Abstract
Background
Bifidobacteria constitute a specific group of commensal bacteria that commonly inhabit the mammalian gastrointestinal tract. Bifidobacterium breve UCC2003 was previously shown to utilize a variety of plant/diet/host-derived carbohydrates, including cellodextrin, starch and galactan, as well as the mucin and HMO-derived monosaccharide, sialic acid. In the current study, we investigated the ability of this strain to utilize parts of a host-derived source of carbohydrate, namely the mucin glycoprotein, when grown in co-culture with the mucin-degrading Bifidobacterium bifidum PRL2010.
Results
B. breve UCC2003 was shown to exhibit growth properties in a mucin-based medium, but only when grown in the presence of B. bifidum PRL2010, which is known to metabolize mucin. A combination of HPAEC-PAD and transcriptome analyses identified some of the possible monosaccharides and oligosaccharides which support this enhanced co-cultivation growth/viability phenotype.
Conclusion
This study describes the potential existence of a gut commensal relationship between two bifidobacterial species. We demonstrate the in vitro ability of B. breve UCC2003 to cross-feed on sugars released by the mucin-degrading activity of B. bifidum PRL2010, thus advancing our knowledge on the metabolic adaptability which allows the former strain to colonize the (infant) gut by its extensive metabolic abilities to (co-)utilize available carbohydrate sources.
Keywords: Bifidobacteria, Probiotic, Fucose, Sialic acid, Galactose
Background
Bifidobacteria are Gram positive, anaerobic, Y-shaped bacteria that have been found in the gastrointestinal tract (GIT) of mammals, birds and insects and have also been isolated from the human oral cavity and sewage [1]. In recent years, bifidobacteria have attracted attention due to the purported health benefits associated with their presence in the gut. Such beneficial contributions include development and modulation of the immune system [2], provision of vitamins [3], and protection against pathogenic bacteria [4]. Bifidobacteria rapidly colonize the infant gut in the first days and weeks of life. In a recent study on the microbial diversity of the infant gut, it was found that members of the Actinobacteria phylum were dominant, with various bifidobacterial species present in high abundance, in particular Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium breve and Bifidobacterium catenulatum [5].
Survival and growth of bifidobacteria in the gastrointestinal tract requires them to employ an arsenal of enzymes to metabolize the complex carbohydrates prevalent in this environment [6]. B. breve UCC2003 has previously been shown to utilize various diet/plant-derived oligo- and poly-saccharides, including melezitose, raffinose, cellodextrins, GOS, starch and galactan [7-12]. Recently, B. breve UCC2003 was shown to utilize the mucin- and human milk oligosaccharide (HMO)-derived monosaccharide sialic acid [13], which is more consistent with this strain’s origin as a nursling stool isolate from a breast-fed infant, where it would be expected to metabolize HMOs and/or the structurally similar oligosaccharides found in mucin glycoproteins. Along with dietary components, host-derived oligosaccharides are believed to form part of the nutrient resource for certain intestinal bacteria.
The original investigations on the (bio)chemical composition of human colonic mucin described twenty-one discrete oligosaccharide structures [14]. Since then, it has been estimated that carbohydrate constitutes approximately 80% of the total mucin mass, and that MUC2, the prominent secretory mucin in the colon, may contain more than 100 different O-linked glycans [15,16]. Glycosylation of the peptide backbone can be N-linked to an asparagine residue or O-linked to serine or threonine residues via N-acetylgalactosamine (GalNAc). Subsequent elongation of this structure results in a number of distinct core structures. While eight mucin type core structures are known, only four regularly occur in human mucins. Core 1 is formed by the addition of galactose (Gal) in a β-(1,3) linkage to GalNAc to produce galacto-N-biose (GNB, also known as the T antigen). Core 3, the most common core structure in the human colon, is formed by the addition of N-acetylglucosamine (GlcNAc) in a β-(1,3) linkage to GalNAc. Core 2 and core 4 structures are formed by the addition of GlcNAc in a β-(1,6) linkage to core 1 and core 3, respectively. Each of the core structures can be further elongated by the addition of Gal, GalNAc and GlcNAc. The oligosaccharide chains can also be substituted with sialic acid, fucose (Fuc) or sulfate residues in terminal or branched positions [14,15,17,18].
Due to the high complexity and variability of the mucin oligosaccharide chains, only a small proportion of the culturable intestinal microbiota is believed to encode enzymes required for (partial) degradation of mucin into free sugars. These include members of the Bifidobacterium, Bacteroides and Ruminococcus genera, as well as a more recently characterized bacterium isolated from human faeces, Akkermansia muciniphila [19,20]. The degradation of mucin occurs sequentially with the removal of component monosaccharides, rather than the removal of the entire polysaccharide structure, thus requiring a multitude of enzymes with various glycosidic specificities [21].
The first observation of mucin-degrading bifidobacteria was described by Hoskins et al., who described the isolation of two bifidobacterial strains that constitutively expressed extracellular enzymes capable of degrading oligosaccharide side chains of gut mucins [22]. Cell surface-anchored glycosyl hydrolases were later characterized, including two α-L-fucosidases, AfcA and AfcB, from B. bifidum JCM1254 [23,24], and an endo-α-N-acetylgalactosaminidase, EngBF, from B. longum JCM1217, which hydrolyzes the linkage between GalNAc of the core 1 disaccharide and the serine or threonine residue of the proteinaceous backbone [25].
As mentioned previously, B. bifidum is one of the most abundant species in the infant intestine [5]. A study in 2010 revealed that some 60% of the glycosyl hydrolases identified on the genome of B. bifidum PRL2010 are linked to the degradation of mucin [26,27]. The identified glycosyl hydrolases include two putative exo-α-sialidases, two putative α-L-fucosidases and a cell wall-anchored endo-α-N-acetylgalactosaminidase [26]. Other mucin-degrading enzymes on the genome of B. bifidum PRL2010 include four N-acetyl-β-hexosaminidases and four β-galactosidases [26,27]. Comparative genome hybridization analysis revealed that most of these genes encoding the above mentioned enzymes are conserved within the examined members of the B. bifidum species [26,27].
It has been suggested that the degradation of mucin by a small number of extracellular glycosidase-producing bacteria may provide nutritional support to other enteric bacteria [22]. A similar concept was more recently suggested in relation to the metabolism of HMO [28] and it has been shown that a number of HMO-derived degradation products remain in the media during vegetative growth of B. bifidum [29]. We recently demonstrated that B. breve UCC2003 can indeed cross-feed on sialic acid derived from the metabolism of 3′ sialyllactose, an abundant HMO, by B. bifidum PRL2010 [13]. The aim of the current study was to establish if B. breve UCC2003 is able to cross-feed on the oligosaccharides released by B. bifidum PRL2010 extracellular activity on mucin, and secondly, to investigate which, if any, particular components of mucin B. breve UCC2003 utilizes under such circumstances.
Methods
Bacterial strains, plasmids, media and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. B. breve UCC2003 and its derivatives were routinely cultured in Reinforced Clostridial Medium (RCM; Oxoid Ltd, Basingstoke, Hampshire, United Kingdom). B. bifidum PRL2010 was routinely cultured in modified deMan Rogosa Sharpe (mMRS) medium made from first principles (but excluding a carbohydrate source) [30], and supplemented with 0.05% (wt/vol) L-cysteine HCl and 1% (wt/vol) lactose (unless otherwise stated). All carbohydrates used in this study were purchased from Sigma Aldrich and were of the highest purity available. To prepare mucin-containing media, a 0.8% (wt/vol) concentration of mucin from porcine stomach (Type III) was prepared in water and autoclaved at 115°C for 10 minutes, in order to optimize mucin dissolution, yet minimizing glycosidic hydrolysis. This was added to an equal volume of twice-concentrated mMRS, resulting in a final concentration of 0.4% mucin in mMRS. Bifidobacterial cultures were incubated under anaerobic conditions in a modular atmosphere-controlled system (Davidson and Hardy, Belfast, Ireland) at 37°C. Escherichia coli was cultured in Luria Bertani (LB) broth at 37°C with agitation [31]. Where appropriate, growth media contained tetracycline (Tet; 10 μg ml−1), chloramphenicol (Cm; 5 μg ml−1 for E. coli, 2.5 μg ml−1 for B. bifidum), erythromycin (Em; 100 μg ml−1) or kanamycin (Kan; 50 μg ml−1).
