA analysis.
Infected THP-1 cells were suspended in SDS-PAGE sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue, 5% 2-mercaptoethanol) and boiled for 3 min. Samples containing the same number of bacteria as determined by qPCR were loaded onto a 12% SDS-PAGE gel. Following separation, proteins were transferred to a nitrocellulose membrane that was then incubated with rabbit anti-CtrA serum (12) at 4°C overnight. After being washed, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MA) at room temperature for 3 h. The membrane was incubated with ECL Western blotting detection reagents. The bands were detected using an LAS-3000 and analyzed using Multi-Gauge v 3.0.
Statistical analysis.
Statistical analyses were performed using analysis of variance and Tukey’s honestly significant differences test or Student’s t test, and P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Zhihua Zhou (Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for kindly providing E. coli strain YMC11 and Preeti Pancholi (the Ohio State University Medical Center) for providing Chlamydia trachomatis human isolate.
This work was supported by the National Institutes of Health grant R01 AI054476.
Footnotes
Citation Cheng Z, Lin M, Rikihisa Y. 2014. Ehrlichia chaffeensis proliferation begins with NtrY/NtrX and PutA/GlnA upregulation and CtrA degradation induced by proline and glutamine uptake. mBio 5(6):e02141-14. doi:10.1128/mBio.02141-14.
REFERENCES
- 1. Adams DA, Jajosky RA, Ajani U, Kriseman J, Sharp P, Onwen DH, Schley AW, Anderson WJ, Grigoryan A, Aranas AE, Wodajo MS, Abellera JP, Centers for Disease Control and Prevention 2014. Summary of notifiable diseases—United States, 2012. MMWR Morb. Mortal. Wkly. Rep. 61:1–121. [PubMed] [Google Scholar]
- 2. Olano JP, Walker DH. 2002. Human ehrlichioses. Med. Clin. North Am. 86:375–392. 10.1016/S0025-7125(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 3. Paddock CD, Childs JE. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37–64. 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rohmer L, Hocquet D, Miller SI. 2011. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19:341–348. 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349–377. 10.1146/annurev.micro.61.080706.093409. [DOI] [PubMed] [Google Scholar]
- 7. Brown ED, Wood JM. 1992. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J. Biol. Chem. 267:13086–13092. [PubMed] [Google Scholar]
- 8. Colombo G, Villafranca JJ. 1986. Amino acid sequence of Escherichia coli glutamine synthetase deduced from the DNA nucleotide sequence. J. Biol. Chem. 261:10587–10591. [PubMed] [Google Scholar]
- 9. Miller RE, Stadtman ER. 1972. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J. Biol. Chem. 247:7407–7419. [PubMed] [Google Scholar]
- 10. Zhou Y, Larson JD, Bottoms CA, Arturo EC, Henzl MT, Jenkins JL, Nix JC, Becker DF, Tanner JJ. 2008. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J. Mol. Biol. 381:174–188. 10.1016/j.jmb.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hervás AB, Canosa I, Little R, Dixon R, Santero E. 2009. NtrC-dependent regulatory network for nitrogen assimilation in Pseudomonas putida. J. Bacteriol. 191:6123–6135. 10.1128/JB.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng Z, Kumagai Y, Lin M, Zhang C, Rikihisa Y. 2006. Intra-leukocyte expression of two-component systems in Ehrlichia chaffeensis and Anaplasma phagocytophilum and effects of the histidine kinase inhibitor closantel. Cell. Microbiol. 8:1241–1252. 10.1111/j.1462-5822.2006.00704.x. [DOI] [PubMed] [Google Scholar]
- 13. Assumpção MC, de Souza EM, Yates MG, de Oliveira Pedrosa F, Benelli EM. 2007. Purification and characterisation of Azospirillum brasilense N-truncated NtrX protein. Protein Expr. Purif. 53:302–308. 10.1016/j.pep.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 14. Carrica MDC, Fernandez I, Martí MA, Paris G, Goldbaum FA. 2012. The NtrY/X two-component system of Brucella spp. acts as a redox sensor and regulates the expression of nitrogen respiration enzymes. Mol. Microbiol. 85:39–50. 10.1111/j.1365-2958.2012.08095.x. [DOI] [PubMed] [Google Scholar]
- 15. Drepper T, Wiethaus J, Giaourakis D, Gross S, Schubert B, Vogt M, Wiencek Y, McEwan AG, Masepohl B. 2006. Cross-talk towards the response regulator NtrC controlling nitrogen metabolism in Rhodobacter capsulatus. FEMS Microbiol. Lett. 258:250–256. 10.1111/j.1574-6968.2006.00228.x. [DOI] [PubMed] [Google Scholar]
- 16. Ishida ML, Assumpção MC, Machado HB, Benelli EM, Souza EM, Pedrosa FO. 2002. Identification and characterization of the two-component NtrY/NtrX regulatory system in Azospirillum brasilense. Braz. J. Med. Biol. Res. 35:651–661. 10.1590/S0100-879X2002000600004. [DOI] [PubMed] [Google Scholar]