Table 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Relevant features | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli strains | ||
| E.coli EC101 | Cloning host; repA + kmr | [37] |
| E.coli EC101-pNZ-M.Bbrll + Bbr11 | EC101 harboring a pNZ8048 derivative containing bbrllM and bbrlllM | [39] |
| Bifidobacterium sp. strains | ||
| B. breve UCC2003 | Isolate from a nursling stool | [36] |
| B. breve UCC2003 - pAM5 | B. breve UCC2003 harboring pAM5 | This study |
| B. breve UCC2003-nanA | pORI19-Tet-nanA (Bbr_0168) insertion mutant of B. breve UCC2003 | [13] |
| B. breve UCC2003-fucP | pORI19-Tet-fucP (Bbr_1742) insertion mutant of B. breve UCC2003 | This study |
| B. breve UCC2003-lnbP | pORI19-Tet-lnbP (Bbr_1587) insertion mutant of B. breve UCC2003 | This study |
| B. breve UCC2003-lacZ7 | pORI19-Tet-lacZ7 (Bbr_1833) insertion mutant of B. breve UCC2003 | This study |
| B. breve UCC2003-galT | Tetr transposon mutant of B. breve UCC2003 | [41] |
| B. bifidum PRL2010 | Isolate from infant faeces | [26] |
| B. bifidum PRL2010-pPKCM7 | B. bifidum PRL2010 harboring pPKCM7 | This study |
| Plasmids | ||
| pAM5 | pBC1-puC19-Tcr | [38] |
| pPKCM7 | pblueCm harboring rep pCIBA089 | [40] |
| pORI19 | Emr, repA−, ori+, cloning vector | [37] |
| pORI19-tetW-fucP | Internal 400 bp fragment of fucP and tetW cloned in pORI19 | This study |
| pORI19-tetW-lnbP | Internal 479 bp fragment of lnbP and tetW cloned in pORI19 | This study |
| pORI19-tetW-lacZ7 | Internal 568 bp fragment of lacZ7 and tetW cloned in pORI19 | This study |
Nucleotide sequence analysis
Sequence data were obtained from the Artemis-mediated genome annotations of B. breve UCC2003 [32,33]. Database searches were performed using non-redundant sequences available at the National Centre for Biotechnology Information internet site (http://www.ncbi.nlm.nih.gov) using BLAST [34]. Sequence analysis was performed using the Seqbuilder and Seqman programs of the DNASTAR software package (DNASTAR, Madison, WI, USA).
DNA manipulations
Chromosomal DNA was isolated from B. breve UCC2003 as previously described [35]. Plasmid DNA was isolated from E. coli, B. breve and B. bifidum using the Roche High Pure plasmid isolation kit (Roche Diagnostics, Basel, Switzerland). An initial lysis step was performed using 30 mg ml−1 of lysozyme for 30 min at 37°C prior to plasmid isolation from bifidobacteria. Single stranded oligonucleotide primers used in this study were synthesized by Eurofins (Ebersberg, Germany) (Table 2). PCRs were performed using Taq PCR master mix (Qiagen GmbH, Hilden, Germany). PCR products were purified using the Roche High Pure PCR purification kit (Roche Diagnostics). Electroporation of plasmid DNA into E. coli or bifidobacteria was performed as described previously [31,36].
Table 2.
Oligonucleotide primers used in this study
| Purpose | Primer | Sequence |
|---|---|---|
| Cloning of 400 bp fragment of fucP (Bbr_1742) in pORI19 | FucPF | TAGCATAAGCTTGGCGAATCGTTCGTATCA |
| FucPR | GATATCTCTAGAGCGCCCCAGTGCTTGAGC | |
| Cloning of 479 bp fragment of lnbP (Bbr_1587) in pORI19 | LnbPF | TAGCATAAGCTTCACACAGGTATTGGGAGGTTG |
| LnbPR | CTAGTCTCTAGAGTTGTAGGCGCCACCATCC | |
| Cloning of 568 bp fragment of lacZ7 (Bbr_1833) in pORI19 | LacZ7F | TAGCATAAGCTTCCAGGCCAAGAACTCCAGTG |
| LacZ7R | CATGATTCTAGACAGCTTGGGCAGGTTGAACG | |
| Amplification of tetW | TetWF | TCAGCTGTCGACATGCTCATGTACGGTAAG |
| TetWR | GCGACGGTCGACCATTACCTTCTGAAACATA | |
| Confirmation of site-specific homologous recombination | FucPconfirm | TGTTCGCCATGTTCGTTATC |
| LnbPconfirm | GATCACTCTGCATATGGACG | |
| LacZ7confirm | GTACCGACATCGACGCGTTC |
Restriction sites incorporated into oligonucleotide primer sequences are indicated in italics.
Construction of B. breve UCC2003 insertion mutants
An internal fragment of Bbr_1742, designated here as fucP (400 base pairs (bp), representing codon numbers 137 through to 270 out of the 421 codons of fucP), Bbr_1587, designated here as lnbP (479 bp, representing codon numbers 108 through to 267 of the 751 codons of lnbP) and Bbr_1833, designated here as lacZ7 (568 bp, representing codon numbers 123 through 312 of the 699 codons of lacZ7), were amplified by PCR using B. breve UCC2003 chromosomal DNA as a template and primer pairs FucPF and FucPR, LnbPF and LnbPR and LacZ7F and LacZ7R, respectively. The generated amplicons were digested with HindIII and XbaI, and ligated to the similarly digested pORI19, an ORI+, RepA− integration plasmid [37]. The ligation mixtures were introduced into E. coli EC101 by electroporation and transformants were selected on LB agar containing Em and supplemented with 40 mg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) and 1 mM IPTG (isopropyl-β-D-galactopyranoside). The plasmid content of a number of Emr transformants was screened by restriction analysis and the integrity of positively identified clones was verified by sequencing. This was followed by subcloning of the Tet antibiotic resistance cassette, tetW from pAM5 [38] as a SacI fragment into the unique SacI site present on each pORI19 derivative. The orientation of the tetW fragment in each of the resulting plasmids, pORI19-tetW-fucP, pORI19-tetW-lnbP and pORI 19-tetW-lacZ7, was verified by restriction analysis, followed by the introduction of the plasmid by electroporation into the E. coli EC101 pNZ-MBbrI-MBbrII strain to methylate the plasmid constructs prior to introduction into B. breve UCC2003 [39]. Methylation of the plasmids was confirmed by their observed resistance to PstI digestion [39]. The methylated plasmids, pORI19-tetW-fucP, pORI19-tetW-lnbP and pORI19-tetW-lacZ7, were introduced into B. breve UCC2003 by electroporation and transformants were selected on Reinforced Clostridial Agar (RCA) supplemented with Tet. Site-specific recombination in potential Tet-resistant mutant isolates was confirmed by colony PCR using primer combinations TetWF and TetWR to confirm tetW gene integration, and the primers FucPconfirm, LnbPconfirm and LacZ7confirm (positioned upstream of the selected internal fragment of fucP, lnbP and lacZ7, respectively) and TetWF to confirm integration at the correct chromosomal location.