- 17. Kumagai Y, Cheng Z, Lin M, Rikihisa Y. 2006. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect. Immun. 74:5014–5022. 10.1128/IAI.00735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergström J, Fürst P, Norée LO, Vinnars E. 1974. Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 36:693–697. [DOI] [PubMed] [Google Scholar]
- 19. Cheng Z, Miura K, Popov VL, Kumagai Y, Rikihisa Y. 2011. Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol. Microbiol. 82:1217–1234. 10.1111/j.1365-2958.2011.07885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 55:1233–1245. 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 21. Menzel R, Roth J. 1981. Enzymatic properties of the purified PutA protein from Salmonella Typhimurium. J. Biol. Chem. 256:9762–9766. [PubMed] [Google Scholar]
- 22. Keuntje B, Masepohl B, Klipp W. 1995. Expression of the putA gene encoding proline dehydrogenase from Rhodobacter capsulatus is independent of NtrC regulation but requires an Lrp-like activator protein. J. Bacteriol. 177:6432–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho K, Winans SC. 1996. The putA gene of Agrobacterium tumefaciens is transcriptionally activated in response to proline by an Lrp-like protein and is not autoregulated. Mol. Microbiol. 22:1025–1033. 10.1046/j.1365-2958.1996.01524.x. [DOI] [PubMed] [Google Scholar]
- 24. Soto MJ, Jiménez-Zurdo JI, van Dillewijn P, Toro N. 2000. Sinorhizobium meliloti putA gene regulation: a new model within the family Rhizobiaceae. J. Bacteriol. 182:1935–1941. 10.1128/JB.182.7.1935-1941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menzel R, Roth J. 1981. Regulation of the genes for proline utilization in Salmonella Typhimurium: autogenous repression by the putA gene product. J. Mol. Biol. 148:21–44. 10.1016/0022-2836(81)90233-3. [DOI] [PubMed] [Google Scholar]
- 26. Zhou Y, Zhu W, Bellur PS, Rewinkel D, Becker DF. 2008. Direct linking of metabolism and gene expression in the proline utilization A protein from Escherichia coli. Amino Acids 35:711–718. 10.1007/s00726-008-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wood JM. 1981. Genetics of l-proline utilization in Escherichia coli. J. Bacteriol. 146:895–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Rooyen JM, Abratt VR, Sewell BT. 2006. Three-dimensional structure of a type III glutamine synthetase by single-particle reconstruction. J. Mol. Biol. 361:796–810. 10.1016/j.jmb.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 29. Shatters RG, Kahn ML. 1989. Glutamine synthetase II in Rhizobium: reexamination of the proposed horizontal transfer of DNA from eukaryotes to prokaryotes. J. Mol. Evol. 29:422–428. 10.1007/BF02602912. [DOI] [PubMed] [Google Scholar]
- 30. Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH. 2000. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 1477:122–145. 10.1016/S0167-4838(99)00270-8. [DOI] [PubMed] [Google Scholar]
- 31. Burroughs AM, Balaji S, Iyer LM, Aravind L. 2007. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol. Direct 2:18. 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li X, Liu T, Wu Y, Zhao G, Zhou Z. 2010. Derepressive effect of NH4+ on hydrogen production by deleting the glnA1 gene in Rhodobacter sphaeroides. Biotechnol. Bioeng. 106:564–572. 10.1002/bit.22722. [DOI] [PubMed] [Google Scholar]
- 33. Manning JM, Moore S, Rowe WB, Meister A. 1969. Identification of l-methionine S-sulfoximine as the diastereoisomer of l-methionine SR-sulfoximine that inhibits glutamine synthetase. Biochemistry 8:2681–2685. 10.1021/bi00834a066. [DOI] [PubMed] [Google Scholar]
- 34. Weisbrod RE, Meister A. 1973. Studies on glutamine synthetase from Escherichia coli. Formation of pyrrolidone carboxylate and inhibition by methionine sulfoximine. J. Biol. Chem. 248:3997–4002. [PubMed] [Google Scholar]
- 35. Harth G, Horwitz MA. 1999. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential novel drug targets. J. Exp. Med. 189:1425–1436. 10.1084/jem.189.9.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aspedon A, Groisman EA. 1996. The antibacterial action of protamine: evidence for disruption of cytoplasmic membrane energization in Salmonella Typhimurium. Microbiology 142(12):3389–3397. 10.1099/13500872-142-12-3389. [DOI] [PubMed] [Google Scholar]
- 37. Deutch CE. 2011. l-Proline nutrition and catabolism in Staphylococcus saprophyticus. Antonie Van Leeuwenhoek 99:781–793. 10.1007/s10482-011-9552-7. [DOI] [PubMed] [Google Scholar]
- 38. Albrecht J, Dolińska M, Hilgier W, Lipkowski AW, Nowacki J. 2000. Modulation of glutamine uptake and phosphate-activated glutaminase activity in rat brain mitochondria by amino acids and their synthetic analogues. Neurochem. Int. 36:341–347. 10.1016/S0197-0186(99)00142-4. [DOI] [PubMed] [Google Scholar]