Evaluation of B. breve UCC2003 growth on mucin
Growth of B. breve UCC2003 using mucin as the sole carbon source was determined both independently and in co-culture with B. bifidum PRL2010. Plasmid-containing derivatives of the wild type strains were used, namely B. breve UCC2003-pAM5, which contains the pAM5 plasmid and is therefore Tet resistant [38] and B. bifidum PRL2010-pPKCM7, which contains the pPKCM7 plasmid and is Cm resistant [40]. Use of plasmid-containing derivatives allowed for selection and enumeration of either B. breve UCC2003 or B. bifidum PRL2010 colonies on RCA containing the appropriate antibiotic. All B. breve UCC2003 mutant strains were also Tet resistant. A 0.001% inoculum of a stationary phase culture of B. breve UCC2003 strains (see Table 1; B. breve UCC2003-pAM5, or the mutant strains, B. breve UCC2003-nanA [13], B. breve UCC2003-fucP, B. breve UCC2003-lnbP, B. breve UCC2003-lacZ7 or B. breve UCC2003-galT [41]) and/or a 0.01% inoculum of B. bifidum PRL2010-pPKCM7, were added to mMRS medium supplemented with 0.05% (wt/vol) L-cysteine HCl and 0.4% (wt/vol) mucin (see above). Growth of the cultures was measured over 72 h, with samples taken every 6 or 12 h. All samples collected were serially 10-fold diluted in sterile Ringers solution and plated onto RCA supplemented with 1% (wt/vol) lactose and the appropriate antibiotic. Viable counts were determined by counting colonies on agar plates using dilutions that yielded between 30 and 300 colony forming units (CFU).
Transcriptome analysis using B. breve UCC2003-based microarrays
B. breve UCC2003-pAM5 was grown in mMRS medium supplemented with 0.05% (wt/vol) L-cysteine HCl and 0.4% ribose to an OD600nm of 0.5 and then harvested by centrifugation at 9,000 × g for 2 min at room temperature. B. breve UCC2003-pAM5 grown in mucin in co-culture with B. bifidum PRL2010-pPKCM7 (see above) was similarly harvested after 30 h of growth. DNA microarrays containing oligonucleotide primers representing each of the 1864 annotated genes on the genome of B. breve UCC2003 were designed by and obtained from Agilent Technologies (Palo Alto, CA, USA). Methods for cell disruption, RNA isolation, RNA quality control, complementary DNA (cDNA) synthesis and labelling were performed as described previously [42]. Labelled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part number 5188–5242) as described in the Agilent Two-Color Microarray-Based Gene Expression Analysis (v4.0) manual (publication number G4140-90050). Following hybridization, microarrays were washed as described in the manual and scanned using Agilent’s DNA microarray scanner G2565A. The scanning results were converted to data files with Agilent’s Feature Extraction software (version 9.5). DNA-microarray data were processed as previously described [43-45]. Differential expression tests were performed with the Cyber-T implementation of a variant of the t-test [46].
Analysis of the monosaccharide composition of fermented and non-fermented mucin
Identification and quantification of the free monosaccharides in non-fermented mMRS supplemented with 0.4% mucin and the media following 30 h fermentation by B. bifidum PRL2010-pPKCM7 was performed according to Dionex technical note 40 (http://www.dionex.com/en-us/webdocs/5052-TN40-IC-Glycoprotein-Monosaccharide-23May2012-LPN1632-01.pdf) and as previously described [47], using high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). In brief, media was pelleted out, sterilized through a 0.2 μm filter and then lyophilized to dryness. The lyophilized powder was dissolved at 1 mg ml−1 in purified water. Dilutions were injected on to a Dionex ICS 3000 system (Dionex, Sunnyvale, CA) equipped with a CarboPac PA20 analytical column (150 mm × 3 mm) with an Amino-trap column (30 mm × 3 mm) and separated as previously described [48]. Resulting peaks were identified and quantified by comparison to a standard curve with a mixture containing the common mammalian residues Fuc, glucosamine (GlcN), galactosamine (GalN), Gal, glucose (Glc) and mannose (Man). Samples were injected three times and the average value is reported for the concentration. Samples were then spiked with a known concentration of Fuc and Gal standards and re-injected to confirm the identification of these residues in the sample.
To further confirm the identity of Fuc in spent medium, samples were labelled with 2-aminobenzamide (2-AB) according to the published method [49]. The 2-AB labelled samples were cleaned on Glycoclean S cartridges (Prozyme) according to manufacturer’s instructions and vacuum centrifuged dry. Samples were analyzed by reverse phase-high performance liquid chromatography (RP-HPLC) injected onto a Waters Alliance 2695 instrument and separated on a Phenomenex Luna 3u C18(2) (150 mm × 4.6 mm) column using previously described conditions [50].
During co-culture experiments samples of cell free supernatants were taken every 6 or 12 h to qualitatively analyze the carbohydrate composition of the media by HPAEC-PAD. Samples (25 μl aliquots) were separated on a CarboPac PA1 analytical-exchange column (250 mm × 4 mm) with a CarboPac PA1 guard column (50 mm × 4 mm). Elution was performed at a constant flow-rate of 1.0 ml min−1 at 30°C using the following eluents for the analysis (A) 200 mM NaOH, (B) 100 mM NaOH, 550 mM sodium acetate, and (C) purified water. The following linear gradient of sodium acetate was used with 100 mM NaOH: from 0 to 50 min, 0 mM; from 50 to 51 min, 16 mM; from 51 to 56 min, 100 mM; from 56 to 61 min, 0 mM. Eluate was monitored with a Dionex ED40 detector in the PAD mode. The chromatogram of non-fermented mucin was used to evaluate mucin utilization by B. bifidum PRL2010-pPKCM7 and B. breve UCC2003-pAM5, and the Chromeleon software v. 6.70 (Dionex Corporation) was used for the integration and evaluation of the chromatograms obtained.
Microarray data accession number
The microarray data obtained in this study have been deposited in NCBI’s Gene Expression Omnibus database and are accessible through GEO series accession number GSE59013.
Ethics statement
No animal or human subjects were involved in this study.
Results
Growth of B. breve UCC2003 on mucin
Growth of B. breve UCC2003-pAM5 and B. bifidum PRL2010-pPKCM7 in mMRS supplemented with 0.4% (wt/vol) mucin, independently or in co-culture, was measured by viable plate counts over 72 h. Monitoring of growth in co-culture required the use of plasmid-containing derivatives of the wild type strains. The plasmids used, pAM5 for B. breve UCC2003 or pPKCM7 for B. bifidum PRL2010, conferred Tet or Cm resistance to the respective strains, thus allowing selection and enumeration of each strain by viable plate count on RCA containing the corresponding antibiotic. Both B. breve UCC2003-pAM5 and B. bifidum PRL2010-pPKCM7 were also cultured in mMRS supplemented with 0.5% lactose, a substrate on which both strains achieve a high level of growth [51,52], and it was found that presence of the plasmid did not impair growth of the strain on this substrate (results not shown). A low inoculum of 0.001% and/or 0.01% of B. breve UCC2003-pAM5 and B. bifidum PRL2010-pPKCM7, respectively, was used to allow the strains to undergo multiple generations of growth. In the absence of an added carbohydrate, no growth was observed for either B. breve UCC2003-pAM5 or B. bifidum PRL2010-pPKCM7 in mMRS medium (results not shown). As expected, the positive control, B. bifidum PRL2010-pPKCM7, was capable of growth in mucin-containing mMRS medium, from a low inoculum of 104 CFU ml−1 it attained cell numbers of approximately 108 CFU ml−1 following 36 h of incubation [26] (Figure 1). However, a notably different growth profile was observed for B. breve UCC2003-pAM5 on this medium. During the first 12 h of incubation in this medium, B. breve UCC2003-pAM5 viable cell numbers increased from 104 CFU ml−1 to almost 107 CFU ml−1, followed by a decline in viable cells to below 104 CFU ml−1 (Figure 1). HPAEC-PAD analysis (see below) revealed the presence of contaminating monosaccharides, namely Glc and Gal, in the non-fermented mucin preparation. It is unknown whether these carbohydrates were released from partial degradation of the mucin during autoclaving, despite efforts made to reduce such degradation (see above), or were present as contaminating monosaccharides in the mucin preparation. Given that B. breve UCC2003 can utilize both of these contaminants as a sole carbon source [51], it is hypothesized that they are responsible for the observed growth during the first 12 h following inoculation. Once these carbon sources were utilized, and as B. breve UCC2003-pAM5 apparently does not have the required enzymes to degrade intact mucin, a substantial drop in viable count was observed. Interestingly, when B. bifidum PRL2010-pPKCM7 was included as a potential carbohydrate-releasing bacterium in co-culture with B. breve UCC2003-pAM5, an improvement in growth and survival abilities of B. breve UCC2003-pAM5 was observed. In co-culture, B. breve UCC2003-pAM5 reached cell numbers of almost 108 CFU ml−1 after 24 h and maintained a viable count of over 106 CFU ml−1 after 72 h (Figure 1). This suggests that B. breve UCC2003-pAM5 is cross-feeding on (some of) the sugars released following mucin breakdown by B. bifidum PRL2010-pPKCM7. The cell numbers of B. bifidum PRL2010-pPKCM7 were moderately lower in co-culture as compared to when the strain was grown independently in this medium and this was attributed to the presence of another acid-producing strain in the culture. However, the growth profile of B. bifidum PRL2010-pPKCM7 in co-culture followed a similar trend to that of the strain growing individually, suggesting that the two strains do not compete for limiting amounts of mucin-derived carbohydrates.
Figure 1.
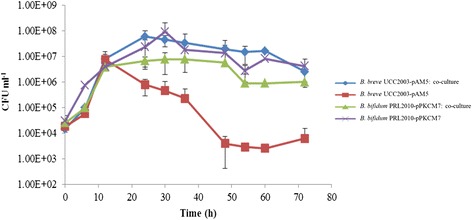
Individual and co-culture growth profiles of B. breve UCC2003-pAM5 and B. bifidum PRL2010-pPKCM7 in mucin. All growth experiments were performed in mMRS supplemented with 0.4% mucin from porcine stomach for 72 h. The results presented are the mean values of duplicate experiments. Error bars represent the standard deviation.
Gene expression analysis using B. breve UCC2003 DNA microarrays
In order to investigate how the transcriptome of B. breve UCC2003-pAM5 is affected by growth in co-culture with B. bifidum PRL2010-pPKCM7 in mucin-containing mMRS, global gene expression was determined by microarray analysis during growth of the strain in co-culture and compared with gene expression when grown on ribose as the sole carbon source. Ribose was considered an appropriate carbohydrate for comparative transcriptome analysis as the genes involved in ribose metabolism are unique to this sugar [53], thus making it an ideal substrate for gene expression analysis on other carbohydrates. It has previously been used as a reference condition in several transcriptome studies in B. breve UCC2003 [7,9,10,13,54]. Furthermore, as a pentose sugar, ribose is suitable for gene expression analysis in mucin, given that the monosaccharide components of mucin are all hexose sugars (Glc, Gal, GalNAc, GlcNAc or Fuc), except sialic acid which is a nine-carbon monosaccharide. In the current study, gene expression was determined following 30 h growth in co-culture. This was to ensure that any observed changes in the transcriptome of B. breve UCC2003-pAM5 could be attributed solely to growth in co-culture with B. bifidum PRL2010-pPKCM7, as opposed to growth on free monosaccharides in the mucin preparation which appeared to be responsible for the initial 12 h of growth of B. breve UCC2003-pAM5 in mucin (see above). Analysis of DNA microarray data showed that a number of stress-related genes were up-regulated, which was attributed to the cells being in co-culture with another bacterium or to the possibility that the cells had entered the stationary phase. Nonetheless, the majority of genes that were shown to be significantly up-regulated (fold change >2.5, P < 0.001) in co-culture are predicted to be involved in the transport and metabolism of carbohydrates. One such cluster, Bbr_1740-1742, is believed to be involved in Fuc metabolism and includes genes predicted to encode a dihydrodipicolinate synthase (Bbr_1740), an enzyme which was previously shown to be involved in Fuc metabolism in Campylobacter jejuni [55], a hypothetical protein (Bbr_1741) and a Fuc permease (fucP, Bbr_1742). Transcription of a predicted β-galactosidase-encoding gene (Bbr_1833, designated lacZ7) was also significantly increased, this enzyme is possibly involved in the removal of Gal from mucin oligosaccharides, suggesting that B. breve UCC2003 may utilize one or more Gal-containing fractions of mucin. The Bbr_1585-1590 cluster was also up-regulated during growth in co-culture. Bbr_1585 encodes a predicted UDP-Glc-4-epimerase, designated galE. Previously, UDP-Glc-4-epimerase enzymes have been shown to be involved in the Leloir pathway for Gal metabolism [56], as well as the GNB/LNB pathway for the metabolism of (ga)lacto-N-biose (GNB/LNB) derived from HMO or mucin [57,58]. Bbr_1586 encodes a predicted N-acetylhexosamine-1-kinase, designated nahK. In B. longum JCM 1217, an N-acetylhexosamine-1-kinase, which shares 90% identity to NahK, was shown to have a role in the previously mentioned GNB/LNB pathway [57]. Bbr_1587 encodes a predicted LNB phosphorylase, designated lnbP, an enzyme which is also involved in the GNB/LNB pathway [57-59]. The LnbP protein from B. breve UCC2003 shares 90% identity with the previously characterized LnbP protein from B. bifidum JCM 1254 [58]. Finally, as regards to this upregulated cluster, Bbr_1588-1590 encode a predicted ABC transport system, including two predicted permease proteins (represented by Bbr_1588 and Bbr_1589), and a solute binding protein (Bbr_1590), which shares 98% identity with the GNB/LNB-specific binding protein of B. longum JCM1217 [60]. Bbr_1884 (designated galT2), encoding a predicted Gal-1-phosphate uridyltransferase, another enzyme required for the Leloir pathway and the GNB/LNB pathway [56-58], was also up-regulated when B. breve UCC2003 was grown in mucin in co-culture with B. bifidum PRL2010. Finally, two gene clusters previously shown to be responsible for the transport and metabolism of sialic acid were significantly up-regulated under these growth conditions (Bbr_0160-0171 and Bbr_1247-1248) [13] (Table 3).
Table 3.
Effect of mucin (in co-culture with B. bifidum PRL2010) on the transcriptome of B. breve UCC2003
| Gene No. | Predicted Function | Level of upregulation ( P < 0.001) |
|---|---|---|
| Bbr_0161_ nanK | Conserved hypothetical protein in ROK family | 4.22 |
| Bbr_0162_ nanE | N-acetylmannosamine-6-phosphate 2-epimerase | 5.46 |
| Bbr_0164_ nanB | ABC transport system, solute binding protein | 14.08 |
| Bbr_0165_ nanC | ABC transport system, permease protein | 11.82 |
| Bbr_0166_ nanD | ABC transport system, ATP-binding protein | 18.92 |
| Bbr_0167_ nanF | ABC transport system, ATP-binding protein | 22.14 |
| Bbr_0168_ nanA | N-acetylneuraminate lyase | 15.17 |
| Bbr_0169_ nagB1 | Glucosamine-6-phosphate isomerase | 17.77 |
| Bbr_0171_ nanH | Sialidase | 7.23 |
| Bbr_0173_ nanR | Transcriptional regulator, GntR family | 4.80 |
| Bbr_1247_ nagA2 | N-acetylglucosamine-6-phosphate deacetylase | 4.59 |
| Bbr_1248_ nagB3 | Glucosamine-6-phosphate isomerase | 6.08 |
| Bbr_1585_ galE | UDP-Glc-4-epimerase | 3.96 |
| Bbr_1586_ nahK | N-acetylhexosamine kinase | 3.67 |
| Bbr_1587_ lnbP | Lacto-N-biose phosphorylase | 3.19 |
| Bbr_1588 | ABC transport system, permease protein | 3.34 |
| Bbr_1589 | ABC transport system, permease protein | 2.88 |
| Bbr_1590 | ABC transport system, solute binding protein | 8.95 |
| Bbr_1740_ dapA4 | Dihydrodipicolinate synthase | 9.91 |
| Bbr_1741 | Conserved hypothetical protein | 8.83 |
| Bbr_1742_ fucP | L-fucose permease | 7.81 |
| Bbr_1833_ lacZ7 | Beta-galactosidase | 4.31 |
| Bbr_1879 | PTS system, glucose-specific IIABC component | 6.97 |
| Bbr_1880 | PTS system, GlcNAc-specific IIBC component | 18.15 |
| Bbr_1884_ galT2 | Gal-1-phosphate uridylyltransferase | 3.86 |
The cut-off point for the level of up-regulation is 2.5-fold with a P-value of <0.001.