- 39. Pawlowski K, Klosse U, de Bruijn FJ. 1991. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol. Gen. Genet. 231:124–138. 10.1007/BF00293830. [DOI] [PubMed] [Google Scholar]
- 40. Lin M, Kikuchi T, Brewer HM, Norbeck AD, Rikihisa Y. 2011. Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front. Microbiol. 2:24. 10.3389/fmicb.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cullen PJ, Bowman WC, Kranz RG. 1996. In vitro reconstitution and characterization of the Rhodobacter capsulatus NtrB and NtrC two-component system. J. Biol. Chem. 271:6530–6536. 10.1074/jbc.271.11.6530. [DOI] [PubMed] [Google Scholar]
- 42. Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 43. Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12:361–365. 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 44. Lin M, Zhu MX, Rikihisa Y. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70:889–898. 10.1128/IAI.70.2.889-898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tullius MV, Harth G, Horwitz MA. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 71:3927–3936. 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harth G, Maslesa-Galić S, Tullius MV, Horwitz MA. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58:1157–1172. 10.1111/j.1365-2958.2005.04899.x. [DOI] [PubMed] [Google Scholar]
- 47. Weiss E, Dasch GA, Kang YH, Westfall HN. 1988. Substrate utilization by Ehrlichia sennetsu and Ehrlichia risticii separated from host constituents by Renografin gradient centrifugation. J. Bacteriol. 170:5012–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Messick JB, Rikihisa Y. 1993. Characterization of Ehrlichia risticii binding, internalization, and proliferation in host cells by flow cytometry. Infect. Immun. 61:3803–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williams JC, Weiss E. 1978. Energy metabolism of Rickettsia typhi: pools of adenine nucleotides and energy charge in the presence and absence of glutamate. J. Bacteriol. 134:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Allan I, Pearce JH. 1983. Differential amino acid utilization by Chlamydia psittaci (strain guinea pig inclusion conjunctivitis) and its regulatory effect on chlamydial growth. J. Gen. Microbiol. 129:1991–2000. [DOI] [PubMed] [Google Scholar]
- 51. Josemans AI, Zweygarth E. 2002. Amino acid content of cell cultures infected with Cowdria ruminantium propagated in a protein-free medium. Ann. N. Y. Acad. Sci. 969:141–146. 10.1111/j.1749-6632.2002.tb04366.x. [DOI] [PubMed] [Google Scholar]
- 52. Gregor J, Zeller T, Balzer A, Haberzettl K, Klug G. 2007. Bacterial regulatory networks include direct contact of response regulator proteins: interaction of RegA and NtrX in Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 13:126–139. 10.1159/000103604. [DOI] [PubMed] [Google Scholar]
- 53. Kamberov ES, Atkinson MR, Feng J, Chandran P, Ninfa AJ. 1994. Sensory components controlling bacterial nitrogen assimilation. Cell. Mol. Biol. Res. 40:175–191. [PubMed] [Google Scholar]
- 54. Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 99:4632–4637. 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Skerker JM, Laub MT. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2:325–337. 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 56. Mohan Kumar D, Yamaguchi M, Miura K, Lin M, Los M, Coy JF, Rikihisa Y. 2013. Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog. 9:e1003666. 10.1371/journal.ppat.1003666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sambrook J, Pollack R. 1974. Basic methodology for cell culture—cell transformation. Methods Enzymol. 32:583–592. 10.1016/0076-6879(74)32058-7. [DOI] [PubMed] [Google Scholar]
- 59. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 60. Hosted TJ, Rochefort DA, Benson DR. 1993. Close linkage of genes encoding glutamine synthetases I and II in Frankia alni CpI1. J. Bacteriol. 175:3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng Z, Wang X, Rikihisa Y. 2008. Regulation of type IV secretion apparatus genes during Ehrlichia chaffeensis intracellular development by a previously unidentified protein. J. Bacteriol. 190:2096–2105. 10.1128/JB.01813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rikihisa Y, Zhang Y, Park J. 1994. Inhibition of infection of macrophages with Ehrlichia risticii by cytochalasins, monodansylcadaverine, and taxol. Infect. Immun. 62:5126–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kenney LJ, Bauer MD, Silhavy TJ. 1995. Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 92:8866–8870. 10.1073/pnas.92.19.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang X, Kikuchi T, Rikihisa Y. 2007. Proteomic identification of a novel Anaplasma phagocytophilum DNA binding protein that regulates a putative transcription factor. J. Bacteriol. 189:4880–4886. 10.1128/JB.00318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]