Analysis of the monosaccharide composition of fermented and non-fermented mucin
HPAEC-PAD was used to quantitatively analyze the carbohydrate profile of non-fermented mucin, compared to the cell free supernatant (CFS) of B. bifidum PRL2010-pPKCM7 grown in mucin for 30 h. Non-fermented mucin was shown to contain 0.13 nmol mg−1 of free Gal and 3.96 nmol mg−1 of free Glc, both of which support growth of B. breve UCC2003 [51], thus presenting a plausible explanation for the initial increase in B. breve UCC2003-pAM5 cell numbers during the first 12 h of incubation in mucin-containing medium (Table 4). However, after 30 h growth of B. bifidum PRL2010-pPKCM7, the carbohydrate profile of the medium was significantly altered. Glc was no longer detected, indicating its uptake by B. bifidum PRL2010-pPKCM7, but the amount of free Gal increased to 51.73 nmol mg−1, while 34.94 nmol mg−1 of free Fuc was also detected, indicative of the extracellular glycosidase activity of B. bifidum PRL2010-pPKCM7 (Table 4). A consistent pattern was observed in regards to the amount of Fuc and Gal released by B. bifidum PRL2010 activity. The presence of Fuc was further verified by fluorescent labelling of the CFS samples and analysis by RP-HPLC (Figure 2A). It was also confirmed that free Fuc was absent in non-fermented mMRS supplemented with 0.4% mucin, which indicated that its presence was a result of B. bifidum PRL2010-pPKCM7 extracellular activity (Figure 2A). To investigate whether these released monosaccharides from mucin support the observed growth and viability of B. breve UCC2003-pAM5 in co-culture, the carbohydrate profile of the CFS of the co-culture was compared to that of B. bifidum PRL2010-pPKCM7 grown independently in mucin (hence any difference observed could be attributed to the presence of B. breve UCC2003-pAM5). The results support the hypothesis that B. breve UCC2003-pAM5 is cross-feeding on at least one carbohydrate released following degradation of mucin by B. bifidum PRL2010-pPKCM7. After 30 h Fuc was present in the carbohydrate profile of B. bifidum PRL2010-pPKCM7, but absent (or at least below the detection level) in that of the co-culture, indicative of its uptake and utilization by B. breve UCC2003-pAM5 (Figure 2B).
Table 4.
Quantification of Fuc, Gal and Glc by HPAEC-PAD
| Fuc | Gal | Glc | |
|---|---|---|---|
| mMRS +0.4% mucin (non-fermented) | n.d. | 0.13 | 3.96 |
| mMRS +0.4% mucin (30 h growth of B. bifidum PRL2010) | 34.94 | 51.73 | n.d. |
Concentrations given in nmol mg−1. n.d. = not detected.
Figure 2.
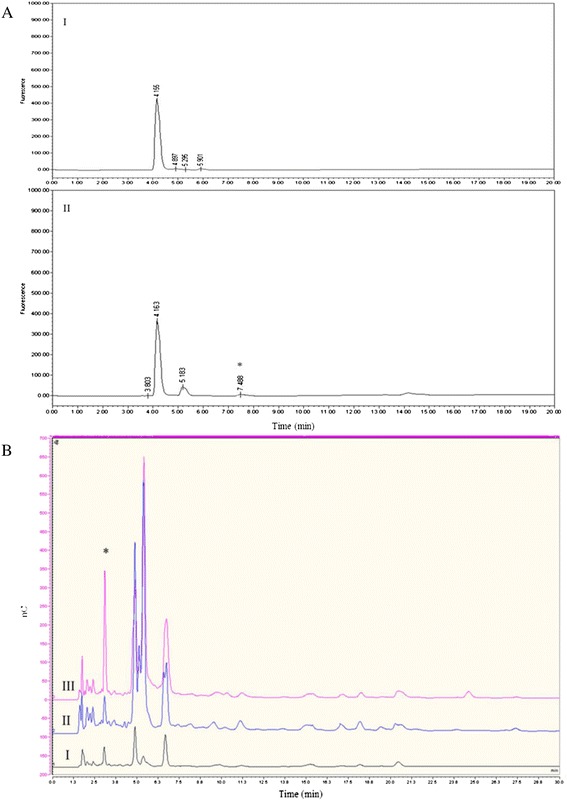
Analysis of the monosaccharide composition of fermented and non-fermented mucin. (A) HPLC profile of 2-AB -labelled (I) mMRS supplemented with 0.4% mucin and (II) the media from (I) after 30 h growth of B. bifidum PRL2010-pPKCM7. The peak for Fuc is marked with *. (B) Qualitative HPAEC-PAD analysis of (I) mMRS supplemented with 0.4% mucin, (II) the media from (I) following 30 h growth of B. bifidum PRL2010-pPKCM7 and B. breve UCC2003-pAM5 in co-culture, (III) the media from (I) after 30 h growth of B. bifidum PRL2010-pPCM7. The peak for Fuc is marked with *. nC, nanocoulombs.
Growth of B. breve UCC2003 mutants in co-culture in mucin with B. bifidum PRL2010-pPKCM7
HPAEC-PAD and transcriptome data suggested that the improved growth and viability of B. breve UCC2003-pAM5 in co-culture was a result of the strain cross-feeding on Fuc, while microarray data suggested sialic acid, Gal, GNB and/or another of the Gal-containing constituents of mucin may also be utilized. To determine which, if any, of these sugars was primarily responsible for growth of the strain in co-culture, insertion mutants were constructed in the fucP, lnbP and lacZ7 genes, resulting in strains B. breve UCC2003-fucP, B. breve UCC2003-lnbP and B. breve UCC2003-lacZ7. Two additional mutants, B. breve UCC2003-galT, which harbours a transposon in a predicted Gal-1-phosphate uridyltransferase-encoding gene, designated galT1 and was shown to be incapable of growth in Gal [41], and the previously described insertion mutant, B. breve UCC2003-nanA, which cannot utilize sialic acid [13], were also tested. The insertion mutants were individually analyzed for their ability to grow in mucin in the presence or absence of B. bifidum PRL2010-pPKCM7. In the absence of B. bifidum PRL2010-pPKCM7, the mutant strains behaved similarly to B. breve UCC2003-pAM5, as initial growth over the first 12 h was followed by a decline in viable cells to 104 CFU ml−1 or lower at 72 h (Figure 3). It was also found that removing the ability of the strain to utilize a particular sugar did not significantly affect its ability to grow in co-culture with B. bifidum PRL2010-pPKCM7. Similar to B. breve UCC2003-pAM5, all mutant strains attained a viable count of between 107 and 108 CFU ml−1 between 12 h and 30 h, and maintained a viable count of 106 CFU ml−1 until 72 h, with the exception of B. breve UCC2003-fucP which dropped to 105 CFU ml−1 (Figure 3). The carbohydrate profiles of each mutant grown in co-culture were also analyzed by HPAEC-PAD and it was found that each mutant, with the exception of B. breve UCC2003-fucP, produced an identical carbohydrate profile to that produced by the B. breve UCC2003-pAM5. B. breve UCC2003-fucP, as expected, was not capable of utilizing/transporting Fuc, as evident from the presence of Fuc in its carbohydrate profile (Figure 4).
Figure 3.
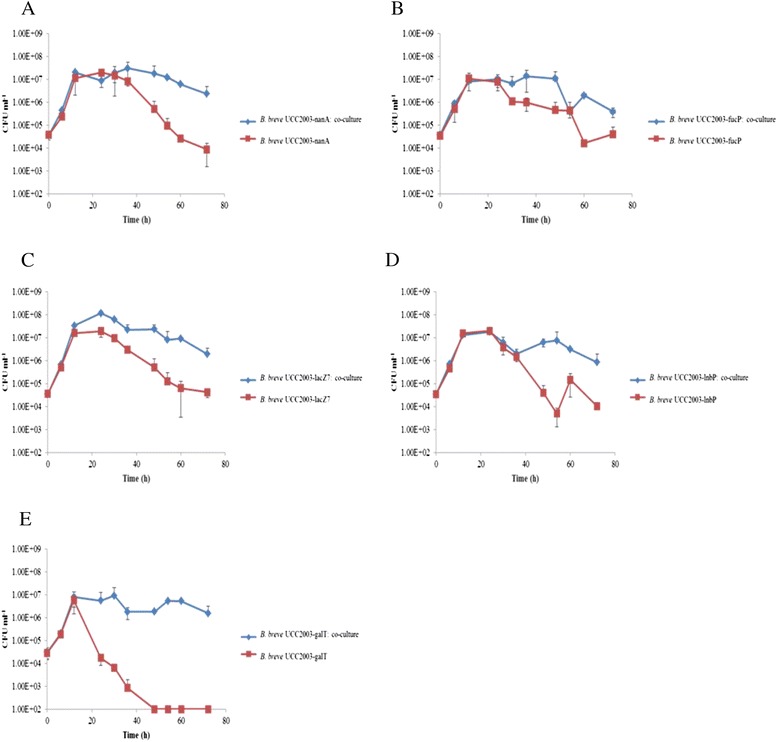
Individual and co-culture growth profiles of B. breve UCC2003 mutant strains in mucin. Growth profiles of the mutants (A) B. breve UCC2003-nanA, (B) B. breve UCC2003-fucP, (C) B. breve UCC2003-lacZ7, (D) B. breve UCC2003-lnbP and (E) B. breve UCC2003-galT in mMRS supplemented with 0.4% mucin, separately or in co-culture with B. bifidum PRL2010-pPKCM7, over 72 h. The results presented are the mean values of duplicate experiments. Error bars represent standard deviation.
Figure 4.
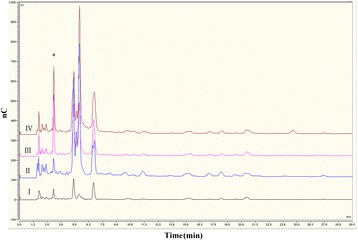
Analysis of the monosaccharide composition of fermented and non-fermented mucin including B. breve UCC2003-fucP. Qualitative HPAEC-PAD analysis of (I) mMRS supplemented with 0.4% mucin, (II) the media from (I) following 36 h growth of B. breve UCC2003-pAM5 and B. bifidum PRL2010-pPKCM7 in co-culture, (III) the media from (I) following 36 h growth of B. bifidum PRL2010-pPKCM7, (IV) the media from (I) following 36 h growth of B. breve UCC2003-fucP and B. bifidum PRL2010-pPKCM7 in co-culture. The peak for Fuc is marked with *. nC, nanocoulombs.
Discussion
Early investigations into the degradation of mucin established the role of extracellular glycosidases produced by a sub-population of bacteria in the gut, suggesting that the remaining enteric bacteria may cross feed on the released oligosaccharides [19,22]. Since then, much of the research into bacterial degradation of mucin has focused on the characterization of such extracellular glycosidases, such as two α-L-fucosidases and an exo-α-sialidase from B. bifidum JCM1254, and an endo-α-N-acetylgalactosaminidase from B. longum JCM1217 [23,24,61,62]. Another study identified, by a genomic as well as a transcriptomic and proteomic approach, a number of extracellular enzymes involved in mucin degradation by B. bifidum PRL2010, including a predicted cell wall-anchored endo-α-N-acetylgalactosamidase, two α-L-fucosidases, two exo-α-sialidases, a β-galactosidase and two putative N-acetyl-β-hexosaminidases, all of which contained a signal peptide [26].
Extracellular enzymes similar to those outlined above were not found in the genome of B. breve UCC2003 [33]. Therefore, it was not surprising that B. breve UCC2003-pAM5 was incapable of high density growth in a medium containing mucin as the sole carbon source. However, when B. bifidum PRL2010-pPKCM7 was included as a co-cultivating, mucin-degrading bacterium, the growth and viability of B. breve UCC2003-pAM5 was improved compared to the control situation. A combination of HPAEC-PAD and transcriptome analyses identified some of the possible monosaccharides and oligosaccharides which could support this enhanced co-cultivation growth/viability phenotype of B. breve UCC2003-pAM5, represented by sialic acid, Fuc, Gal and/or Gal-containing constituents of mucin.
HPAEC-PAD analysis identified two monosaccharides, namely Fuc and Gal, which were released from mucin by B. bifidum PRL2010-pPKCM7 activity. Fuc was shown to be internalized by B. breve UCC2003-pAM5 during growth in co-culture, suggesting a role in the enhanced growth/viability phenotype of B. breve UCC2003-pAM5 in co-culture. Transcriptome and mutagenesis data supports this hypothesis, as shown by increased transcription of a cluster (Bbr_1740-1742) involved in the uptake and utilization of Fuc and by the inability of a mutant in the fucP gene to internalize Fuc from the growth medium.
Gal has previously been shown to support the growth of B. breve UCC2003 [51]. However, given that Gal is ubiquitous in mucin, it may be internalized by B. breve UCC2003-pAM5 as a monosaccharide and/or as a constituent of a larger oligosaccharide. Interestingly, transcriptome data revealed increased transcription of the Bbr_1585-1590 gene cluster, which includes predicted GalE, NahK and LnbP-encoding genes, all of which are required for the metabolism of GNB [57], as well as an adjacent predicted ABC transport system. Outside of this cluster, the predicted galT2 gene (Bbr_1884) was also up-regulated. A previous study in B. bifidum hypothesized that GalT2 is specifically required for the metabolism of GNB from mucin [63]. Up-regulation of each of the required genes for the complete GNB pathway, as well as a predicted transport system, suggests that GNB might be the preferred source of Gal for B. breve UCC2003 in co-culture. A predicted β-galactosidase-encoding gene, lacZ7, was also up-regulated in co-culture, implying that B. breve UCC2003 may utilize another Gal-containing oligosaccharide. However, the precise substrate or preferred linkage of this enzyme is still a matter of speculation. Gal-containing oligosaccharides and GNB were not identified in the HPAEC-PAD data. However, the presence of four predicted β-galactosidases on the genome of B. bifidum PRL2010, as well as a clear homolog of the GNB pathway [26], suggests that B. bifidum PRL2010 also utilizes these sugars, and it is therefore possible that in co-culture the two strains compete for such sugars.
Interestingly, transcriptome data also revealed increased transcription of two clusters previously shown to be up-regulated in the presence of sialic acid [13]. This is inconsistent with the HPAEC-PAD data, in which sialic acid was not identified; however, it should be noted that the mucin from porcine stomach used in this study contains only 0.5%- 1.5% bound sialic acid, and it is thus possible that the released sialic acid is below the detection level of the HPAEC-PAD system. However, since B. bifidum PRL2010 has been shown to be incapable of using sialic acid [13,26], it seems likely that B. breve UCC2003 would utilize this sugar, even if available in very low quantities.
While accepting the HPAEC-PAD and transcriptome data are not definitive, the results suggest that B. breve UCC2003-pAM5 utilizes a combination of sugars in co-culture, a suggestion supported by the fact that none of the mutants tested displayed a different phenotype to B. breve UCC2003-pAM5 in co-culture. Construction of double or triple mutant strains is expected to result in strains that are impaired in growth/viability under co-culture conditions, however, at present such multiple mutant construction is not technically feasible for B. breve UCC2003 (and have to the best of our knowledge not been described for any bifidobacterial species/strain).
Our results highlight the different approaches taken by two species of bifidobacteria to proliferate and survive in the gut. B. bifidum PRL2010 has been shown to utilize mucin oligosaccharides (and structurally similar HMOs), a highly complex, yet ubiquitous carbohydrate source in the gut. Aside from host glycans such as mucin and HMO, however, the fermentation ability of B. bifidum PRL2010 is rather limited [26,52]. B. breve UCC2003, on the other hand, is a more versatile bacterium from a metabolic perspective, capable of utilizing a number of host and diet-derived carbohydrates. As seen in this study it can scavenge constituents of mucin released by the extracellular glycosidase activity of other bifidobacteria, as well as cross-feed on certain HMO [13], a characteristic which reflects the abundance of representatives of this species in the infant gut [5]. However, B. breve UCC2003 also has the ability to utilize a number of plant-derived carbohydrates such as starch, galactan, cellodextrins and raffinose [7,8,10,11]. B. breve strains have been identified in the adult human bifidobacterial population, although the relative abundance is lower than in infants [5,64]. The similar phenotype of B. breve UCC2003-pAM5 and the mutant strains in co-culture also highlights the adaptability of the strain, emphasizing its ability to switch to a different carbon source, depending on the carbohydrates available. This versatility is reflected on the genome of B. breve UCC2003, which contains a large number of carbohydrate utilization clusters [33], suggesting an ability to alternate between and/or co-utilize diet- and host- derived carbohydrates depending on availability. The gut commensal Bacteroides thetaiotaomicron, displays a remarkable metabolic flexibility, whereby in the absence of a dietary-derived fibre, this bacterium will shift its metabolic activities towards the degradation of mucin [65]. These results suggest that B. breve UCC2003 may also be capable of such flexibility, although further study is required.
The ability to degrade mucin seems to be limited to particular gut commensals, such as certain species of Bacteroides, Bifidobacterium, Ruminococcus and Akkermansia [20,26,66-68]. Pathogenic bacteria appear to be poorly adapted to mucin degradation [69], however, multiple studies have shown pathogens utilizing constituents of mucin released by commensal glycosidases. For example, C. jejuni has been shown to utilize Fuc as a substrate for growth [55], while enterohaemorrhagic E. coli uses Fuc as a signal to induce virulence [70]. Similarly, Salmonella typhimurium and Clostridium difficile have been shown to utilize sialic acid released by the sialidase activity of Bacteroides thetaotaomicron in a gnotobiotic mouse [71]. The uptake of mucin-derived Fuc by B. breve UCC2003, as well as the presumable utilization of mucin-derived sialic acid, suggests a potential role for B. breve UCC2003 in “mopping up” the released constituents of mucin, providing competition to potential pathogens and inhibiting or limiting their proliferation, although further study is required to expand on this hypothesis.
The mutually beneficial relationship between the host and the intestinal bacteria has been well established [72], however, it is of equal importance to acknowledge the symbiotic relationships formed between genera and species of the intestinal bacteria. Cross-feeding between B. breve UCC2003 and B. bifidum PRL2010 has now been shown for mucin in the present work and previously for 3′ sialyllactose [13], and given the structural similarity between mucin oligosaccharides and HMOs, it may be assumed that B. breve UCC2003 has the ability to cross feed on other HMO-derived constituents that are released by B. bifidum PRL2010. These results highlight the compatibility of these two species of bifidobacteria, especially when viewed in contrast to the strategy employed by B. longum subsp. infantis, which assimilates HMOs in their intact form, leaving none for potential cross-feeding with another species [73]. Interestingly, another type of cross-feeding has been described, whereby Eubacterium halii utilizes lactate produced by Bifidobacterium adolescentis during growth on starch, resulting in butyrate production by Eu. halii [74]. Butyrate is a major source of energy for colonocytes and has also been implicated in protection against colonic carcinogenesis [75,76]. Understanding such complex interactions between different members of the intestinal bacteria is of crucial importance when attempting to influence the activity or composition of the intestinal microbiota, such as in the use of probiotics.
Conclusion
This study provides in vitro proof for the existence of a commensal relationship between two species of bifidobacteria in the large intestine, namely B. breve and B. bifidum, in which B. breve UCC2003 benefits from the carbohydrates released by the extracellular glycosidase activities of B. bifidum PRL2010. To our knowledge, this is the first study to describe the molecular details of such cross-feeding, with particular emphasis on the carbohydrate components which support the improved growth and survival in co-culture. The results shown here improve our knowledge on how B. breve UCC2003 colonizes the (infant) gut in the absence of dietary-derived carbohydrates, and also emphasize this strain’s ability to switch between carbohydrate sources depending on availability. This is an advantageous characteristic in terms of enhanced survival and colonization ability in both the infant and adult gut, and may present an advantage to the host in limiting opportunities for pathogenic microbes to proliferate in the gut.
Acknowledgments
The Alimentary Pharmabiotic Centre is a research centre funded by Science Foundation Ireland (SFI), through the Irish Government’s National Development Plan. The authors and their work were supported by SFI (Grant Nos. 07/CE/B1368, and SFI/12/RC/2273 and 08/SRC/B1393), the EU FP7 programme (Grant no. 260600) and a HRB postdoctoral fellowship (Grant No. PDTM/20011/9) awarded to MOCM.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ME participated in the design and coordination of the study, performed growth experiments, qualitative HPAEC-PAD, microarray analysis and insertion mutagenesis, analyzed data and drafted the manuscript. MOCM participated in the design and coordination of the study, constructed the plasmid-containing derivatives of B. breve UCC2003 and B. bifidum PRL2010, assisted in HPAEC-PAD and microarray analysis, analyzed data and helped draft the manuscript. MK, MK and JL performed quantitative HPAEC-PAD and HPLC analysis and helped draft the manuscript. MV analyzed data and helped draft the manuscript. DvS conceived the study, participated in the design and coordination of the study, analyzed data and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Muireann Egan, Email: muireann.egan@umail.ucc.ie.
Mary O’Connell Motherway, Email: m.oconnellmotherway@ucc.ie.
Michelle Kilcoyne, Email: michelle.kilcoyne@nuigalway.ie.
Marian Kane, Email: marian.kane@nuigalway.ie.
Lokesh Joshi, Email: lokesh.joshi@nuigalway.ie.
Marco Ventura, Email: marco.ventura@unipr.it.
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
References
- 1.Ventura M, Canchaya C, Fitzgerald G, Gupta R, van Sinderen D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie van Leeuwenhoek. 2007;92(2):265–265. doi: 10.1007/s10482-007-9182-2. [DOI] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24(2):160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28(4):405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7(5):e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pokusaeva K, Fitzgerald G, van Sinderen D. Carbohydrate metabolism in bifidobacteria. Genes Nutr. 2011;6(3):285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Connell Motherway M, Fitzgerald GF, van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol. 2011;4(3):403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pokusaeva K, O’Connell-Motherway M, Zomer A, MacSharry J, Fitzgerald GF, van Sinderen D. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2011;77(5):1681–1690. doi: 10.1128/AEM.01786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003. Microb Biotechnol. 2013;6(1):67–79. doi: 10.1111/1751-7915.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell KJ, O’Connell Motherway M, O’Callaghan J, Fitzgerald GF, Ross RP, Ventura M, Stanton C, van Sinderen D. Metabolism of four α-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2013;79(20):6280–6292. doi: 10.1128/AEM.01775-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell Motherway M, Fitzgerald G, Neirynck S, Ryan S, Steidler L, van Sinderen D. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74(20):6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan S, Fitzgerald G, van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial Strains. Appl Environ Microbiol. 2006;72(8):5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80(14):4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Podolsky DK. Oligosaccharide structures of human colonic mucin. J Biol Chem. 1985;260(14):8262–8271. [PubMed] [Google Scholar]
- 15.Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. 2009;19(7):756–766. doi: 10.1093/glycob/cwp048. [DOI] [PubMed] [Google Scholar]
- 16.Tytgat KM, Buller HA, Opdam FJ, Kim YS, Einerhand AW, Dekker J. Biosynthesis of human colonic mucin: Muc2 is the prominent secretory mucin. Gastroenterol. 1994;107(5):1352–1363. doi: 10.1016/0016-5085(94)90537-1. [DOI] [PubMed] [Google Scholar]
- 17.Capon C, Maes E, Michalski J, Leffler H, Kim YS. Sd(a)-antigen-like structures carried on core 3 are prominent features of glycans from the mucin of normal human descending colon. Biochem J. 2001;358(3):657–664. doi: 10.1042/bj3580657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbe C, Capon C, Coddeville B, Michalski J. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoskins LC, Boulding ET. Mucin degradation in human colon ecosystems. Evidence for the existence and role of bacterial subpopulations producing glycosidases as extracellular enzymes. J Clin Invest. 1981;67(1):163–172. doi: 10.1172/JCI110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Micr. 2004;54(5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 21.Corfield AP, Wagner SA, Clamp J, Kriaris M, Hoskins L. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60(10):3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest. 1985;75(3):944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19(9):1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 24.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-L-fucosidase (AfcA), a novel inverting glycosidase (Glycoside Hydrolase family 95) J Bacteriol. 2004;186(15):4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-α-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280(45):37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- 26.Turroni F, Bottacini F, Foroni E, Mulder I, Kim J-H, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald G, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turroni F, Milani C, van Sinderen D, Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010. Aging. 2010;107:19514–19519. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 28.Ward R, Niñonuevo M, Mills D, Lebrilla C, German B. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51(11):1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 29.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286(40):34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Bacteriol. 1960;23(1):130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 33.O’Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O’Brien F, Flynn K, Casey PG, Moreno Munoz JA, Kearney B, Houston AM, O’Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O’Toole PW, van Sinderen D. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A. 2011;108(27):11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Riordan O: Studies on antimicrobial activity and genetic diversity ofBifidobacteriumspecies: molecular characterization of a 5.75 kb plasmid and a chromosomally encoded recA gene homologue fromBifidobacterium breve.PhD thesis. National University of Ireland, Cork; 1998
- 36.Maze A, O’Connell-Motherway M, Fitzgerald G, Deutscher J, van Sinderen D. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73(2):545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol. 1995;177(24):7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Álvarez-Martín P, O’Connell-Motherway M, van Sinderen D, Mayo B. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl Microbiol Biotechnol. 2007;76(6):1395–1402. doi: 10.1007/s00253-007-1115-5. [DOI] [PubMed] [Google Scholar]
- 39.O’Connell Motherway M, O’Driscoll J, Fitzgerald GF, Van Sinderen D. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb Biotechnol. 2009;2(3):321–332. doi: 10.1111/j.1751-7915.2008.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cronin M, Knobel M, O’Connell-Motherway M, Fitzgerald GF, van Sinderen D. Molecular dissection of a bifidobacterial replicon. Appl Environ Microbiol. 2007;73(24):7858–7866. doi: 10.1128/AEM.01630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz L, Motherway MOC, Lanigan N, van Sinderen D. Transposon mutagenesis in Bifidobacterium breve: construction and characterization of a Tn5 transposon mutant library for Bifidobacterium breve UCC2003. PLoS One. 2013;8(5):e64699. doi: 10.1371/journal.pone.0064699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191(22):7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.García de la Nava J, Santaella DF, Alba JC, Carazo JM, Trelles O, Pascual-Montano A. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics. 2003;19(5):657–658. doi: 10.1093/bioinformatics/btg028. [DOI] [PubMed] [Google Scholar]
- 44.van Hijum S, de Jong A, Baerends R, Karsens H, Kramer N, Larsen R, den Hengst C, Albers C, Kok J, Kuipers O. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics. 2005;6(1):77. doi: 10.1186/1471-2164-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Hijum SAFT, Garcia De La Nava J, Trelles O, Kok J, Kuipers OP. MicroPreP: a cDNA microarray data pre-processing framework. ApplBioinformatics. 2003;2(4):241–244. [PubMed] [Google Scholar]
- 46.Long AD, Mangalam HJ, Chan BYP, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework: analysis of global gene expression in Escherichia coli K12. J Biol Chem. 2001;276(23):19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- 47.Kilcoyne M, Shah M, Gerlach JQ, Bhavanandan V, Nagaraj V, Smith AD, Fujiyama K, Sommer U, Costello CE, Olszewski N, Joshi L. O-glycosylation of protein subpopulations in alcohol-extracted rice proteins. J Plant Physiol. 2009;166(3):219–232. doi: 10.1016/j.jplph.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Kilcoyne M, Gerlach JQ, Gough R, Gallagher ME, Kane M, Carrington SD, Joshi L. Construction of a natural mucin microarray and interrogation for biologically relevant glyco-epitopes. Anal Chem. 2012;84(7):3330–3338. doi: 10.1021/ac203404n. [DOI] [PubMed] [Google Scholar]
- 49.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230(2):229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 50.Hardy MR. Techniques in Glycobiology. New York: Marcel Dekker Inc; 1997. Glycan labeling with the fluorophores 2-aminobenzamide and anthranilic acid; pp. 359–376. [Google Scholar]
- 51.Watson D, O’Connell Motherway M, Schoterman MHC, van Neerven RJJ, Nauta A, van Sinderen D. Selective carbohydrate utilization by lactobacilli and bifidobacteria. J Appl Microbiol. 2013;114(4):1132–1146. doi: 10.1111/jam.12105. [DOI] [PubMed] [Google Scholar]
- 52.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol. 2012;78(14):5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pokusaeva K, Neves AR, Zomer A, O’Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol. 2010;3(3):311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connell KJ, O’Connell Motherway M, Liedtke A, Fitzgerald GF, Ross RP, Stanton C, Zomer A, van Sinderen D. Transcription of two adjacent carbohydrate utilization gene clusters in Bifidobacterium breve UCC2003 is controlled by LacI- and Repressor Open reading frame Kinase (ROK)-type regulators. Appl Environ Microbiol. 2014;80(12):3604–3614. doi: 10.1128/AEM.00130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahl M, Friis LM, Nothaft H, Liu X, Li J, Szymanski CM, Stintzi A. l-Fucose utilization provides Campylobacter jejuni with a competitive advantage. ProcNatl Acad Sci U S A. 2011;108(17):7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. The FASEB Journal. 1996;10(4):461–470. [PubMed] [Google Scholar]
- 57.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol. 2007;73(20):6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71(6):3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derensy-Dron D, Krzewinski F, Brassart C, Bouquelet S. β-1,3-Galactosyl-N-acetylhexosamine phosphorylase from Bifidobacterium bifidum DSM 20082: characterization, partial purification and relation to mucin degradation. Biotechnol Appl Biochem. 1999;29(1):3–10. [PubMed] [Google Scholar]
- 60.Suzuki R, Wada J, Katayama T, Fushinobu S, Wakagi T, Shoun H, Sugimoto H, Tanaka A, Kumagai H, Ashida H, Kitaoka M, Yamamoto K. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283(19):13165–13173. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 61.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21(4):437–447. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 62.Katayama T, Fujita K, Yamamoto K. Novel bifidobacterial glycosidases acting on sugar chains of mucin glycoproteins. J Biosci Bioeng. 2005;99(5):457–465. doi: 10.1263/jbb.99.457. [DOI] [PubMed] [Google Scholar]
- 63.De Bruyn F, Beauprez J, Maertens J, Soetaert W, De Mey M. Unraveling the Leloir pathway of Bifidobacterium bifidum: significance of the uridylyltransferases. Appl Environ Microbiol. 2013;79(22):7028–7035. doi: 10.1128/AEM.02460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de’Angelis GL, Shanahan F, van Sinderen D, Ventura M. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75(6):1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sonnenburg JL, Xu J, Leip DD, Chen C-H, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 66.Salyers AA, West SE, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34(5):529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salyers AA, Palmer JK, Wilkins TD. Degradation of polysaccharides by intestinal bacterial enzymes. Am J Clin Nutr. 1978;31(10):S128–S130. doi: 10.1093/ajcn/31.10.S128. [DOI] [PubMed] [Google Scholar]
- 68.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8(10):e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23(9):1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492(7427):113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502(7469):96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 73.Sela D, Mills D. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18(7):298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72(5):3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Velázquez O, Lederer H, Rombeau J. Butyrate and the colonocyte. Digest Dis Sci. 1996;41(4):727–739. doi: 10.1007/BF02213129. [DOI] [PubMed] [Google Scholar]
- 76.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21(9):793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]


